Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens
Abstract
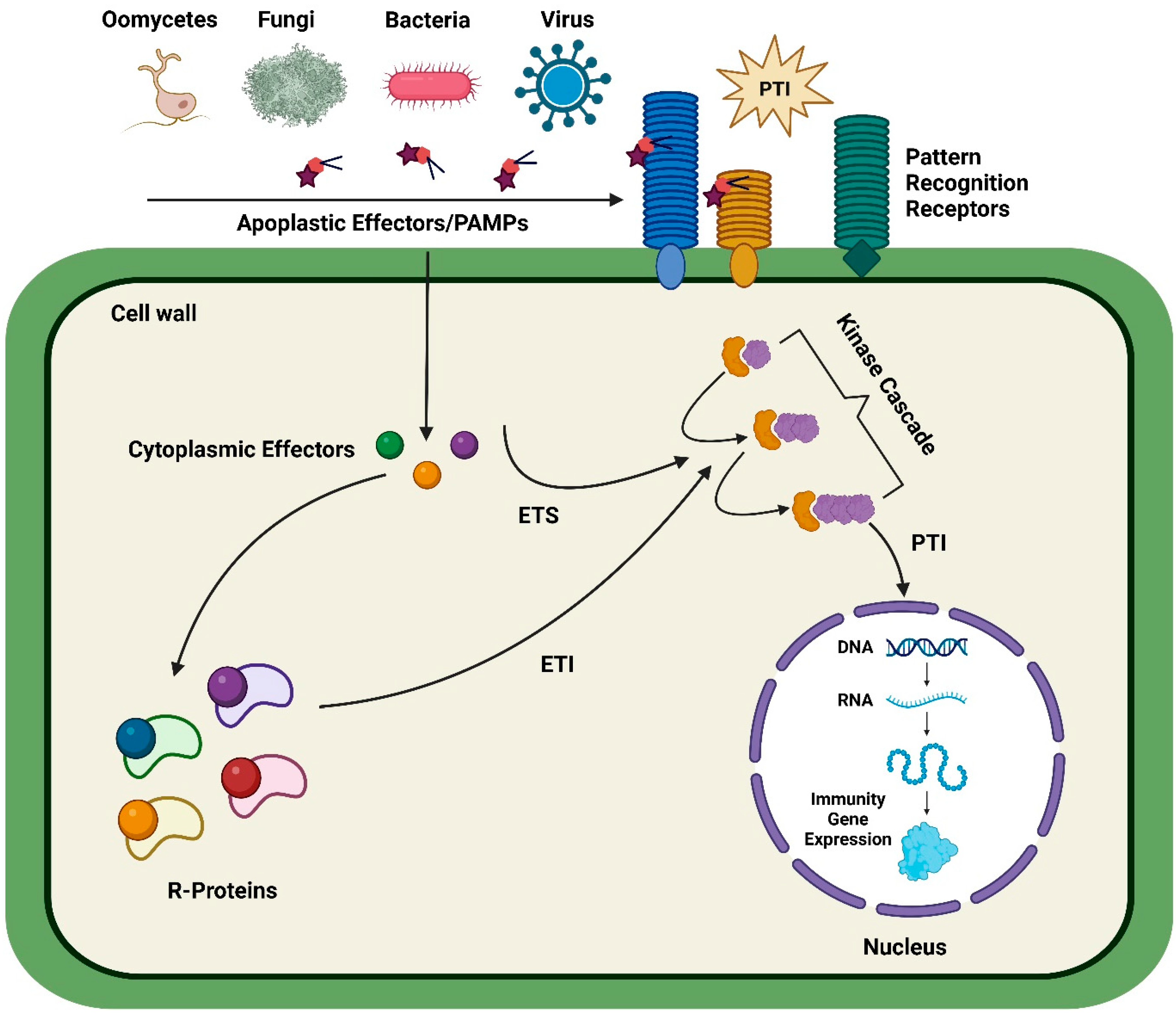
1. Introduction
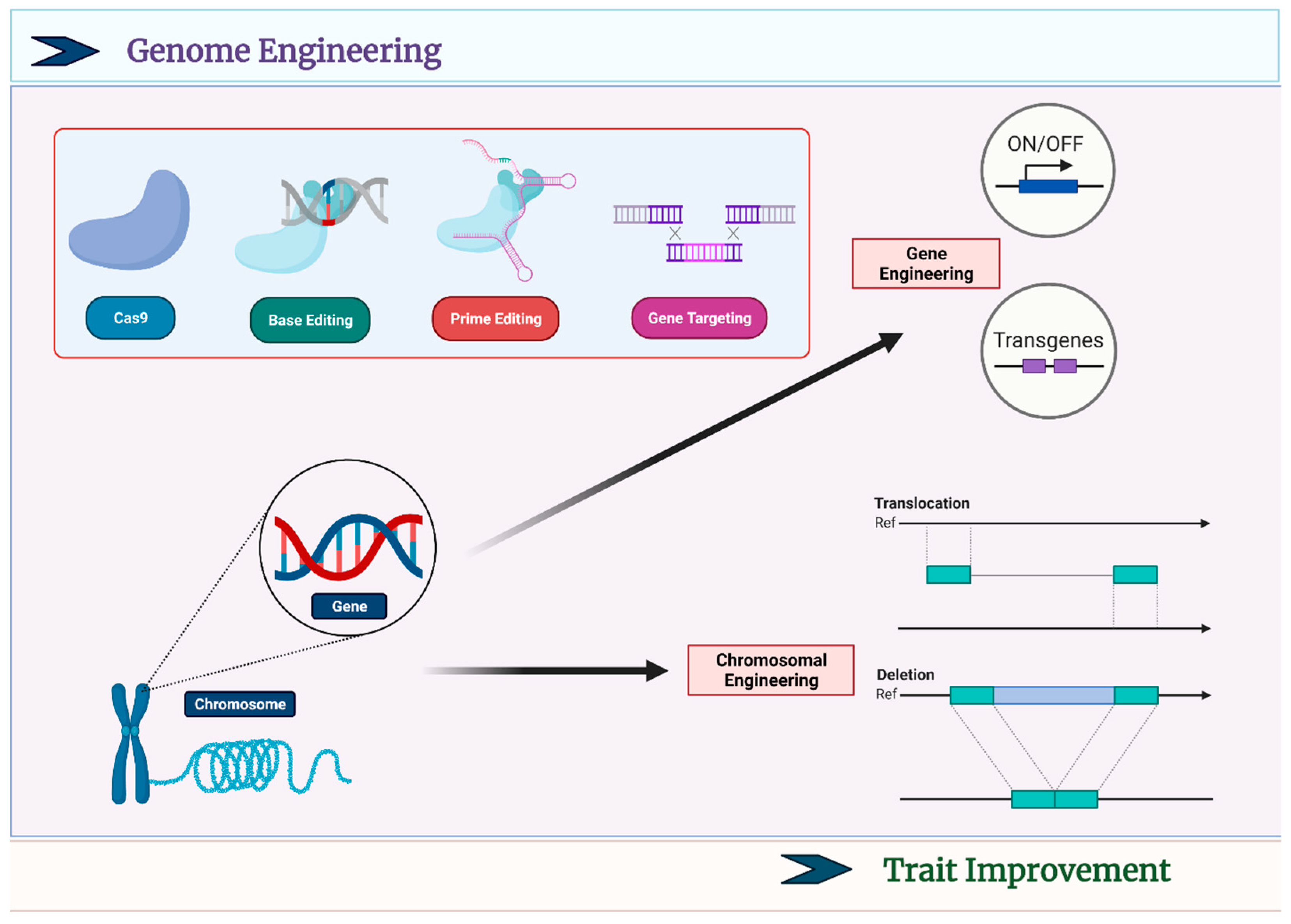
2. Different CRISPR-Based Tools
3. CRISPR-Based Genome Editing of Plants for Disease Resistance against Pathogens
3.1. Disease Resistance against Bacteria
3.2. Disease Resistance against Fungi
3.3. Diseases Resistance against Oomycetes
3.4. Disease Resistance against Plant Viruses
4. CRISPR-Based Crop Breeding
5. Limitations of CRISPR-Based Tools for Plant Pathogen Resistance
6. Future Research Directions
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bahadur Kc, K.; Dias, G.M.; Veeramani, A.; Swanton, C.J.; Fraser, D.; Steinke, D.; Lee, E.; Wittman, H.; Farber, J.M.; Dunfield, K.; et al. When too much isn’t enough: Does current food production meet global nutritional needs? PLoS ONE 2018, 13, e0205683. [Google Scholar] [CrossRef]
- Kleyn, F.J.; Ciacciariello, M. Future demands of the poultry industry: Will we meet our commitments sustainably in developed and developing economies? Worlds Poult. Sci. J. 2021, 77, 267–278. [Google Scholar] [CrossRef]
- Akoijam, N.; Joshi, S.R.; Akoijam, N. Conservation Metagenomics: Understanding Microbiomes for Biodiversity Sustenance and Conservation. In Molecular Genetics and Genomics Tools in Biodiversity Conservation; Springer: Singapore, 2022; pp. 31–61. [Google Scholar] [CrossRef]
- Lata, S.; Bhardwaj, S.; Garg, R. Nanomaterials for sensing and biosensing: Applications in agri-food diagnostics. Int. J. Environ. Anal. Chem. 2022, 1–12. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Genetic engineering for disease resistance in plants: Recent progress and future perspectives. Plant Physiol. 2019, 180, 26–38. [Google Scholar] [CrossRef]
- Gupta, R.; Pizarro, L.; Leibman-Markus, M.; Marash, I.; Bar, M. Cytokinin response induces immunity and fungal pathogen resistance, and modulates trafficking of the PRR LeEIX2 in tomato. Mol. Plant Pathol. 2020, 21, 1287–1306. [Google Scholar] [CrossRef]
- Sulima, A.S.; Zhukov, V.A. War and Peas: Molecular Bases of Resistance to Powdery Mildew in Pea (Pisum sativum L.) and Other Legumes. Plants 2022, 11, 339. [Google Scholar] [CrossRef]
- Goode, K.; Mitchum, M.G. Pattern-triggered immunity against root-knot nematode infection: A minireview. Physiol. Plant. 2022, 174, e13680. [Google Scholar] [CrossRef]
- Min, C.W.; Jang, J.W.; Lee, G.H.; Gupta, R.; Yoon, J.; Park, H.J.; Cho, H.S.; Park, S.R.; Kwon, S.W.; Cho, L.H.; et al. TMT-based quantitative membrane proteomics identified PRRs potentially involved in the perception of MSP1 in rice leaves. J. Proteom. 2022, 267, 104687. [Google Scholar] [CrossRef]
- Ke, X.; Wang, J.; Xu, X.; Guo, Y.; Zuo, Y.; Yin, L. Histological and molecular responses of Vigna angularis to Uromyces vignae infection. BMC Plant Biol. 2022, 22, 489. [Google Scholar] [CrossRef]
- Yang, C.; Dolatabadian, A.; Fernando, W.G.D. The wonderful world of intrinsic and intricate immunity responses in plants against pathogens. Can. J. Plant Pathol. 2021, 44, 1–20. [Google Scholar] [CrossRef]
- Waheed, A.; Haxim, Y.; Islam, W.; Kahar, G.; Liu, X.; Zhang, D. Role of pathogen’s effectors in understanding host-pathogen interaction. Mol. Cell Res. 2022, 1869, 119347. [Google Scholar] [CrossRef]
- Chen, D.; Hao, F.; Mu, H.; Ahsan, N.; Thelen, J.J.; Stacey, G. S-acylation of P2K1 mediates extracellular ATP-induced immune signaling in Arabidopsis. Nat. Commun. 2021, 12, 2750. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef]
- Koseoglou, E.; van der Wolf, J.M.; Visser, R.G.F.; Bai, Y. Susceptibility reversed: Modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci. 2022, 27, 69–79. [Google Scholar] [CrossRef]
- Laflamme, B.; Dillon, M.M.; Martel, A.; Almeida, R.N.D.; Desveaux, D.; Guttman, D.S. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 2020, 367, 763–768. [Google Scholar] [CrossRef]
- Gaj, T.; Sirk, S.J.; Shui, S.; Liu, J. Genome-Editing Technologies: Principles and Applications. Cold Spring Harb. Perspect. Biol. 2016, 8, 023754. [Google Scholar] [CrossRef]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Huang, J.; Zhou, Y.; Li, J.; Lu, A.; Liang, C. CRISPR/Cas systems: Delivery and application in gene therapy. Front. Bioeng. Biotechnol. 2022, 10, 2168. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Wang, B.; Lou, J.; Ni, P.; Jin, Y.; Chen, S.; Duan, G.; Zhang, R. Application of CRISPR/Cas Systems in the Nucleic Acid Detection of Infectious Diseases. Diagnostics 2022, 12, 2455. [Google Scholar] [CrossRef]
- Lee, H.; Sashital, D.G. Creating memories: Molecular mechanisms of CRISPR adaptation. Trends Biochem. Sci. 2022, 47, 464–476. [Google Scholar] [CrossRef]
- Saber Sichani, A.; Ranjbar, M.; Baneshi, M.; Torabi Zadeh, F.; Fallahi, J. A Review on Advanced CRISPR-Based Genome-Editing Tools: Base Editing and Prime Editing. Mol. Biotechnol. 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Otoupal, P.B.; Cress, B.F.; Doudna, J.A.; Schoeniger, J.S. CRISPR-RNAa: Targeted activation of translation using dCas13 fusions to translation initiation factors. Nucleic Acids Res. 2022, 50, 8986–8998. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Das, P.; Panda, D.; Xie, K.; Baig, M.J.; Molla, K.A. A detailed landscape of CRISPR-Cas-mediated plant disease and pest management. Plant Sci. 2022, 323, 111376. [Google Scholar] [CrossRef] [PubMed]
- Bonagiri, G.; Gurjar, D.; Kuri, A.; Kumar Yella, V. CRISPR/cas9 gene editing tool for diseases resistant varieties. Pharma Innov. J. 2022, SP-11, 2731–2738. [Google Scholar]
- Krishnaswami, V.; Kumar, M.; Vijayaraghavalu, S.; Krishnaswami, V.; Kumar, M.; Vijayaraghavalu, S. Advances in Epigenetics for Crop Improvement and Sustainable Agriculture. In Plant Genomics for Sustainable Agriculture; Springer: Singapore, 2022; pp. 351–370. [Google Scholar] [CrossRef]
- Villalobos-López, M.A.; Arroyo-Becerra, A.; Quintero-Jiménez, A.; Iturriaga, G. Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. Int. J. Mol. Sci. 2022, 23, 12053. [Google Scholar] [CrossRef]
- Delplace, F.; Huard-Chauveau, C.; Berthomé, R.; Roby, D. Network organization of the plant immune system: From pathogen perception to robust defense induction. Plant J. 2022, 109, 447–470. [Google Scholar] [CrossRef]
- Sharma, Y.; Sharma, A.; Madhu; Shumayla; Singh, K.; Upadhyay, S.K. Long Non-Coding RNAs as Emerging Regulators of Pathogen Response in Plants. Non-Coding RNA 2022, 8, 4. [Google Scholar] [CrossRef]
- Wang, Y.; Zafar, N.; Ali, Q.; Manghwar, H.; Wang, G.; Yu, L.; Ding, X.; Ding, F.; Hong, N.; Wang, G.; et al. CRISPR/Cas Genome Editing Technologies for Plant Improvement against Biotic and Abiotic Stresses: Advances, Limitations, and Future Perspectives. Cells 2022, 11, 3928. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Classification and Nomenclature of CRISPR-Cas Systems: Where from Here? Cris. J. 2018, 1, 325–336. [Google Scholar] [CrossRef]
- Ji, X.; Wang, D.; Gao, C. CRISPR editing-mediated antiviral immunity: A versatile source of resistance to combat plant virus infections. Sci. China Life Sci. 2019, 62, 1246–1249. [Google Scholar] [CrossRef]
- Murovec, J.; Pirc, Ž.; Yang, B. New variants of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol. J. 2017, 15, 917–926. [Google Scholar] [CrossRef]
- Nozawa, T.; Furukawa, N.; Aikawa, C.; Watanabe, T.; Haobam, B.; Kurokawa, K.; Maruyama, F.; Nakagawa, I. CRISPR Inhibition of Prophage Acquisition in Streptococcus pyogenes. PLoS ONE 2011, 6, e19543. [Google Scholar] [CrossRef]
- Le Rhun, A.; Escalera-Maurer, A.; Bratovič, M.; Charpentier, E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019, 16, 380–389. [Google Scholar] [CrossRef]
- Rozhdestvensky, S.; Kondrashov, A.; Kuzmin, A.A.; Tomilin, A.N. Building Blocks of Artificial CRISPR-Based Systems beyond Nucleases. Int. J. Mol. Sci. 2022, 24, 397. [Google Scholar] [CrossRef]
- Wang, H.X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.W.; Bao, G.; Leong, K.W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef]
- Wheatley, M.S.; Yang, Y. Versatile applications of the CRISPR/cas toolkit in plant pathology and disease management. Phytopathology 2021, 111, 1080–1090. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]
- Ren, Q.; Sretenovic, S.; Liu, S.; Tang, X.; Huang, L.; He, Y.; Liu, L.; Guo, Y.; Zhong, Z.; Liu, G.; et al. PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat. Plants 2021, 7, 25–33. [Google Scholar] [CrossRef]
- Xu, Z.; Kuang, Y.; Ren, B.; Yan, D.; Yan, F.; Spetz, C.; Sun, W.; Wang, G.; Zhou, X.; Zhou, H. SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol. 2021, 22, 6. [Google Scholar] [CrossRef]
- Sukegawa, S.; Saika, H.; Toki, S. Plant genome editing: Ever more precise and wide reaching. Plant J. 2021, 106, 1208–1218. [Google Scholar] [CrossRef]
- Gehrke, J.M.; Cervantes, O.; Clement, M.K.; Wu, Y.; Zeng, J.; Bauer, D.E.; Pinello, L.; Joung, J.K. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 2018, 36, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, Z.; Gosavi, G.; Ren, B.; Cao, Y.; Kuang, Y.; Zhou, C.; Spetz, C.; Yan, F.; Zhou, X.; et al. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol. J. 2020, 18, 1645–1647. [Google Scholar] [CrossRef] [PubMed]
- Schindele, P.; Puchta, H. Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 2020, 18, 1118. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Kuang, Y.; Ren, B.; Wang, J.; Zhang, D.; Lin, H.; Yang, B.; Zhou, X.; Zhou, H. Highly Efficient A·T to G·C Base Editing by Cas9n-Guided tRNA Adenosine Deaminase in Rice. Mol. Plant 2018, 11, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, R.; Qin, R.; Liu, X.; Kong, F.; Wei, P. Genome editing mediated by SpCas9 variants with broad non-canonical PAM compatibility in plants. Mol. Plant 2021, 14, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Wada, N.; Miyaji, T.; Murakami, E.; Marui, K.; Ueta, R.; Hashimoto, R.; Abe-Hara, C.; Kong, B.; Yano, K.; et al. Genome editing in plants using CRISPR type I-D nuclease. Commun. Biol. 2020, 3, 648. [Google Scholar] [CrossRef]
- Xu, R.; Qin, R.; Xie, H.; Li, J.; Liu, X.; Zhu, M.; Sun, Y.; Yu, Y.; Lu, P.; Wei, P. Genome editing with type II-C CRISPR-Cas9 systems from Neisseria meningitidis in rice. Plant Biotechnol. J. 2022, 20, 350–359. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Khan, M.Z.; Haider, S.; Mansoor, S.; Amin, I. Targeting Plant ssDNA Viruses with Engineered Miniature CRISPR-Cas14a. Trends Biotechnol. 2019, 37, 800–804. [Google Scholar] [CrossRef]
- Sukegawa, S.; Toki, S.; Saika, H. Genome Editing Technology and Its Application to Metabolic Engineering in Rice. Rice 2022, 15, 21. [Google Scholar] [CrossRef]
- Guzmán-Benito, I.; Achkar, N.P.; Bologna, N.G.; Ursache, R. CRISPR/Cas-mediated in planta gene targeting: Current advances and challenges. J. Exp. Bot. 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Huang, T.K.; Armstrong, B.; Schindele, P.; Puchta, H. Efficient gene targeting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a. Plant Biotechnol. J. 2021, 19, 1314–1324. [Google Scholar] [CrossRef]
- Agudelo, D.; Carter, S.; Velimirovic, M.; Duringer, A.; Levesque, S.; Rivest, J.F.; Loehr, J.; Mouchiroud, M.; Cyr, D.; Waters, P.J.; et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020, 30, 107–117. [Google Scholar] [CrossRef]
- Negishi, K.; Mikami, M.; Toki, S.; Endo, M. Enhanced FnCas12a-Mediated Targeted Mutagenesis Using crRNA with Altered Target Length in Rice. Front. Genome Ed. 2020, 2, 608563. [Google Scholar] [CrossRef]
- Zhang, Y.; Iaffaldano, B.; Qi, Y. CRISPR ribonucleoprotein-mediated genetic engineering in plants. Plant Commun. 2021, 2, 100168. [Google Scholar] [CrossRef]
- Wang, Q.; Alariqi, M.; Wang, F.; Li, B.; Ding, X.; Rui, H.; Li, Y.; Xu, Z.; Qin, L.; Sun, L.; et al. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol. J. 2020, 18, 2436–2443. [Google Scholar] [CrossRef]
- Wu, F.; Qiao, X.; Zhao, Y.; Zhang, Z.; Gao, Y.; Shi, L.; Du, H.; Wang, L.; Zhang, Y.J.; Zhang, Y.; et al. Targeted mutagenesis in Arabidopsis thaliana using CRISPR-Cas12b/C2c1. J. Integr. Plant Biol. 2020, 62, 1653–1658. [Google Scholar] [CrossRef]
- Li, B.; Rui, H.; Li, Y.; Wang, Q.; Alariqi, M.; Qin, L.; Sun, L.; Ding, X.; Wang, F.; Zou, J.; et al. Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol. J. 2019, 17, 1862. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Mukhtar, M.S.; Mansoor, S. Genome Editing: Targeting Susceptibility Genes for Plant Disease Resistance. Trends Biotechnol. 2018, 36, 898–906. [Google Scholar] [CrossRef]
- Kim, Y.A.; Moon, H.; Park, C.J. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice 2019, 12, 67. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [PubMed]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Ortigosa, A.; Gimenez-Ibanez, S.; Leonhardt, N.; Solano, R. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2019, 17, 665–673. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, Z.; Feng, Z.; Wei, P.; Zhang, H.; Botella, J.R.; Zhu, J.K. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 2016, 14, 519–532. [Google Scholar] [CrossRef]
- Paul, N.C.; Park, S.W.; Liu, H.; Choi, S.; Ma, J.; MacCready, J.S.; Chilvers, M.I.; Sang, H. Plant and Fungal Genome Editing to Enhance Plant Disease Resistance Using the CRISPR/Cas9 System. Front. Plant Sci. 2021, 12, 1534. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 482. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef]
- Langner, T.; Kamoun, S.; Belhaj, K. CRISPR Crops: Plant Genome Editing Toward Disease Resistance. Annu. Rev. Phytopathol. 2018, 11, 479–512. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar] [CrossRef]
- Sharma, S.; Sundaresha, S.; Bhardwaj, V. Biotechnological approaches in management of oomycetes diseases. 3 Biotech 2021, 11, 274. [Google Scholar] [CrossRef]
- Ho, H.H. The taxonomy and biology of Phytophthora and Pythium. J. Bacteriol. Mycol. 2018, 6, 40–45. [Google Scholar] [CrossRef]
- De Lange, J.; Nalley, L.L.; Yang, W.; Shew, A.; de Steur, H. The future of CRISPR gene editing according to plant scientists. iScience 2022, 25, 105012. [Google Scholar] [CrossRef]
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 2018, 173, 1454–1467.e10. [Google Scholar] [CrossRef]
- Sung, Y.C.; Lin, C.P.; Hsu, H.J.; Chen, Y.L.; Chen, J.C. Silencing of CrNPR1 and CrNPR3 Alters Plant Susceptibility to Periwinkle Leaf Yellowing Phytoplasma. Front. Plant Sci. 2019, 10, 1183. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef]
- German-Retana, S.; Walter, J.; Doublet, B.; Roudet-Tavert, G.; Nicaise, V.; Lecampion, C.; Houvenaghel, M.-C.; Robaglia, C.; Michon, T.; Gall, O. Le Mutational Analysis of Plant Cap-Binding Protein eIF4E Reveals Key Amino Acids Involved in Biochemical Functions and Potyvirus Infection. J. Virol. 2008, 82, 7601. [Google Scholar] [CrossRef]
- Gosavi, G.; Yan, F.; Ren, B.; Kuang, Y.; Yan, D.; Zhou, X.; Zhou, H. Applications of CRISPR technology in studying plant-pathogen interactions: Overview and perspective. Phytopathol. Res. 2020, 2, 21. [Google Scholar] [CrossRef]
- Juma, B.S.; Mukami, A.; Mweu, C.; Ngugi, M.P.; Mbinda, W. Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava. Front. Plant Sci. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhou, H.; Zhou, X.; Li, F. Control of Plant Viruses by CRISPR/Cas System-Mediated Adaptive Immunity. Front. Microbiol. 2020, 11, 2613. [Google Scholar] [CrossRef] [PubMed]
- Gomez, M.A.; Lin, Z.D.; Moll, T.; Chauhan, R.D.; Hayden, L.; Renninger, K.; Beyene, G.; Taylor, N.J.; Carrington, J.C.; Staskawicz, B.J.; et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 2019, 17, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-Debast, A.; Nogué, F.; Robaglia, C.; Gallois, J.L. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019, 17, 1736–1750. [Google Scholar] [CrossRef]
- Beam, K.; Ascencio-Ibáñez, J.T. Geminivirus Resistance: A Minireview. Front. Plant Sci. 2020, 11, 1131. [Google Scholar] [CrossRef]
- Khatodia, S.; Bhatotia, K.; Tuteja, N. Development of CRISPR/Cas9 mediated virus resistance in agriculturally important crops. Bioengineered 2017, 8, 274. [Google Scholar] [CrossRef]
- Shukla, A.; Pagán, I.; García-Arenal, F. Effective tolerance based on resource reallocation is a virus-specific defence in Arabidopsis thaliana. Mol. Plant Pathol. 2018, 19, 1454. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Park, J.; Kim, M.J.; Hwang, I.; Park, Y.; Kim, J.Y. CRISPR/Cas9-Mediated Generation of Pathogen-Resistant Tomato against Tomato Yellow Leaf Curl Virus and Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 1878. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X.; et al. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185. [Google Scholar] [CrossRef]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415. [Google Scholar] [CrossRef]
- Tripathi, S.; Khatri, P.; Fatima, Z.; Pandey, R.P.; Hameed, S. A Landscape of CRISPR/Cas Technique for Emerging Viral Disease Diagnostics and Therapeutics: Progress and Prospects. Pathogens 2022, 12, 56. [Google Scholar] [CrossRef]
- Mehta, D.; Stürchler, A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. CRISPR-Cas9 interference in cassava linked to the evolution of editing-resistant geminiviruses. Genome Biol. 2018, 20, 80. [Google Scholar] [CrossRef]
- Hamdan, M.F.; Karlson, C.K.S.; Teoh, E.Y.; Lau, S.-E.; Tan, B.C.; Carimi, F.; Hamdan, M.F.; Khai, C.; Karlson, S.; Teoh, E.Y.; et al. Genome Editing for Sustainable Crop Improvement and Mitigation of Biotic and Abiotic Stresses. Plants 2022, 11, 2625. [Google Scholar] [CrossRef]
- Zaidi, S.S.A.; Mahas, A.; Vanderschuren, H.; Mahfouz, M.M. Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 2020, 21, 289. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Kwon, C.T.; Heo, J.; Lemmon, Z.H.; Capua, Y.; Hutton, S.F.; Van Eck, J.; Park, S.J.; Lippman, Z.B. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 2020, 38, 182–188. [Google Scholar] [CrossRef]
- Zhang, H.; Si, X.; Ji, X.; Fan, R.; Liu, J.; Chen, K.; Wang, D.; Gao, C. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 2018, 36, 894–900. [Google Scholar] [CrossRef]
- Zhu, G.; Zhu, H. Modified Gene Editing Systems: Diverse Bioengineering Tools and Crop Improvement. Front. Plant Sci. 2022, 13, 491. [Google Scholar] [CrossRef]
- Son, S.; Park, S.R. Challenges Facing CRISPR/Cas9-Based Genome Editing in Plants. Front. Plant Sci. 2022, 13, 1627. [Google Scholar] [CrossRef] [PubMed]
- Nethery, M.A.; Hidalgo-Cantabrana, C.; Roberts, A.; Barrangou, R. CRISPR-based engineering of phages for in situ bacterial base editing. Proc. Natl. Acad. Sci. USA 2022, 119, e2206744119. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, A.; Taufiqa, S.; Baig, M.J.; Molla, K.A. Increasing disease resistance in host plants through genome editing. Proc. Indian Natl. Sci. Acad. 2022, 88, 417–429. [Google Scholar] [CrossRef]
- Deb, S.; Choudhury, A.; Kharbyngar, B.; Satyawada, R.R. Applications of CRISPR/Cas9 technology for modification of the plant genome. Genetica 2022, 150, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hillary, V.E.; Ceasar, S.A. Prime editing in plants and mammalian cells: Mechanism, achievements, limitations, and future prospects. BioEssays 2022, 44, 2200032. [Google Scholar] [CrossRef]
- Shin, J.; Miller, M.; Wang, Y.C. Recent advances in CRISPR-based systems for the detection of foodborne pathogens. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3010–3029. [Google Scholar] [CrossRef]
- Sánchez, E.; Ali, Z.; Islam, T.; Mahfouz, M. A CRISPR-based lateral flow assay for plant genotyping and pathogen diagnostics. Plant Biotechnol. J. 2022, 20, 2418–2429. [Google Scholar] [CrossRef]
- Mohammad, N.; Katkam, S.S.; Wei, Q. Recent Advances in CRISPR-Based Biosensors for Point-of-Care Pathogen Detection. Cris. J. 2022, 5, 500–516. [Google Scholar] [CrossRef]
- Lim, C.K.W.; McCallister, T.X.; Saporito-Magriña, C.; McPheron, G.D.; Krishnan, R.; Zeballos, C.M.A.; Powell, J.E.; Clark, L.V.; Perez-Pinera, P.; Gaj, T. CRISPR base editing of cis-regulatory elements enables the perturbation of neurodegeneration-linked genes. Mol. Ther. 2022, 30, 3619–3631. [Google Scholar] [CrossRef]
- Roy, R.K.; Debashree, I.; Srivastava, S.; Rishi, N.; Srivastava, A. CRISPR/Cas9 Off-targets: Computational Analysis of Causes, Prediction, Detection, and Overcoming Strategies. Curr. Bioinform. 2021, 17, 119–132. [Google Scholar] [CrossRef]
- Secgin, Z.; Uluisik, S.; Yıldırım, K.; Abdulla, M.F.; Mostafa, K.; Kavas, M. Genome-Wide Identification of the Aconitase Gene Family in Tomato (Solanum lycopersicum) and CRISPR-Based Functional Characterization of SlACO2 on Male-Sterility. Int. J. Mol. Sci. 2022, 23, 13963. [Google Scholar] [CrossRef]
- Editors, A.; Zhang, B.; Alok, A.; Awasthi, P.; Min, T.; Hwarari, D.; Li, D.A.; Movahedi, A.; Yang, L. CRISPR-Based Genome Editing and Its Applications in Woody Plants. Int. J. Mol. Sci. 2022, 23, 10175. [Google Scholar] [CrossRef]
- Roueinfar, M.; Templeton, H.N.; Sheng, J.A.; Hong, K.L. An Update of Nucleic Acids Aptamers Theranostic Integration with CRISPR/Cas Technology. Molecules 2022, 27, 1114. [Google Scholar] [CrossRef]
- Kantor, A.; McClements, M.E.; Maclaren, R.E. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef]
- Bashor, C.J.; Hilton, I.B.; Bandukwala, H.; Smith, D.M.; Veiseh, O. Engineering the next generation of cell-based therapeutics. Nat. Rev. Drug Discov. 2022, 21, 655–675. [Google Scholar] [CrossRef]
- Weeks, D.P.; Spalding, M.H.; Yang, B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 2016, 14, 483–495. [Google Scholar] [CrossRef]
- Faber, N.R.; McFarlane, G.R.; Gaynor, R.C.; Pocrnic, I.; Whitelaw, C.B.A.; Gorjanc, G. Novel combination of CRISPR-based gene drives eliminates resistance and localises spread. Sci. Rep. 2021, 11, 3791. [Google Scholar] [CrossRef]
- Gurr, G.M.; Johnson, A.C.; Ash, G.J.; Wilson, B.A.L.; Ero, M.M.; Pilotti, C.A.; Dewhurst, C.F.; You, M.S. Coconut lethal yellowing diseases: A phytoplasma threat to palms of global economic and social significance. Front. Plant Sci. 2016, 7, 1521. [Google Scholar] [CrossRef]
- Touzdjian Pinheiro Kohlrausch Távora, F.; de Assis dos Santos Diniz, F.; de Moraes Rêgo-Machado, C.; Chagas Freitas, N.; Barbosa Monteiro Arraes, F.; Chumbinho de Andrade, E.; Furtado, L.L.; Osiro, K.O.; Lima de Sousa, N.; Cardoso, T.B.; et al. CRISPR/Cas- and Topical RNAi-Based Technologies for Crop Management and Improvement: Reviewing the Risk Assessment and Challenges Towards a More Sustainable Agriculture. Front. Bioeng. Biotechnol. 2022, 10, 28. [Google Scholar] [CrossRef]
| Sr. No. | Cas Nucleases | Targeted Plants | PAM Sequences 5′-3′ | Organisms | References |
|---|---|---|---|---|---|
| 1 | SpRY | Rice | NGD and NAN | Streptococcus pyogenes | [41] |
| 2 | SpG | Rice | NGD | Streptococcus pyogenes | [41] |
| 3 | SpCas9 | Many plants | NGG | Streptococcus pyogenes | [47] |
| 4 | Cas3d/Cas5d/Cas6d/Cas7d/Cas10d | Rice and Tomato | GTH | Microcystis aeruginosa | [48] |
| 5 | NmeCas9 | Rice | NNNNGATT | Neisseria meningitidis | [49] |
| 6 | CjCas9 | Various plants | NNNNRYAC | Campylobacter jejuni | [50] |
| 7 | Cas14 | - | T-rich PAM sequences, eg. TTTA for dsDNA cleavage, no PAM sequence requirement for ssDNA | Uncultivated archea | [51] |
| 8 | Cas3 | - | No PAM sequence needed | in silico analysis of various prokaryotic genomes | [52] |
| 9 | ScCas9 | Rice | NNG | Streptococcus canis | [44] |
| 10 | LbCpf1 (Cas12a) | Rice and Arabidopsis | TTTN (TTTV) (V = A/G/C) | Lachnospiraceae bacterium ND2006 | [53] |
| 11 | SaCas9 | Arabidopsis, Rice, and Tobacco | NNGRRT | Staphylococcus aureus | [54] |
| 12 | St1Cas9 | Arabidopsis | NNAGAAW | Streptococcus thermophiles | [55] |
| 13 | FnCas12a | Rice and Tobacco | TTN | Francisella novicida | [56] |
| 14 | AsCas12a | Rice | TTTN | Acidaminococcus sp. BV3L6 | [57] |
| 15 | AacCas12b | Cotton | TTN | Alicyclobacillus acidiphilus | [58] |
| 16 | BhCas12b v4 | Arabidopsis | ATTN, TTTN, and GTTN | Bacillus hisashii | [59] |
| 17 | AsCpf1 (Cas12a) | Cotton | TTTV | Acidaminococcus sp. | [60] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ijaz, M.; Khan, F.; Zaki, H.E.M.; Khan, M.M.; Radwan, K.S.A.; Jiang, Y.; Qian, J.; Ahmed, T.; Shahid, M.S.; Luo, J.; et al. Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens. Plants 2023, 12, 1911. https://doi.org/10.3390/plants12091911
Ijaz M, Khan F, Zaki HEM, Khan MM, Radwan KSA, Jiang Y, Qian J, Ahmed T, Shahid MS, Luo J, et al. Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens. Plants. 2023; 12(9):1911. https://doi.org/10.3390/plants12091911
Chicago/Turabian StyleIjaz, Munazza, Fahad Khan, Haitham E. M. Zaki, Muhammad Munem Khan, Khlode S. A. Radwan, Yugen Jiang, Jiahui Qian, Temoor Ahmed, Muhammad Shafiq Shahid, Jinyan Luo, and et al. 2023. "Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens" Plants 12, no. 9: 1911. https://doi.org/10.3390/plants12091911
APA StyleIjaz, M., Khan, F., Zaki, H. E. M., Khan, M. M., Radwan, K. S. A., Jiang, Y., Qian, J., Ahmed, T., Shahid, M. S., Luo, J., & Li, B. (2023). Recent Trends and Advancements in CRISPR-Based Tools for Enhancing Resistance against Plant Pathogens. Plants, 12(9), 1911. https://doi.org/10.3390/plants12091911









