Root–Soil Interactions for Pepper Accessions Grown under Organic and Conventional Farming
Abstract
1. Introduction
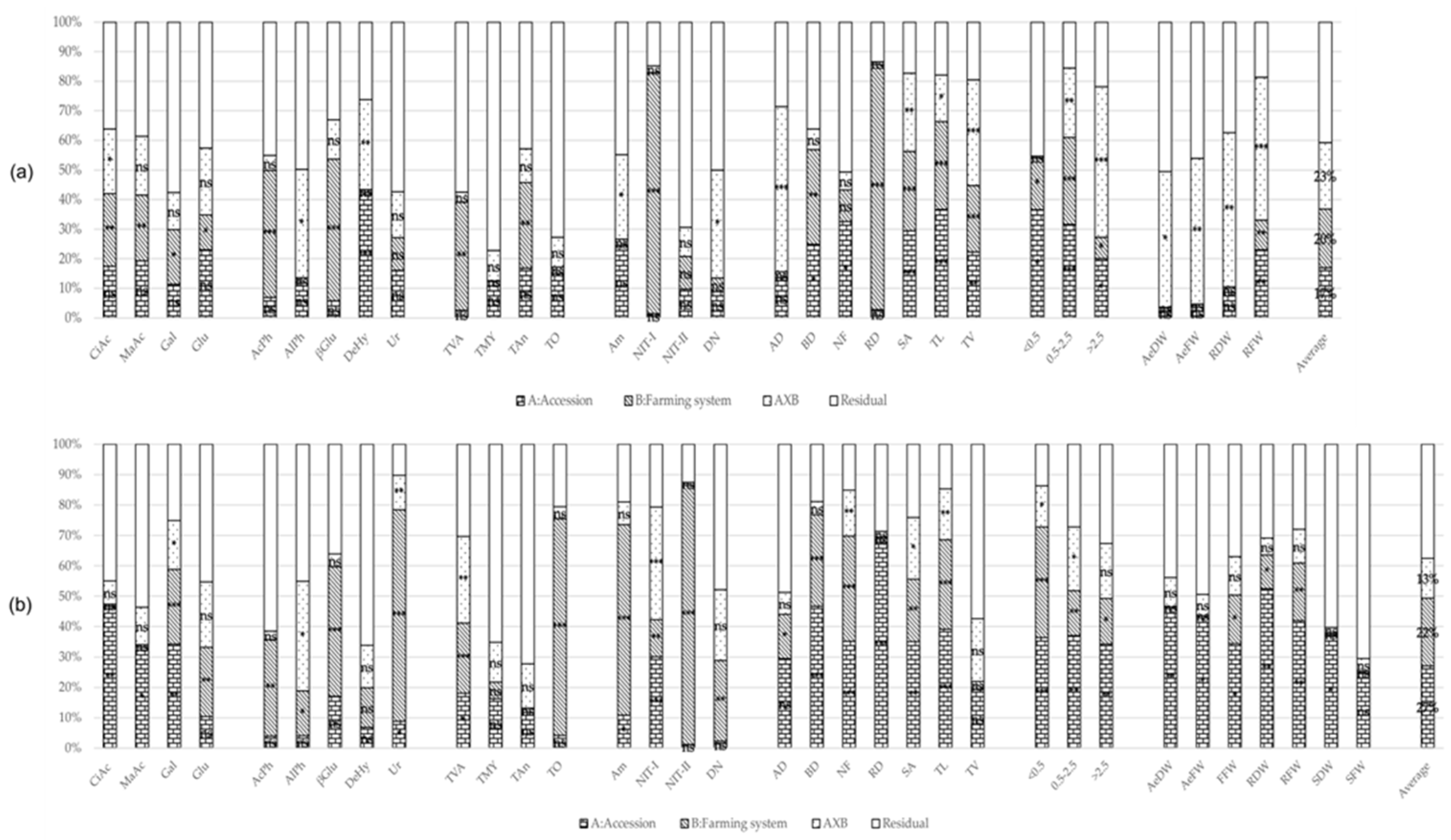
2. Results
2.1. Rhizosphere’s Traits
2.1.1. Substrate Induced Respiration
2.1.2. Soil Enzymatic Activity
2.1.3. Microbial Counts
2.1.4. Nitrogen Catabolism Potential (N-Cycle)
2.2. Plant’s Traits
2.2.1. Biomass and Yield
2.2.2. Root Parameters
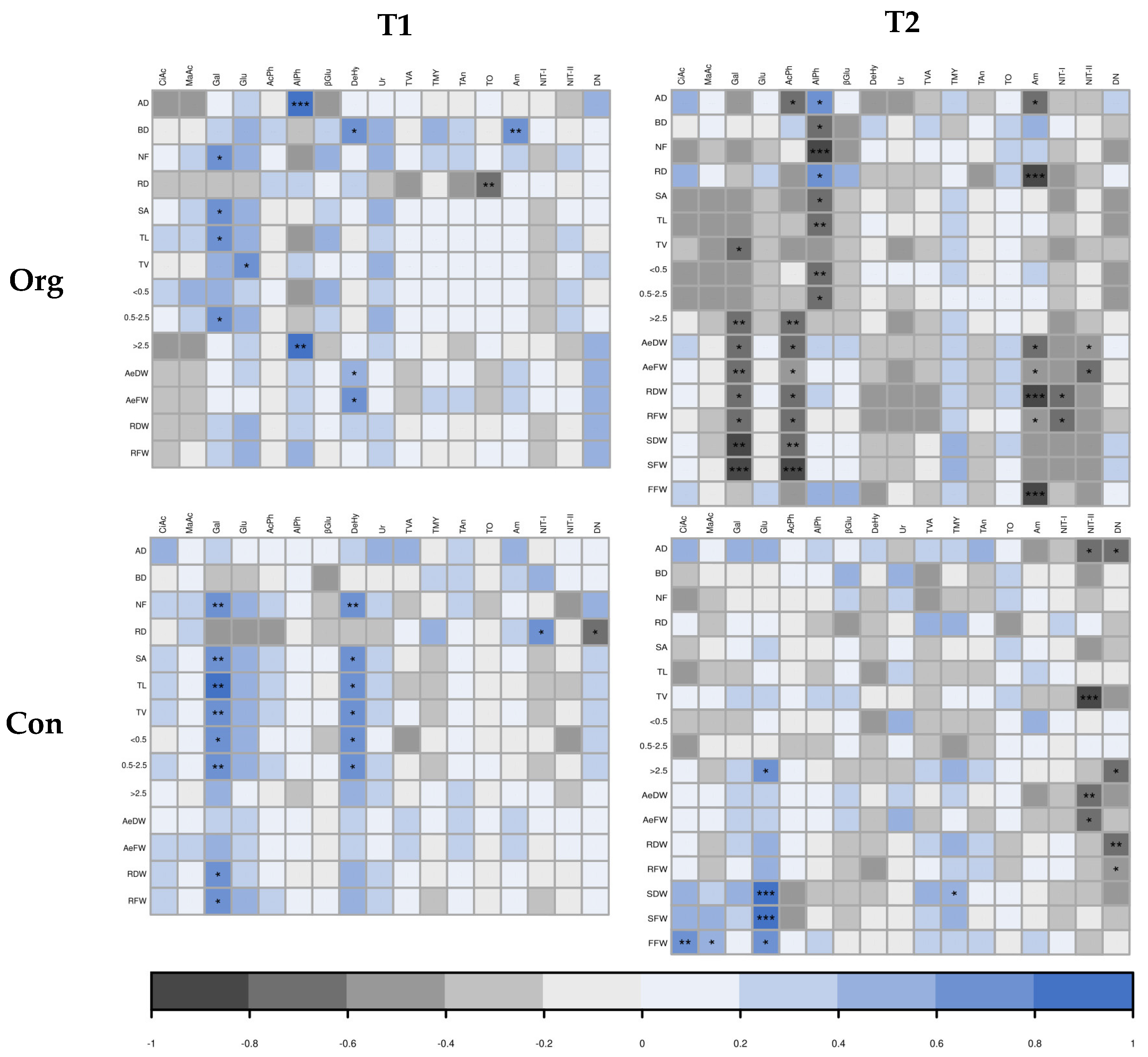
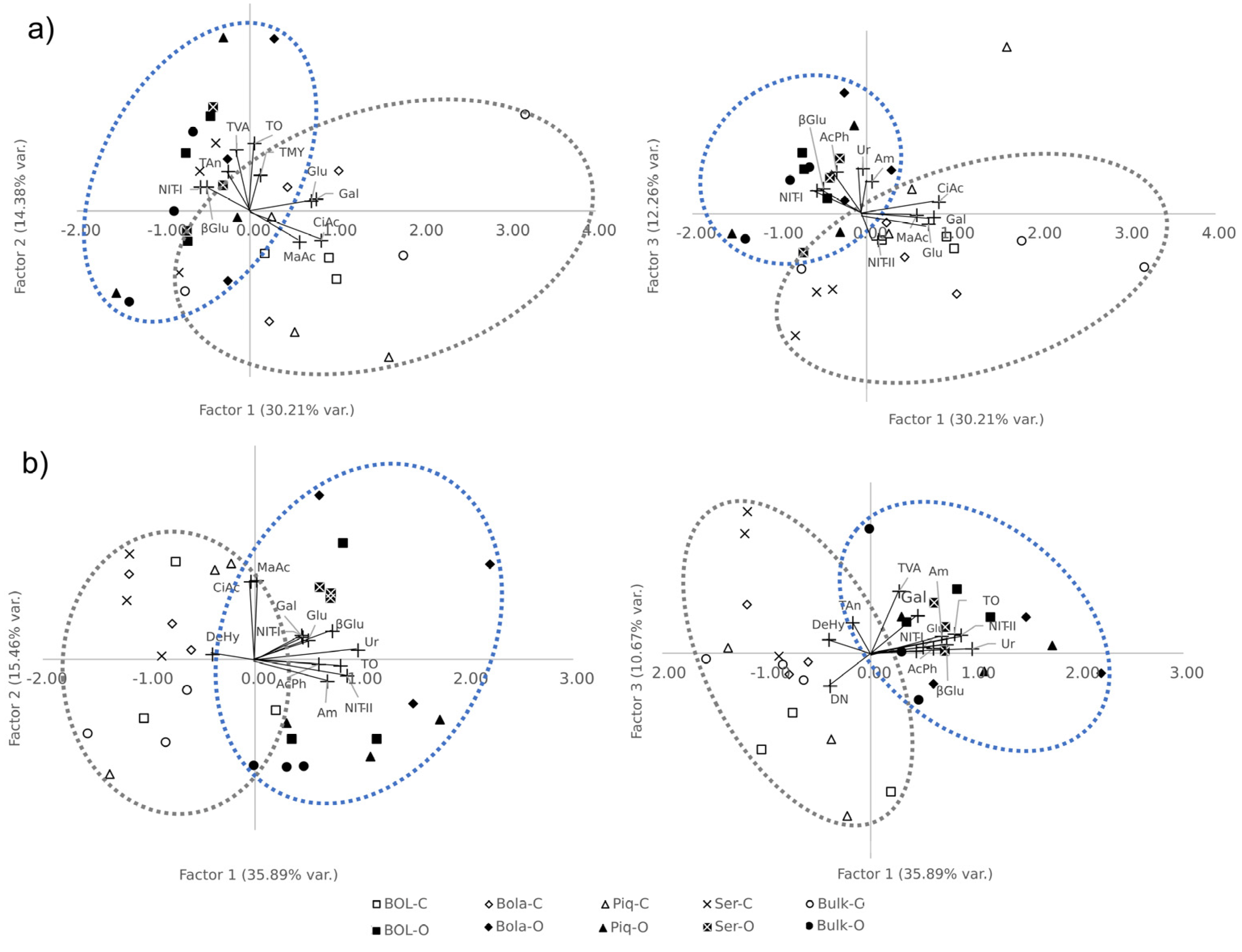
2.3. Correlations and Exploratory Factor Analysis
2.3.1. Correlation among Root, Biomass, and Rhizosphere Traits
2.3.2. Factor Analysis of Rhizosphere and Bulk Soil
3. Discussion
3.1. The Farming System Conditions the Status of the Soil
3.2. Rhizosphere Performance Depends on the Genotypes as the Crop Evolves
3.3. Plant-Soil Interactions Are Complex and Multifactorial
4. Materials and Methods
4.1. Plant Material, Experimental Design, and Sampling
4.2. Induced Respiration (IR)
4.3. Enzymatic Activities (EA) Analysis
4.4. Microbial Count (MC)
4.5. N2 Cycle (NC)
4.6. Evaluation of Plant Samples
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rodriguez-Burruezo, A.; González-Mas, M.D.C.; Nuez, F. Carotenoid composition and vitamin A value in ají (Capsicum baccatum L.) and rocoto (C. pubescens R. & P.), 2 pepper species from the Andean region. J. Food Sci. 2010, 75, S446–S453. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT Statistic Division, Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/faostat/ (accessed on 26 October 2022).
- MAPA Ministerio de Agricultura, Pesca y Alimentación. 2021. Available online: https://www.mapa.gob.es/es/ (accessed on 26 October 2022).
- Willer, H.; Trávníček, J.; Meier, C.; Schlatter, B. (Eds.) The World of Organic Agriculture 2021-Statistics and Emerging Trends; Research Institute of Organic Agriculture (FiBL) and IFOAM–Organics International: Bonn, Germany, 2021. [Google Scholar]
- Ribes-Moya, A.M.; Raigon, M.D.; Moreno-Peris, E.; Fita, A.; Rodriguez-Burruezo, A. Response to organic cultivation of heirloom Capsicum peppers: Variation in the level of bioactive compounds and effect of ripening. PLoS ONE 2018, 13, e0207888. [Google Scholar] [CrossRef] [PubMed]
- Ribes-Moya, A.M.; Adalid, A.M.; Raigon, M.D.; Hellín, P.; Fita, A.; Rodriguez-Burruezo, A. Variation in flavonoids in a collection of peppers (Capsicum sp.) under organic and conventional cultivation: Effect of the genotype, ripening stage, and growing system. J. Sci. Food Agric. 2020, 100, 2208–2223. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, P.; Cardi, T.; Bianchi, G.; Migliori, C.A.; Schiavi, M.; Rotino, G.L.; Lo Scalzo, R. Genetic and environmental factors underlying variation in yield performance and bioactive compound content of hot pepper varieties (Capsicum annuum) cultivated in two contrasting Italian locations. Eur. Food Res. Technol. 2018, 244, 1555–1567. [Google Scholar] [CrossRef]
- Zheng, Q.; Hu, Y.; Zhang, S.; Noll, L.; Böckle, T.; Dietrich, M.; Herbold, C.W.; Eichorst, S.A.; Woebken, D.; Richter, A.; et al. Soil multifunctionality is affected by the soil environment and by microbial community composition and diversity. Soil Biol. Biochem. 2019, 136, 107521. [Google Scholar] [CrossRef]
- Yang, J.; Kloepper, J.W.; Ryu, C.M. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef]
- Kandasamy, S.; Loganathan, K.; Muthuraj, R.; Duraisamy, S.; Seetharaman, S.; Thiruvengadam, R.; Ponnusamy, B.; Ramasamy, S. Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci. 2009, 7, 47. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Ryu, C.M.; Zhang, S. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 2004, 94, 1259–1266. [Google Scholar] [CrossRef]
- Singh, B.K.; Millard, P.; Whiteley, A.S.; Murrell, J.C. Unravelling rhizosphere–microbial interactions: Opportunities and limitations. Trends Microbiol. 2004, 12, 386–393. [Google Scholar] [CrossRef]
- Wang, X.; Whalley, W.R.; Miller, A.J.; White, P.J.; Zhang, F.; Shen, J. Sustainable Cropping Requires Adaptation to a Heterogeneous Rhizosphere. Trends Plant Sci. 2020, 25, 1194–1202. [Google Scholar] [CrossRef]
- Pereira-Dias, L.; Gil-Villar, D.; Castell-Zeising, V.; Quiñones, A.; Calatayud, Á.; Rodríguez-Burruezo, A.; Fita, A. Main root adaptations in pepper germplasm (Capsicum spp.) to phosphorus low-input conditions. Agronomy 2020, 10, 637. [Google Scholar] [CrossRef]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef]
- Aira, M.; Gomez-Brandon, M.; Lazcano, C.; Bååth, E.; Dominguez, J. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 2010, 42, 2276–2281. [Google Scholar] [CrossRef]
- Toljander, J.F.; Lindahl, B.D.; Paul, L.R.; Elfstrand, M.; Finlay, R.D. Influence of arbuscular mycorrhizal mycelial exudates on soil bacterial growth and community structure. FEMS Microbiol. Ecol. 2007, 61, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.A. Earthworm Ecology, 2nd ed.; CRC Press: London, UK, 2004. [Google Scholar]
- Tate, R.L. Soil Microbiology; John Wiley & Sons Ltd.: New York, NY, USA, 2000. [Google Scholar]
- Nelkner, J.; Henke, C.; Lin, T.W.; Pätzold, W.; Hassa, J.; Jaenicke, S.; Grosch, R.; Pühler, A.; Sczyrba, A.; Schlüter, A. Effect of Long-Term Farming Practices on Agricultural Soil Microbiome Members Represented by Metagenomically Assembled Genomes (MAGs) and Their Predicted Plant-Beneficial Genes. Genes 2019, 10, 424. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Micro.-Biol. 2013, 37, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Degens, B.P.; Harris, J.A. Development of a physiological approach to measuring the catabolic diversity of soil microbial communities. Soil Biol. Biochem. 1997, 29, 1309–1320. [Google Scholar] [CrossRef]
- Nkongolo, K.K.; Narendrula-Kotha, R. Advances in monitoring soil microbial community dynamic and function. J. Appl. Genet. 2020, 61, 249–263. [Google Scholar] [CrossRef]
- Degens, B.P.; Schipper, L.A.; Sparling, G.P.; Vojvodic-Vukovic, M. Decreases in organic C reserves in soils can reduce the catabolic diversity of soil microbial communities. Soil Biol. Biochem. 2000, 32, 189–196. [Google Scholar] [CrossRef]
- Aon, M.A.; Cabello, M.N.; Sarena, D.E.; Colaneri, A.C.; Franco, M.G.; Burgos, J.L.; Cortassa, S.I. Spatio-temporal patterns of soil microbial and enzymatic activities in an agricultural soil. Appl. Soil Ecol. 2001, 18, 239–254. [Google Scholar] [CrossRef]
- Lin, Q.; Brookes, P.C. An evaluation of the substrate-induced respiration method. Soil Biol. Biochem. 1999, 31, 1969–1983. [Google Scholar] [CrossRef]
- Campbell, C.D.; Chapman, S.J.; Cameron, C.M.; Davidson, M.S.; Potts, J.M. A rapid microtiter plate method to measure carbon dioxide evolved from carbon substrate amendments so as to determine the physiological profiles of soil microbial communities by using whole soil. Appl. Env. Microbiol. 2003, 69, 3593–3599. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Wood, S.A.; de Mesquita, C.P.B. How microbes can, and cannot, be used to assess soil health. Soil Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Kaiser, H.F. An index of factorial simplicity. Psychometrika 1974, 39, 31–36. [Google Scholar] [CrossRef]
- De Brogniez, D.; Ballabio, C.; Stevens, A.; Jones, R.J.A.; Montanarella, L.; van Wesemael, B. A map of the topsoil organic carbon content of Europe generated by a generalized additive model. Eur. J. Soil Sci. 2015, 66, 121–134. [Google Scholar] [CrossRef]
- Olayemi, O.P.; Schneekloth, J.P.; Wallenstein, M.D.; Trivedi, P.; Calderón, F.J.; Corwin, J.; Fonte, S.J. Soil macrofauna and microbial communities respond in similar ways to management drivers in an irrigated maize system of Colorado (USA). Appl. Soil Ecol. 2022, 178, 104562. [Google Scholar] [CrossRef]
- Rowell, M.J. Colorimetric method for CO2 measurement in soils. Soil Biol. Biochem. 1995, 27, 373–375. [Google Scholar] [CrossRef]
- Cookson, W.R.; Murphy, D.V.; Roper, M.M. Characterizing the relationships between soil organic matter components and microbial function and composition along a tillage disturbance gradient. Soil Biol. Biochem. 2008, 40, 763–777. [Google Scholar] [CrossRef]
- Sandén, T.; Zavattaro, L.; Spiegel, H.; Grignani, C.; Sandén, H.; Baumgarten, A.; Tiirola, M.; Mikkonen, A. Out of sight: Profiling soil characteristics, nutrients and bacterial communities affected by organic amendments down to one meter in a long-term maize experiment. Appl. Soil Ecol. 2019, 134, 54–63. [Google Scholar] [CrossRef]
- Swallow, M.J.; Quideau, S.A. A method for determining community level physiological profiles of organic soil horizons. Soil Sci. Soc. Am. J. 2015, 79, 536–542. [Google Scholar] [CrossRef]
- Babur, E.; Dindaroğlu, T.; Riaz, M.; Uslu, O.S. Seasonal variations in litter layers’ characteristics control microbial respiration and microbial carbon utilization under mature pine, cedar, and beech forest stands in the Eastern Mediterranean Karstic Ecosystems. Microb. Ecol. 2022, 84, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Dacal, M.; Bradford, M.A.; Plaza, C.; Maestre, F.T.; García-Palacios, P. Soil microbial respiration adapts to ambient temperature in global drylands. Nat. Ecol. Evol. 2019, 3, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.A.; McCulley, R.L.; Crowther, T.; Oldfield, E.E.; Wood, S.A.; Fierer, N. Cross-biome patterns in soil microbial respiration predictable from evolutionary theory on thermal adaptation. Nat. Ecol. Evol. 2019, 3, 223–231. [Google Scholar] [CrossRef]
- Ali, R.S.; Poll, C.; Kandeler, E. Dynamics of soil respiration and microbial communities: Interactive controls of temperature and substrate quality. Soil Biol. Biochem. 2018, 127, 60–70. [Google Scholar] [CrossRef]
- Wang, C.; Morrissey, E.M.; Mau, R.L.; Hayer, M.; Piñeiro, J.; Mack, M.C.; Marks, J.C.; Bell, S.L.; Miller, S.N.; Schwartz, E.; et al. The temperature sensitivity of soil: Microbial biodiversity, growth, and carbon mineralization. ISME J. 2021, 15, 2738–2747. [Google Scholar] [CrossRef] [PubMed]
- Creamer, R.E.; Stone, D.; Berry, P.; Kuiper, I. Measuring respiration profiles of soil microbial communities across Europe using MicroResp™ method. Appl. Soil Ecol. 2016, 97, 36–43. [Google Scholar] [CrossRef]
- Attademo, A.M.; Sanchez-Hernandez, J.C.; Lajmanovich, R.C.; Repetti, M.R.; Peltzer, P.M. Enzyme Activities as Indicators of Soil Quality: Response to Intensive Soybean and Rice Crops. Water Air Soil Pollut. 2021, 232, 295. [Google Scholar] [CrossRef]
- Dotaniya, M.L.; Aparna, K.; Dotaniya, C.K.; Singh, M.; Regar, K.L. Role of soil enzymes in sustainable crop production. In Enzymes in Food Biotechnology; Academic Press: Cambridge, MA, USA, 2019. [Google Scholar]
- Jat, H.S.; Datta, A.; Choudhary, M.; Sharma, P.C.; Dixit, B.; Jat, M.L. Soil enzymes activity: Effect of climate smart agriculture on rhizosphere and bulk soil under cereal based systems of north-west India. Eur. J. Soil Biol. 2021, 103, 103292. [Google Scholar] [CrossRef]
- Sun, Y.; Goll, D.S.; Ciais, P.; Peng, S.; Margalef, O.; Asensio, D.; Sardans, J.; Peñuelas, J. Spatial Pattern and Environmental Drivers of Acid Phosphatase Activity in Europe. Front. Big Data 2020, 2, 51. [Google Scholar] [CrossRef]
- Bandick, A.K.; Dick, R.P. Field management effects on soil enzyme activities. Soil Biol. Biochem. 1999, 31, 1471–1479. [Google Scholar] [CrossRef]
- Mangalassery, S.; Mooney, S.J.; Sparkes, D.L.; Fraser, W.T.; Sjögersten, S. Impacts of zero tillage on soil enzyme activities, microbial characteristics and organic matter functional chemistry in temperate soils. Eur. J. Soil Biol. 2015, 68, 9–17. [Google Scholar] [CrossRef]
- Margalef, O.; Sardans, J.; Maspons, J.; Molowny-Horas, R.; Fernández-Martínez, M.; Janssens, I.A.; Richter, A.; Ciais, P.; Obersteiner, M.; Peñuelas, J. The effect of global change on soil phosphatase activity. Glob. Chang. Biol. 2021, 27, 5989–6003. [Google Scholar] [CrossRef] [PubMed]
- Habteselassie, M.; Woodruff, L.; Norton, J.; Ouyang, Y.; Sintim, H. Changes in microbial communities in soil treated with organic or conventional N sources. J. Environ. Qual. 2022, 51, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.; Zhang, R. Responses of Bacterial Communities in Arable Soils in a Rice-Wheat Cropping System to Different Fertilizer Regimes and Sampling Times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.R.; Delgado-Baquerizo, M.; Wang, J.T.; Hu, H.W.; Yang, Z.; He, J.Z. New insights into the role of microbial community composition in driving soil respiration rates. Soil Biol. Biochem. 2018, 118, 35–41. [Google Scholar] [CrossRef]
- Tkacz, A.; Bestion, E.; Bo, Z.; Hortala, M.; Poole, P.S. Influence of plant fraction, soil, and plant species on microbiota: A multikingdom comparison. Mbio 2020, 11, e02785-19. [Google Scholar] [CrossRef]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The Evolution and Future of Earth’s Nitrogen Cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef]
- Xiao, D.; He, X.; Wang, G.; Xu, X.; Hu, Y.; Chen, X.; Zhang, W.; Su, Y.; Wang, K.; Soromotin, A.V.; et al. Network analysis reveals bacterial and fungal keystone taxa involved in straw and soil organic matter mineralization. Appl. Soil Ecol. 2022, 173, 104395. [Google Scholar] [CrossRef]
- Henneron, L.; Kardol, P.; Wardle, D.A.; Cros, C.; Fontaine, S. Rhizosphere control of soil nitrogen cycling: A key component of plant economic strategies. New Phytol. 2020, 228, 1269–1282. [Google Scholar] [CrossRef]
- Yue, H.; Banerjee, S.; Liu, C.; Ren, Q.; Zhang, W.; Zhang, B.; Tian, X.; Wei, G.; Shu, D. Fertilizing-induced changes in the nitrifying microbiota associated with soil nitrification and crop yield. Sci. Total Environ. 2022, 841, 156752. [Google Scholar] [CrossRef]
- Lin, H.-C.; Huber, J.A.; Gerl, G.; Hülsbergen, K.-J. Nitrogen balances and nitrogen-use efficiency of different organic and conventional farming systems. Nutr. Cycl. Agroecosystems 2016, 105, 1–23. [Google Scholar] [CrossRef]
- Masson-Delmotte, V.; Zhai, P.; Pirani, A.; Connors, S.L.; Péan, C.; Berger, S.; Caud, N.; Chen, Y.; Goldfarb, L.; Gomis, M.I.; et al. (Eds.) IPCC, 2021: Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021. [Google Scholar]
- Lazcano, C.; Zhu-Barker, X.; Decock, C. Effects of Organic Fertilizers on the Soil Microorganisms Responsible for N2O Emissions: A Review. Microorganisms 2021, 9, 983. [Google Scholar] [CrossRef] [PubMed]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2019, 39, 3–17. [Google Scholar] [CrossRef]
- Brolsma, K.M.; Vonk, J.A.; Mommer, L.; Van Ruijven, J.; Hoffland, E.; De Goede, R.G. Microbial catabolic diversity in and beyond the rhizosphere of plant species and plant genotypes. Pedobiologia 2017, 61, 43–49. [Google Scholar] [CrossRef]
- Hirte, J.; Leifeld, J.; Abiven, S.; Oberholzer, H.R.; Mayer, J. Below ground carbon inputs to soil via root biomass and rhizodeposition of field-grown maize and wheat at harvest are independent of net primary productivity. Agric. Ecosyst. Environ. 2018, 265, 556–566. [Google Scholar] [CrossRef]
- Bergstrom, D.W.; Monreal, C.M.; Tomlin, A.D.; Miller, J.J. Interpretation of soil enzyme activities in a comparison of tillage practices along a topographic and textural gradient. Can. J. Soil Sci. 2000, 80, 71–79. [Google Scholar] [CrossRef]
- Choudhary, M.; Jat, H.S.; Datta, A.; Yadav, A.K.; Sapkota, T.B.; Mondal, S.; Meena, R.P.; Sharma, P.C.; Jat, M.L. Sustainable ntensification influences soil quality, biota, and productivity in cereal-based agroecosystems. Appl. Soil Ecol. 2018, 126, 189–198. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Factors affecting rhizosphere priming effects. J. Plant Nutr. Soil Sci. 2002, 165, 382–396. [Google Scholar] [CrossRef]
- Fita, A.; Garcia-Martinez, M.D.; Raigon, M.D.; Lerma, M.D.; Moreno, E.; Rodriguez-Burruezo, A. Peppers: Soil dynamics, root architecture and fruit quality. In International Congress. STRATEGIES for Organic and Low Input Agricultures and Their Food Systems; Solibam Congress: Nantes, France, 2014. [Google Scholar]
- Ribes-Moya, A.M.; Morales-Manzo, I.I.; Aguilar, C.L.; Raigón, M.D.; Rodríguez-Burruezo, A. Estudio preliminar de la actividad enzimática fosfatasa alcalina y catalasa en cultivos ecológico y convencional de ecotipos de pimiento (Capsicum sp.). In XXVII Jornadas Técnicas de SEAE. VI Congreso Valenciano de Agricultura Ecológica; Sociedad Española de Agricultura Ecológica: Gandía, España, 2019; pp. 244–254. [Google Scholar]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Moreau, D.; Bardgett, R.D.; Finlay, R.D.; Jones, D.L.; Philippot, L. A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 2019, 33, 540–552. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudín, A.C.M. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Oburger, E.; Jones, D.L. Sampling root exudates—Mission impossible? Rhizosphere 2018, 6, 116–133. [Google Scholar] [CrossRef]
- Wen, Z.; White, P.J.; Shen, J.; Lambers, H. Linking root exudation to belowground economic traits for resource acquisition. New Phytol. 2021, 233, 1620–1635. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Li, H.; Shen, J.; Rengel, Z. Maize responds to low shoot P concentration by altering root morphology rather than increasing root exudation. Plant Soil 2017, 416, 377–389. [Google Scholar] [CrossRef]
- Iannucci, A.; Canfora, L.; Nigro, F.; De Vita, P.; Beleggia, R. Relationships between root morphology, root exudate compounds and rhizosphere microbial community in durum wheat. Appl. Soil Ecol. 2021, 158, 103781. [Google Scholar] [CrossRef]
- Microresp. James Hutton Ltd. Scotland, UK, V3.2. 2022. Available online: www.microresp.com (accessed on 6 October 2022).
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenyl phosphate for assay of soil phosphate activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis, Part 2, Chemical and Microbiological Properties, 2nd ed.; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy—Soil Science Society of America: Madison, WI, USA, 1982; pp. 903–947. [Google Scholar]
- Trevors, J.T.; Mayfield, C.I.; Innis, W.E. Measurement of electron transport systema (ETS) activity in soil. Microb. Ecol. 1982, 8, 163–168. [Google Scholar] [CrossRef]
- García, C.; Hernandez, T.; Costa, F.; Ceccanti, B.; Masciandaro, G. The dehydrogenase activity of soil as an ecological marker in process of perturbed system regeneration. In Proceedings of the XI International Symposium of Environmental Biochemistry; Gallardo-Lancho, J., Ed.; CSIC: Salamanca, Spain, September 1993; pp. 89–100. [Google Scholar]
- Kandeler, E.; Gerber, H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fert. Soils. 1988, 6, 68–72. [Google Scholar] [CrossRef]
- Kandeler, E.; Stemmer, M.; Klimanek, E.M. Response of soil microbial biomass, urease and xylanase within particle size fraction to long-term soil management. Soil. Biol. Biochem. 1999, 31, 261–273. [Google Scholar] [CrossRef]
- Alef, K.; Nannipieri, P. Methods in Applied Soil Microbiology and Biochemistry; Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- Capitán, L.F.; Avidad, R.; Fernández, M.D.; Ariza, A. Sensor de Un Solo Uso Para la Detección y Determinación de Nitrito En Aguas. Patent 2185462, 19 June 2004. [Google Scholar]
- Wei, T.; Simko, V. R Package ‘Corrplot’: Visualization of a Correlation Matrix (Version 0.92). 2021. Available online: https://github.com/taiyun/corrplot (accessed on 13 March 2022).
- Wickham, H.; Henry, L.; Pedersen, T.; Luciani, T.; Decorde, M.; Lise, V. Svglite: An ‘SVG’ Graphics Device. R Package Version 2.1.0. 2022. Available online: https://CRAN.R-project.org/package=svglite (accessed on 14 March 2022).
- Revelle, W. Psych: Procedures for Personality and Psychological Research. R Package Version 2.2.9. 2022. Available online: https://CRAN.R-project.org/package=psych (accessed on 13 March 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Jarek, S. Mvnormtest: Normality Test for Multivariate Variables. R Package Version 0.1-9. 2012. Available online: https://CRAN.R-project.org/package=mvnormtest (accessed on 13 March 2022).
- Raiche, G.; Magis, D. nFactors: Parallel Analysis and Other Non Graphical Solutions to the Cattell Scree Test. R Package Version 2.4.1.1. 2022. Available online: https://CRAN.R-project.org/package=nFactors (accessed on 15 March 2023).
- Navarro-Gonzalez, D.; Lorenzo-Seva, U. EFA.MRFA: Dimensionality Assessment Using Minimum Rank Factor Analysis. R Package Version 1.1.2. 2021. Available online: https://CRAN.R-project.org/package=EFA.MRFA (accessed on 15 March 2023).
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation. R Package Version 1.0.10. 2022. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 1 February 2023).
| Sample | Substrate | Farming System | BOL-58 | Serrano | Bola | Piquillo | Bulk | Average |
|---|---|---|---|---|---|---|---|---|
| T1 | CiAc | Org | 08.35 a | 08.09 a | 45.82 b | 15.58 a | 10.65 a | 17.70 |
| Con | 60.92 ns | 06.17 ns | 38.19 ns | 82.02 ns | 77.80 ns | 53.02 * | ||
| MaAc | Org | 08.86 a | 08.88 a | 46.58 b | 18.66 a | 24.35 a | 21.47 | |
| Con | 70.90 ns | 13.51 ns | 43.07 ns | 61.08 ns | 46.54 ns | 47.02 * | ||
| Gal | Org | 01.69 ns | 01.66 ns | 01.89 ns | 01.36 ns | 01.29 ns | 01.58 | |
| Con | 02.75 ns | 01.07 ns | 04.02 ns | 02.91 ns | 04.06 ns | 02.96 * | ||
| Glu | Org | 03.68 ns | 04.33 ns | 04.00 ns | 03.92 ns | 04.16 ns | 04.02 | |
| Con. | 06.14 ns | 02.46 ns | 12.58 ns | 07.67 ns | 40.16 ns | 13.80 * | ||
| T2 | CiAc | Org. | 25.75 ab | 58.76 b | 37.33 ab | 08.13 a | 12.24 a | 28.44 NS |
| Con. | 22.62 ns | 51.84 ns | 42.64 ns | 35.99 ns | 06.87 ns | 31.99 NS | ||
| MaAc | Org. | 29.80 ns | 43.62 ns | 50.53 ns | 08.06 ns | 05.51 ns | 27.50 NS | |
| Con. | 27.98 ns | 30.28 ns | 43.57 ns | 32.39 ns | 22.37 ns | 31.32 NS | ||
| Gal | Org. | 01.81 ns | 01.80 ns | 02.22 ns | 01.64 ns | 01.66 ns | 01.83 * | |
| Con. | 01.13 ab | 02.08 b | 01.66 ab | 00.41 a | 00.47 a | 01.15 | ||
| Glu | Org. | 04.01 ns | 05.87 ns | 06.56 ns | 03.41 ns | 03.82 ns | 04.73 * | |
| Con. | 01.69 ns | 02.39 ns | 01.77 ns | 02.12 ns | 05.10 ns | 02.61 |
| Sampling | Enzyme | Farming System | BOL-58 | Serrano | Bola | Piquillo | Bulk | Average |
|---|---|---|---|---|---|---|---|---|
| T1 | AcPh | Org. | 90.16 ns | 89.34 ns | 98.49 ns | 87.59 ns | 89.85 ns | 91.09 * |
| Con. | 76.15 ns | 65.68 ns | 76.81 ns | 80.23 ns | 70.56 ns | 73.89 | ||
| AlPh | Org. | 455.69 a | 563.92 b | 450.40 a | 429.01 a | 467.92 a | 473.39 NS | |
| Con. | 460.04 ns | 402.87 ns | 435.10 ns | 459.57 ns | 526.41 ns | 456.80 NS | ||
| βGlu | Org. | 00.72 ns | 00.61 ns | 00.80 ns | 00.68 ns | 00.79 ns | 00.72 * | |
| Con. | 00.40 ns | 00.60 ns | 00.55 ns | 00.44 ns | 00.39 ns | 00.48 | ||
| DeHy | Org. | 01.18 b | 00.95 a | 00.91 a | 00.99 a | 00.94 a | 00.99 NS | |
| Con. | 01.08 b | 00.78 a | 01.14 b | 01.07 b | 01.02 b | 01.02 NS | ||
| Ur | Org. | 00.16 ns | 00.17 ns | 00.17 ns | 00.18 ns | 00.20 ns | 00.18 NS | |
| Con. | 00.14 ns | 00.13 ns | 00.15 ns | 00.20 ns | 00.14 ns | 00.15 NS | ||
| T2 | AcPh | Org. | 93.99 ns | 85.00 ns | 95.04 ns | 87.75 ns | 87.60 ns | 89.88 * |
| Con. | 76.11 ns | 69.38 ns | 72.79 ns | 76.03 ns | 78.22 ns | 74.51 | ||
| AlPh | Org. | 477.13 a | 569.41 b | 472.26 a | 449.11 a | 458.76 a | 485.33 * | |
| Con. | 406.98 ns | 350.58 ns | 420.99 ns | 447.58 ns | 510.30 ns | 427.29 | ||
| βGlu | Org. | 00.62 ns | 00.70 ns | 00.67 ns | 00.60 ns | 00.52 ns | 00.62 * | |
| Con. | 00.53 ns | 00.43 ns | 00.50 ns | 00.44 ns | 00.34 ns | 00.45 | ||
| DeHy | Org. | 01.04 ns | 00.94 ns | 00.96 ns | 00.99 ns | 00.98 ns | 00.98 NS | |
| Con. | 01.02 ns | 01.03 ns | 01.14 ns | 00.99 ns | 01.07 ns | 01.05 NS | ||
| Ur | Org. | 00.31 a | 00.36 a | 00.47 b | 00.35 a | 00.30 a | 00.36 * | |
| Con. | 00.23 ns | 00.19 ns | 00.19 ns | 00.20 ns | 00.18 ns | 00.20 |
| Sampling | Microbial Count | Farming System | BOL-58 | Serrano | Bola | Piquillo | Bulk | Average |
|---|---|---|---|---|---|---|---|---|
| T1 | TVA | Org | 7.34 ns | 7.51 ns | 7.53 ns | 7.37 ns | 7.27 ns | 7.40 * |
| Con | 6.88 ns | 7.00 ns | 6.78 ns | 7.04 ns | 6.89 ns | 6.92 | ||
| TMY | Org | 8.30 ns | 4.67 ns | 5.57 ns | 5.12 ns | 4.45 ns | 5.62 NS | |
| Con | 5.64 ns | 5.78 ns | 5.59 ns | 5.06 ns | 5.40 ns | 5.50 NS | ||
| TAn | Org | 7.29 ns | 5.98 ns | 6.67 ns | 6.57 ns | 5.74 ns | 6.45 * | |
| Con | 5.64 ns | 5.69 ns | 5.53 ns | 5.90 ns | 5.39 ns | 5.63 | ||
| TO | Org | 7.18 ns | 7.33 ns | 7.55 ns | 7.33 ns | 7.23 ns | 7.33 NS | |
| Con. | 7.18 ns | 7.49 ns | 7.38 ns | 6.76 ns | 7.24 ns | 7.21 NS | ||
| T2 | TVA | Org | 7.62 ns | 7.40 ns | 7.43 ns | 7.46 ns | 7.36 ns | 7.45 * |
| Con | 6.19 a | 7.79 b | 7.05 ab | 6.42 a | 7.01 ab | 6.89 | ||
| TMY | Org | 4.76 ns | 5.47 ns | 5.43 ns | 5.33 ns | 5.44 ns | 5.329 NS | |
| Con | 5.10 ns | 5.32 ns | 5.11 ns | 5.05 ns | 4.97 ns | 5.11 NS | ||
| TAn | Org | 5.81 ns | 5.66 ns | 6.18 ns | 5.82 ns | 5.68 ns | 5.83 NS | |
| Con | 5.42 ns | 6.86 ns | 6.13 ns | 5.68 ns | 5.94 ns | 6.01 NS | ||
| TO | Org | 7.41 ns | 7.29 ns | 7.19 ns | 7.31 ns | 7.22 ns | 7.28 * | |
| Con. | 6.73 ns | 6.58 ns | 6.85 ns | 6.64 ns | 6.48 ns | 6.66 |
| Sampling | N-Cycle Stage | Farming System | BOL-58 | Serrano | Bola | Piquillo | Bulk | Average |
|---|---|---|---|---|---|---|---|---|
| T1 | Am | Org | 232.8 ns | 119 ns | 206.9 ns | 144.8 ns | 75.0 ns | 155.7 NS |
| Con | 100.9 a | 119 a | 31.0 a | 310.4 b | 51.7 a | 122.6 NS | ||
| NIT-I | Org | 24.32 ns | 16.21 ns | 20.27 ns | 20.27 ns | 20.27 ns | 20.27 * | |
| Con | 2.28 ns | 3.24 ns | 1.72 ns | 2.03 ns | 3.55 ns | 2.56 | ||
| NIT-II | Org | 22.60 ns | 15.07 ns | 18.83 ns | 18.83 ns | 18.83 ns | 18.83 NS | |
| Con | 22.60 ns | 22.60 ns | 18.83 ns | 22.60 ns | 22.60 ns | 21.85 NS | ||
| DN | Org | 2.50 ns | 3.33 ns | 1.00 ns | 2.33 ns | 3.67 ns | 2.57 NS | |
| Con. | 3.50 ns | 2.67 ns | 3.33 ns | 3.33 ns | 2.33 ns | 3.03 NS | ||
| T2 | Am | Org. | 77.60 ns | 38.80 ns | 77.60 ns | 67.25 ns | 103.5 ns | 72.94 * |
| Con. | 6.73 a | 3.10 a | 4.91 a | 31.04 b | 15.52 a | 12.26 | ||
| NIT-I | Org. | 0.20 a | 0.76 a | 2.53 b | 0.20 a | 0.20 a | 0.78 * | |
| Con. | 0.15 ns | 0.22 ns | 0.15 ns | 0.30 ns | 0.35 ns | 0.24 | ||
| NIT-II | Org. | 18.83 ns | 16.95 ns | 18.83 ns | 18.83 ns | 22.60 ns | 19.21 * | |
| Con. | 1.51 ns | 1.88 ns | 2.26 ns | 2.26 ns | 2.26 ns | 2.03 | ||
| DN | Org. | 1.33 ns | 2.50 ns | 1.67 ns | 1.00 ns | 2.33 ns | 1.77 | |
| Con. | 3.00 ns | 2.00 ns | 3.67 ns | 3.67 ns | 2.67 ns | 3.00 * |
| Sample | Biomass Traits | Farming System | BOL-58 | Serrano | Bola | Piquillo | Average |
|---|---|---|---|---|---|---|---|
| T1 | AeDW | Org | 15.40 ns | 14.74 ns | 9.73 ns | 8.69 ns | 12.14 NS |
| Con | 5.97 ns | 7.95 ns | 17.27 ns | 15.91 ns | 11.78 NS | ||
| AeFW | Org | 106.15 ns | 84.86 ns | 56.36 ns | 49.56 ns | 74.23 NS | |
| Con | 44.85 ns | 48.75 ns | 118.65 ns | 116.83 ns | 82.27 NS | ||
| RDW | Org | 1.79 ns | 1.98 ns | 1.35 ns | 1.22 ns | 1.58 NS | |
| Org | 0.83 a | 0.69 a | 2.24 b | 1.85 b | 1.40 NS | ||
| RFW | Org | 7.98 ns | 10.72 ns | 6.94 ns | 5.48 ns | 7.78 | |
| Con | 4.90 a | 4.84 a | 21.49 b | 15.63 b | 11.72 * | ||
| T2 | AeDW | Org | 239.67 ns | 329.00 ns | 117.67 ns | 175.00 ns | 215.3 NS |
| Con | 369.33 b | 262.00 ab | 112.33 a | 122.67 a | 216.6 NS | ||
| AeFW | Org | 1103.7 ns | 1173.0 ns | 407.00 ns | 723.33 ns | 851.7 NS | |
| Con | 1533.00 b | 883.00 ab | 471.18 a | 574.57 a | 865.4 NS | ||
| RDW | Org | 7.60 ns | 15.06 ns | 3.45 ns | 12.13 ns | 9.56 * | |
| Con | 6.30 b | 10.66 c | 2.62 a | 5.68 b | 6.32 | ||
| RFW | Org | 19.91 ab | 33.69 ab | 13.43 a | 43.68 b | 27.68 * | |
| Con | 14.83 ab | 23.90 b | 9.64 a | 18.86 ab | 16.80 | ||
| FFW | Org | 557.00 ns | 1849.0 ns | 419.33 ns | 714.67 ns | 885.0 | |
| Con | 1047.4 ns | 1629.5 ns | 1261.0 ns | 1657.3 ns | 1398.8 * | ||
| SDW | Org | 17.61 ns | 25.10 ns | 10.89 ns | 19.45 ns | 18.26 NS | |
| Con | 16.48 ns | 26.41 ns | 15.24 ns | 19.84 ns | 19.49 NS | ||
| SFW | Org | 47.94 ns | 69.60 ns | 35.82 ns | 58.58 ns | 52.99 NS | |
| Con | 44.72 ns | 65.79 ns | 52.14 ns | 61.64 ns | 56.08 NS |
| Sample | Root Traits | Farming System | BOL-58 | Serrano | Bola | Piquillo | Average |
|---|---|---|---|---|---|---|---|
| T1 | AD | Org | 0.88 a | 1.06 b | 0.77 a | 0.80 a | 0.88 NS |
| Con | 0.75 ns | 0.79 ns | 0.88 ns | 0.96 ns | 0.85 NS | ||
| BD | Org | 01.08 ns | 00.84 ns | 00.93 ns | 00.94 ns | 00.95 * | |
| Con | 00.90 b | 00.76 ab | 00.59 a | 00.74 ab | 00.75 | ||
| NF | Org | 896.50 ns | 539.00 ns | 1001.00 ns | 715.33 ns | 787.96 NS | |
| Con | 987.00 ab | 588.00 a | 1375.67 b | 1230.67 b | 1045.33 NS | ||
| RD | Org | 0.36 ns | 0.35 ns | 0.30 ns | 0.31 ns | 0.33 * | |
| Con | 0.16 ns | 0.17 ns | 0.15 ns | 0.16 ns | 0.16 | ||
| SA | Org | 229.96 ns | 214.40 ns | 249.69 ns | 182.88 ns | 219.23 | |
| Con | 263.43 a | 190.60 a | 658.16 b | 499.66 b | 402.96 * | ||
| TL | Org | 827.97 ns | 634.98 ns | 1042.61 ns | 728.04 ns | 808.40 | |
| Con | 1097.45 a | 767.85 a | 2356.56 c | 1650.75 b | 1468.15 * | ||
| TV | Org | 5.09 ns | 5.85 ns | 4.76 ns | 3.70 ns | 4.85 | |
| Con | 5.15 a | 3.80 a | 14.91 b | 12.20 b | 9.01 * | ||
| <0.5 | Org. | 411.63 ns | 258.57 ns | 558.74 ns | 353.67 ns | 395.65 | |
| Con. | 550.34 ab | 386.05 a | 732.84 b | 550.65 ab | 554.97 * | ||
| 0.5–2.5 | Org. | 403.25 ns | 350.15 ns | 473.91 ns | 365.93 ns | 398.31 | |
| Con. | 539.32 a | 374.29 a | 1608.41 c | 1088.20 b | 902.56 * | ||
| >2.5 | Org. | 12.43 a | 25.11 b | 07.62 a | 06.78 a | 12.98 * | |
| Con. | 06.43 a | 07.32 a | 13.29 b | 10.79 ab | 09.45 | ||
| T2 | AD | Org | 0.77 a | 0.99 b | 0.69 a | 0.81 a | 0.82 |
| Con | 0.93 ns | 1.06 ns | 0.93 ns | 0.83 ns | 0.94 * | ||
| BD | Org | 01.84 b | 01.15 a | 01.63 b | 01.60 b | 01.56 * | |
| Con | 01.60 b | 00.87 a | 01.01 a | 01.14 a | 01.15 | ||
| NF | Org | 5567.33 a | 2408.50 a | 4940.33 a | 9037.67 b | 5488.46 * | |
| Con | 3913.67 ns | 1373.50 ns | 1759.33 ns | 2862.00 ns | 2477.13 | ||
| RD | Org | 0.50 a | 0.88 b | 0.28 a | 0.40 a | 0.52 NS | |
| Con | 0.40 a | 0.74 b | 0.29 a | 0.40 a | 0.45 NS | ||
| SA | Org | 742.55 a | 645.90 a | 668.90 a | 1450.86 b | 877.05 * | |
| Con | 695.48 ns | 506.02 ns | 465.67 ns | 646.71 ns | 578.47 | ||
| TL | Org | 2976.85 a | 2102.00 a | 3034.65 a | 5622.49 b | 3434.00 * | |
| Con | 2449.62 ns | 1554.31 ns | 1592.53 ns | 2452.94 ns | 2012.35 | ||
| TV | Org | 14.88 a | 16.51 a | 12.17 a | 30.60 b | 18.54 NS | |
| Con | 22.23 ns | 14.40 ns | 10.97 ns | 14.62 ns | 15.56 NS | ||
| <0.5 | Org. | 1860.15 a | 1212.32 a | 1935.92 a | 3267.68 b | 2069.02 * | |
| Con. | 1378.57 ns | 841.44 ns | 702.66 ns | 1426.58 ns | 1087.31 | ||
| 0.5–2.5 | Org. | 1006.41 a | 771.91 a | 1020.79 a | 2151.75 b | 1237.71 * | |
| Con. | 997.92 ns | 580.75 ns | 839.86 ns | 937.97 ns | 839.12 | ||
| >2.5 | Org. | 107.48 ab | 117.57 ab | 74.77 a | 198.18 b | 124.50 * | |
| Con. | 69.95 ab | 129.53 b | 48.82 a | 85.88 ab | 83.55 |
| T1 | T2 | |||||
|---|---|---|---|---|---|---|
| Trait | F1.1 | F2.1 | F3.1 | F1.2 | F2.2 | F3.2 |
| CiAc | 0.83 | −0.36 | 0.19 | −0.04 | 0.91 | 0.16 |
| MaAc | 0.57 | −0.38 | −0.03 | 0.01 | 0.92 | −0.03 |
| Gal | 0.77 | 0.14 | −0.06 | 0.45 | 0.26 | 0.55 |
| Glu | 0.71 | 0.12 | −0.17 | 0.50 | 0.22 | 0.09 |
| AcPh | −0.35 | 0.19 | 0.66 | 0.60 | −0.06 | 0.07 |
| βGlu | −0.50 | 0.29 | 0.40 | 0.73 | 0.33 | 0.12 |
| DeHy | - | - | - | −0.40 | 0.06 | 0.19 |
| Ur | −0.05 | −0.04 | 0.72 | 0.97 | 0.10 | 0.06 |
| TVA | −0.15 | 0.73 | 0.37 | 0.27 | −0.03 | 0.91 |
| TMY | 0.12 | 0.43 | 0.32 | 0.24 | 0.18 | 0.14 |
| TAn | −0.25 | 0.47 | 0.32 | −0.17 | 0.19 | 0.45 |
| TO | 0.05 | 0.81 | −0.16 | 0.81 | −0.08 | 0.28 |
| Am | 0.06 | 0.01 | 0.51 | 0.68 | −0.26 | 0.24 |
| NIT-I | −0.57 | 0.29 | 0.37 | 0.44 | 0.28 | 0.03 |
| NIT-II | 0.13 | −0.14 | −0.42 | 0.87 | −0.19 | 0.26 |
| DN | 0.05 | −0.16 | −0.01 | −0.39 | 0.20 | −0.48 |
| % of Var. | 30.21 | 14.38 | 12.26 | 35.89 | 15.46 | 10.67 |
| Eigenvalues | 4.53 | 2.16 | 1.84 | 5.74 | 2.47 | 1.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Manzo, I.I.; Ribes-Moya, A.M.; Pallotti, C.; Jimenez-Belenguer, A.; Moro, C.P.; Raigón, M.D.; Rodríguez-Burruezo, A.; Fita, A. Root–Soil Interactions for Pepper Accessions Grown under Organic and Conventional Farming. Plants 2023, 12, 1873. https://doi.org/10.3390/plants12091873
Morales-Manzo II, Ribes-Moya AM, Pallotti C, Jimenez-Belenguer A, Moro CP, Raigón MD, Rodríguez-Burruezo A, Fita A. Root–Soil Interactions for Pepper Accessions Grown under Organic and Conventional Farming. Plants. 2023; 12(9):1873. https://doi.org/10.3390/plants12091873
Chicago/Turabian StyleMorales-Manzo, Ivan I., Ana M. Ribes-Moya, Claudia Pallotti, Ana Jimenez-Belenguer, Clara Pérez Moro, María Dolores Raigón, Adrián Rodríguez-Burruezo, and Ana Fita. 2023. "Root–Soil Interactions for Pepper Accessions Grown under Organic and Conventional Farming" Plants 12, no. 9: 1873. https://doi.org/10.3390/plants12091873
APA StyleMorales-Manzo, I. I., Ribes-Moya, A. M., Pallotti, C., Jimenez-Belenguer, A., Moro, C. P., Raigón, M. D., Rodríguez-Burruezo, A., & Fita, A. (2023). Root–Soil Interactions for Pepper Accessions Grown under Organic and Conventional Farming. Plants, 12(9), 1873. https://doi.org/10.3390/plants12091873









