Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Six Ipomoea Species and the Identification of Anthocyanin-Related Members in Sweet Potatoes
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of R2R3-MYB Family Members in Six Ipomoea Species
2.2. Phylogenetic Analysis and Classification of R2R3-MYB Genes in Six Ipomoea Species
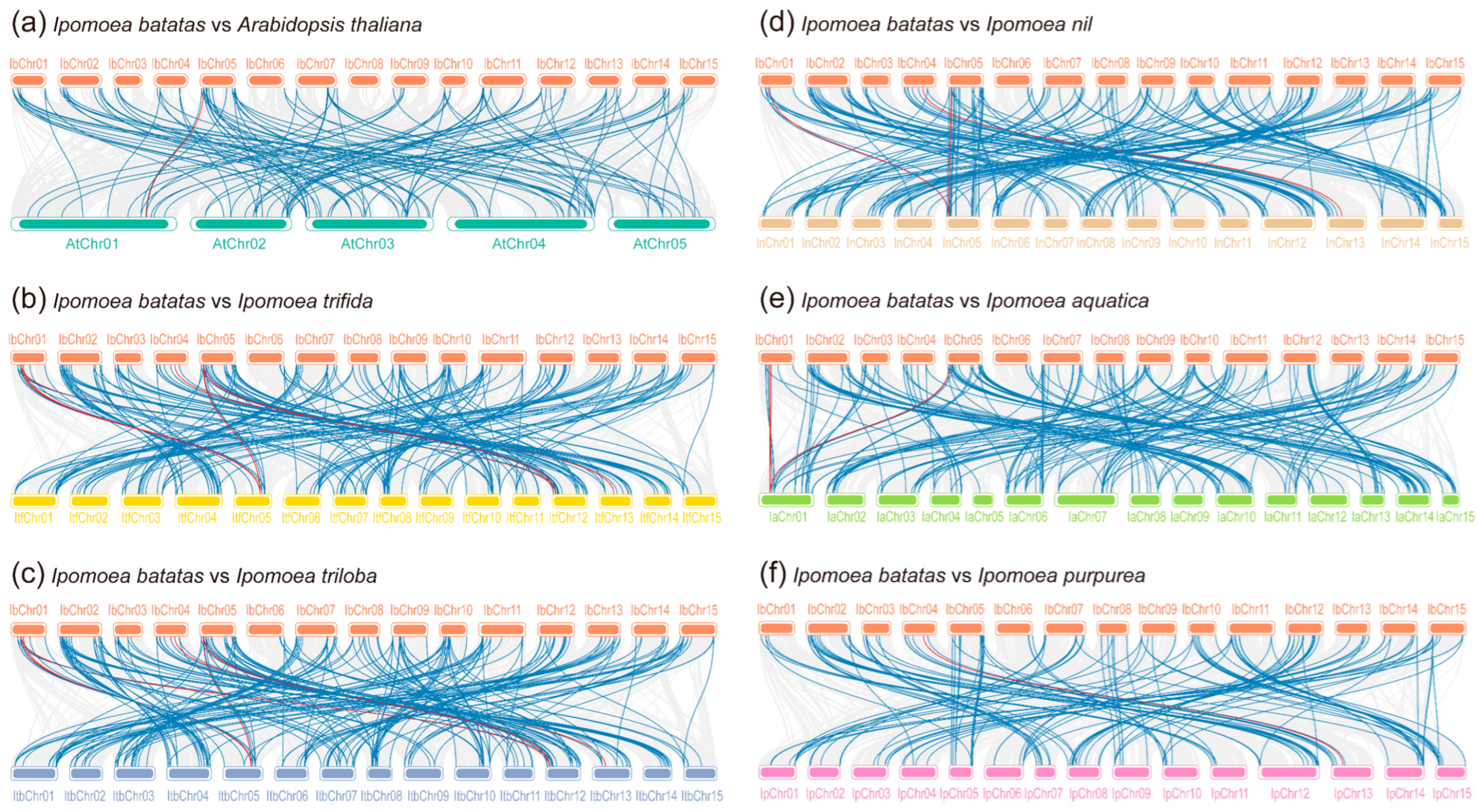
2.3. Chromosomal Distribution and Synteny Analysis of R2R3-MYBs in Six Ipomoea Species
2.4. Gene Duplication Events and Ka/Ks Analysis of R2R3-MYB Genes in Six Ipomoea
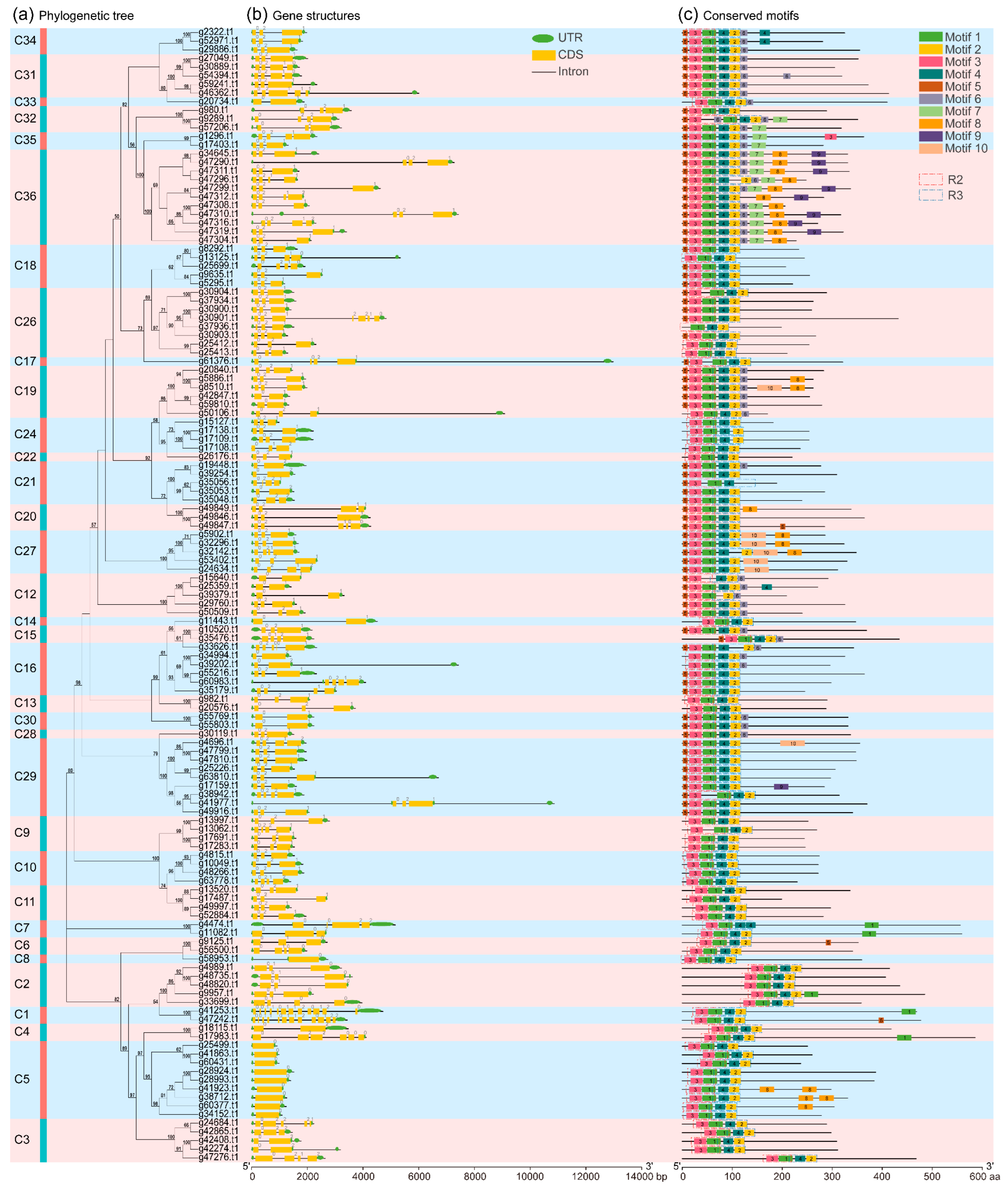
2.5. Phylogenetic Relationships, Gene Structures, and Conserved Motifs Analysis of 131 IbR2R3-MYB Genes
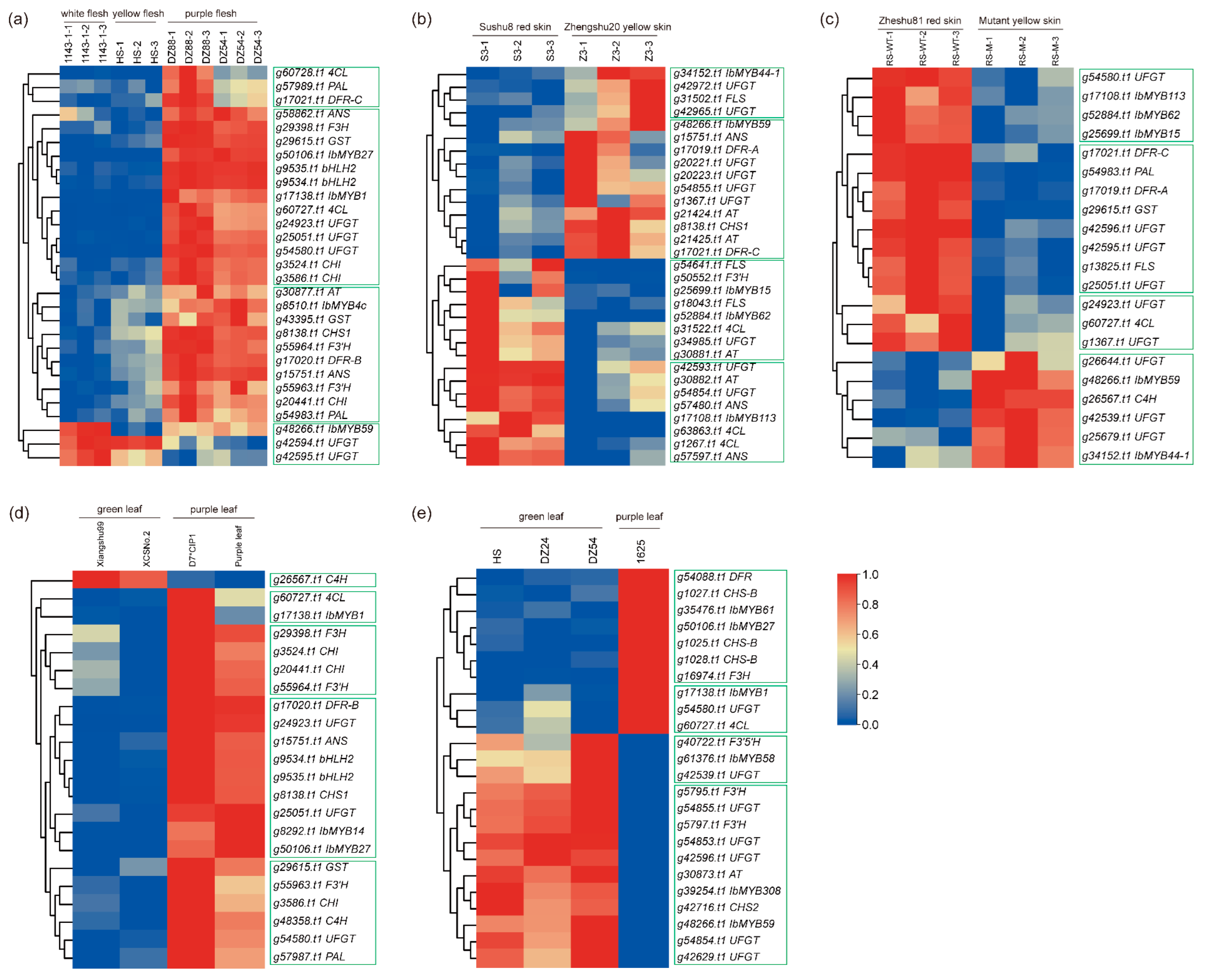
2.6. Identification of R2R3-MYBs Related to Anthocyanin Biosynthesis in Sweet Potato
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of R2R3-MYB Genes in Six Ipomoea Species
4.2. Multiple Sequences Alignment and Phylogenetic Evolution Analysis of R2R3-MYBs
4.3. Chromosome Distribution and Syntenic Analysis of R2R3-MYBs in Six Ipomoea Species
4.4. Gene Duplication Events and Ka/Ks Analysis of R2R3-MYBs in Six Ipomoea Species
4.5. Gene Structure and Conserved Protein Motif Analysis of R2R3-MYBs in Sweet potato
4.6. Identification of Anthocyanin-Related R2R3-MYBs Via RNA-Seq Data in Sweet potato
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Q. Improvement for agronomically important traits by gene engineering in sweetpotato. Breed Sci. 2017, 67, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Nie, H.; Wang, Y.; Wang, X.; Jarret, R.; Zhao, J.; Wang, H.; Yang, J. Exploring and exploiting genetics and genomics for sweetpotato improvement: Status and perspectives. Plant Commun. 2022, 3, 100332. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. Food and Agricultural Organization from the United Nations. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 14 April 2023).
- Mohanraj, R. Sweet Potato: Bioactive Compounds and Health Benefits. In Bioactive Molecules in Food; Mérillon, J.-M., Ramawat, K.G., Eds.; Reference Series in Phytochemistry; Springer International Publishing: Cham, Switerland, 2018; pp. 1–16. [Google Scholar]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Mano, H.; Ogasawara, F.; Sato, K.; Higo, H.; Minobe, Y. Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol. 2007, 143, 1252–1268. [Google Scholar] [CrossRef]
- Zhang, D.; Tan, Y.; Dong, F.; Zhang, Y.; Huang, Y.; Zhou, Y.; Zhao, Z.; Yin, Q.; Xie, X.; Gao, X.; et al. The Expression of IbMYB1 Is Essential to Maintain the Purple Color of Leaf and Storage Root in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Plant Sci. 2021, 12, 688707. [Google Scholar] [CrossRef]
- Zhao, D.; Zhao, L.; Liu, Y.; Zhang, A.; Xiao, S.; Dai, X.; Yuan, R.; Zhou, Z.; Cao, Q. Metabolomic and Transcriptomic Analyses of the Flavonoid Biosynthetic Pathway for the Accumulation of Anthocyanins and Other Flavonoids in Sweetpotato Root Skin and Leaf Vein Base. J. Agric. Food Chem. 2022, 70, 2574–2588. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends. Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jiang, C.K.; Rao, G.Y. Insights into the Diversification and Evolution of R2R3-MYB Transcription Factors in Plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef]
- Ogata, K.; Hojo, H.; Aimoto, S.; Nakai, T.; Nakamura, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structure of a DNA-binding unit of Myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. USA 1992, 89, 6428–6432. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Du, H.; Liang, Z.; Zhao, S.; Nan, M.G.; Tran, L.S.; Lu, K.; Huang, Y.B.; Li, J.N. The Evolutionary History of R2R3-MYB Proteins Across 50 Eukaryotes: New Insights Into Subfamily Classification and Expansion. Sci. Rep. 2015, 5, 11037. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef] [PubMed]
- Pratyusha, D.S.; Sarada, D.V.L. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 2022, 41, 2245–2260. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends. Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Y.; Xiao, Z.; Shen, H.; Li, Y.; Wang, Y. Multilevel regulation of anthocyanin-promoting R2R3-MYB transcription factors in plants. Front. Plant Sci. 2022, 13, 1008829. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Osbourn, A.; Ma, P. MYB Transcription Factors as Regulators of Phenylpropanoid Metabolism in Plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef]
- Cone, K.C.; Burr, F.A.; Burr, B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc. Natl. Acad. Sci. USA 1986, 83, 9631–9635. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2394. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Xiong, A.S. Changing Carrot Color: Insertions in DcMYB7 Alter the Regulation of Anthocyanin Biosynthesis and Modification. Plant Physiol. 2019, 181, 195–207. [Google Scholar] [CrossRef]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Yu, X.; Xiong, A.S. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 2020, 18, 1585–1597. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Schwinn, K.E.; Jameson, P.E.; Davies, K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011, 65, 771–784. [Google Scholar] [CrossRef]
- Aharoni, A.; De Vos, C.H.; Wein, M.; Sun, Z.; Greco, R.; Kroon, A.; Mol, J.N.; O’Connell, A.P. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001, 28, 319–332. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Li, Y.; Liu, Z.; Zhou, P.; Zeng, L.; Zhang, X.; Zhang, J.; et al. StMYB44 negatively regulates anthocyanin biosynthesis at high temperatures in tuber flesh of potato. J. Exp. Bot. 2019, 70, 3809–3824. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Guo, J.; Zhou, W.; Lu, Z.; Li, H.; Li, H.; Gao, F. Isolation and Functional Analysis of Chalcone Isomerase Gene from Purple-Fleshed Sweet Potato. Plant Mol. Biol. Report. 2015, 33, 1451–1463. [Google Scholar] [CrossRef]
- Zhou, W.; Gong, Y.; Lu, X.; Huang, C.; Gao, F. Molecular cloning and characterization of a flavonoid 3’-hydroxylase gene from purple-fleshed sweet potato (Ipomoea batatas). Mol. Biol. Rep. 2012, 39, 295–302. [Google Scholar] [CrossRef]
- Wang, H.; Fan, W.; Li, H.; Yang, J.; Huang, J.; Zhang, P. Functional characterization of Dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE 2013, 8, e78484. [Google Scholar] [CrossRef]
- Zhou, W.; Huang, C.; Gong, Y.; Feng, Q.; Gao, F. Molecular Cloning and Expression Analysis of an ANS Gene Encoding Anthocyanidin Synthase from Purple-Fleshed Sweet Potato [Ipomoea batatas (L.) Lam]. Plant Mol. Biol. Rep. 2009, 28, 112–121. [Google Scholar] [CrossRef]
- Wang, H.; Wang, C.; Fan, W.; Yang, J.; Appelhagen, I.; Wu, Y.; Zhang, P. A novel glycosyltransferase catalyses the transfer of glucose to glucosylated anthocyanins in purple sweet potato. J. Exp. Bot. 2018, 69, 5444–5459. [Google Scholar] [CrossRef]
- Kou, M.; Liu, Y.J.; Li, Z.Y.; Zhang, Y.G.; Tang, W.; Yan, H.; Wang, X.; Chen, X.G.; Su, Z.X.; Arisha, M.H.; et al. A novel glutathione S-transferase gene from sweetpotato, IbGSTF4, is involved in anthocyanin sequestration. Plant Physiol. Biochem. 2019, 135, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Takahata, Y.; Kurata, R.; Nakayama, H.; Yoshinaga, M. Structural and functional characterization of IbMYB1 genes in recent Japanese purple-fleshed sweetpotato cultivars. Mol. Breed. 2011, 29, 565–574. [Google Scholar] [CrossRef]
- Chu, H.; Jeong, J.C.; Kim, W.J.; Chung, D.M.; Jeon, H.K.; Ahn, Y.O.; Kim, S.H.; Lee, H.S.; Kwak, S.S.; Kim, C.Y. Expression of the sweetpotato R2R3-type IbMYB1a gene induces anthocyanin accumulation in Arabidopsis. Physiol. Plant 2013, 148, 189–199. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, Y.; Shi, T.; Kou, M.; Sun, J.; Xu, T.; Li, Q.; Wu, S.; Cao, Q.; Hou, W.; et al. Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals the regulatory architecture of gene expression variation in the storage roots of sweet potato. Hortic. Res. 2020, 7, 90. [Google Scholar] [CrossRef]
- Wei, Z.Z.; Hu, K.D.; Zhao, D.L.; Tang, J.; Huang, Z.Q.; Jin, P.; Li, Y.H.; Han, Z.; Hu, L.Y.; Yao, G.F.; et al. MYB44 competitively inhibits the formation of the MYB340-bHLH2-NAC56 complex to regulate anthocyanin biosynthesis in purple-fleshed sweet potato. BMC Plant Biol. 2020, 20, 258. [Google Scholar] [CrossRef]
- Deng, J.; Wu, D.; Shi, J.; Balfour, K.; Wang, H.; Zhu, G.; Liu, Y.; Wang, J.; Zhu, Z. Multiple MYB Activators and Repressors Collaboratively Regulate the Juvenile Red Fading in Leaves of Sweetpotato. Front. Plant Sci. 2020, 11, 941. [Google Scholar] [CrossRef]
- Kang, L.; Teng, Y.; Cen, Q.; Fang, Y.; Tian, Q.; Zhang, X.; Wang, H.; Zhang, X.; Xue, D. Genome-Wide Identification of R2R3-MYB Transcription Factor and Expression Analysis under Abiotic Stress in Rice. Plants 2022, 11, 1928. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin-Wang, K.; Liu, Z.; Allan, A.C.; Qin, S.; Zhang, J.; Liu, Y. Genome-wide analysis and expression profiles of the StR2R3-MYB transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 148, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Peng, R.; Tian, Y.; Han, H.; Xu, J.; Yao, Q. Genome-Wide Identification and Analysis of the MYB Transcription Factor Superfamily in Solanum lycopersicum. Plant Cell Physiol. 2016, 57, 1657–1677. [Google Scholar] [CrossRef]
- Yin, Y.; Guo, C.; Shi, H.; Zhao, J.; Ma, F.; An, W.; He, X.; Luo, Q.; Cao, Y.; Zhan, X. Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Five Solanaceae Species and Identification of Members Regulating Carotenoid Biosynthesis in Wolfberry. Int. J. Mol. Sci. 2022, 23, 2259. [Google Scholar] [CrossRef]
- Yang, Y.; Yuan, Z.; Ning, C.; Zhao, B.; Wang, R.; Zheng, X.; Liu, Y.; Chen, J.; He, L. The Pea R2R3-MYB Gene Family and Its Role in Anthocyanin Biosynthesis in Flowers. Front. Genet. 2022, 13, 936051. [Google Scholar] [CrossRef] [PubMed]
- Austin, D.F.; Huáman, Z. A synopsis of Ipomoea (Convolvulaceae) in the Americas. Taxon 2019, 45, 3–38. [Google Scholar] [CrossRef]
- Yang, J.; Moeinzadeh, M.H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.; Zheng, J.; Sun, Z.; Fan, W.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef]
- Wu, S.; Lau, K.H.; Cao, Q.; Hamilton, J.P.; Sun, H.; Zhou, C.; Eserman, L.; Gemenet, D.C.; Olukolu, B.A.; Wang, H.; et al. Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat. Commun. 2018, 9, 4580. [Google Scholar] [CrossRef]
- Hoshino, A.; Jayakumar, V.; Nitasaka, E.; Toyoda, A.; Noguchi, H.; Itoh, T.; Shin, I.T.; Minakuchi, Y.; Koda, Y.; Nagano, A.J.; et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat. Commun. 2016, 7, 13295. [Google Scholar] [CrossRef]
- Gupta, S.; Harkess, A.; Soble, A.; Van Etten, M.; Leebens-Mack, J.; Baucom, R.S. Interchromosomal linkage disequilibrium and linked fitness cost loci associated with selection for herbicide resistance. New Phytol. 2023, 238, 1263–1277. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Bao, W.; Li, G.; Gagoshidze, Z.; Shu, H.; Yang, Z.; Cheng, S.; Zhu, G.; Wang, Z. The chromosome-based genome provides insights into the evolution in water spinach. Sci. Hortic. 2021, 289, 110501. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shi, D.; Wang, Y.; Zhang, L.; Chen, X.; Yang, X.; Xiong, H.; Bhattarai, G.; Ravelombola, W.; Olaoye, D.; et al. Transcript profiling for regulation of sweet potato skin color in Sushu8 and its mutant Zhengshu20. Plant Physiol. Biochem. 2020, 148, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Xu, X.; Li, M.; Wu, X.; Guo, H. Regulatory network characterization of anthocyanin metabolites in purple sweetpotato via joint transcriptomics and metabolomics. Front. Plant Sci. 2023, 14, 1030236. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, J.Z.; He, P.W.; Xu, X.M.; Lu, Z.F.; Cui, P.; George, M.S.; Lu, G.Q. Genome-Wide Identification and Expression Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family in Sweet Potato [Ipomoea batatas (L.) Lam]. Int. J. Mol. Sci. 2023, 24, 775. [Google Scholar] [CrossRef]
- Si, Z.; Wang, L.; Qiao, Y.; Roychowdhury, R.; Ji, Z.; Zhang, K.; Han, J. Genome-wide comparative analysis of the nucleotide-binding site-encoding genes in four Ipomoea species. Front. Plant Sci. 2022, 13, 960723. [Google Scholar] [CrossRef]
- Li, X.; Zhao, L.; Zhang, H.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; He, S. Genome-Wide Identification and Characterization of CDPK Family Reveal Their Involvements in Growth and Development and Abiotic Stress in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2022, 23, 3088. [Google Scholar] [CrossRef]
- Dai, Z.; Yan, P.; He, S.; Jia, L.; Wang, Y.; Liu, Q.; Zhai, H.; Zhao, N.; Gao, S.; Zhang, H. Genome-Wide Identification and Expression Analysis of SWEET Family Genes in Sweet Potato and Its Two Diploid Relatives. Int. J. Mol. Sci. 2022, 23, 15848. [Google Scholar] [CrossRef]
- Sun, H.; Mei, J.; Zhao, W.; Hou, W.; Zhang, Y.; Xu, T.; Wu, S.; Zhang, L. Phylogenetic Analysis of the SQUAMOSA Promoter-Binding Protein-Like Genes in Four Ipomoea Species and Expression Profiling of the IbSPLs During Storage Root Development in Sweet Potato (Ipomoea batatas). Front. Plant Sci. 2021, 12, 801061. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, P.; Wu, S.; Lu, Y.; Sun, J.; Cao, Q.; Li, Z.; Xu, T. Identification and expression analysis of GRAS transcription factors in the wild relative of sweet potato Ipomoea trifida. BMC Genom. 2019, 20, 911. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sun, J.; Chen, Y.; Zhu, P.; Zhang, L.; Wu, S.; Ma, D.; Cao, Q.; Li, Z.; Xu, T. Genome-wide identification, structural and gene expression analysis of the bZIP transcription factor family in sweet potato wild relative Ipomoea trifida. BMC Genet. 2019, 20, 41. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Zhu, P.; Cao, Q.; Sun, J.; Li, Z.; Xu, T. Genome-wide identification, characterisation and functional evaluation of WRKY genes in the sweet potato wild ancestor Ipomoea trifida (H.B.K.) G. Don. under abiotic stresses. BMC Genet. 2019, 20, 90. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, C.; Guo, F.; Sun, Q.; Yu, J.; Dong, T.; Wang, X.; Song, W.; Li, Z.; Meng, X.; et al. A systematical genome-wide analysis and screening of WRKY transcription factor family engaged in abiotic stress response in sweetpotato. BMC Plant Biol. 2022, 22, 616. [Google Scholar] [CrossRef]
- Shao, Z.; He, M.; Zeng, Z.; Chen, Y.; Hanna, A.D.; Zhu, H. Genome-Wide Identification and Expression Analysis of the MADS-Box Gene Family in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Genet. 2021, 12, 750137. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Altaf, M.A.; Hao, Y.; Zhou, G.; Li, X.; Zhu, J.; Ma, W.; Wang, Z.; Bao, W. Genome-wide identification of myeloblastosis gene family and its response to cadmium stress in Ipomoea aquatica. Front. Plant Sci. 2022, 13, 979988. [Google Scholar] [CrossRef]
- Komatsuzaki, A.; Hoshino, A.; Otagaki, S.; Matsumoto, S.; Shiratake, K. Genome-wide analysis of R2R3-MYB transcription factors in Japanese morning glory. PLoS ONE 2022, 17, e0271012. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, B.; Gu, G.; Yuan, J.; Shen, S.; Jin, L.; Lin, Z.; Lin, J.; Xie, X. Genome-wide identification and expression analysis of the R2R3-MYB gene family in tobacco (Nicotiana tabacum L.). BMC Genom. 2022, 23, 432. [Google Scholar] [CrossRef]
- Isobe, S.; Shirasawa, K.; Hirakawa, H. Challenges to genome sequence dissection in sweetpotato. Breed Sci. 2017, 67, 35–40. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends. Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic. Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Gill, N.; Dhillon, B. RNA-seq Data Analysis for Differential Expression. In Fusarium wilt: Methods and Protocols; Coleman, J., Ed.; Springer: New York, NY, USA, 2022; pp. 45–54. [Google Scholar]





| Scientific Name | Common Name | Ploidy | Chromosome Number | Genome Size | Genome Gene Number | R2r3-Myb Genes Number |
|---|---|---|---|---|---|---|
| I. batatas | Sweet potato | hexaploid | 15 | 451 Mb | 64,295 | 131 |
| I. trifida | diploid | 15 | 477 Mb | 32,301 | 133 | |
| I. triloba | Trilobed morning glory | diploid | 15 | 447 Mb | 31,426 | 129 |
| I. nil | Japanese morning glory | diploid | 15 | 707 Mb | 42,783 | 127 |
| I. purpurea | Common morning glory | diploid | 15 | 581 Mb | 53,979 | 51 |
| I. aquatica | Water spinach | diploid | 15 | 494 Mb | 29,606 | 124 |
| Clade | Classification in Arabidopsis | Function Annotation | At | Ib | Itf | Itb | In | Ia | Ip | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| C1 | S28 | development/stomatal formation | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 14 |
| C2 | S25 | development/female gametogenesis | 7 | 5 | 5 | 3 | 8 | 6 | 11 | 45 |
| C3 | S21 | development/specialized metabolism/cell wall thickening | 8 | 5 | 6 | 5 | 5 | 5 | 4 | 38 |
| C4 | S23 | abiotic stress/salt stress | 3 | 2 | 2 | 2 | 2 | 2 | 1 | 14 |
| C5 | S22 | abiotic stress/drought, salt, and cold stress | 4 | 9 | 8 | 8 | 8 | 8 | 10 | 55 |
| C6 | S26 | development/sperm cell formation/pollen formation | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 13 |
| C7 | S18 and S77 | development/stamen development | 7 | 2 | 4 | 4 | 4 | 3 | 3 | 27 |
| C8 | S27 | development/leaf patterning | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 |
| C9 | S17 | abiotic stress | 3 | 4 | 3 | 3 | 3 | 3 | 1 | 20 |
| C10 | S78 | abiotic stress/potassium stress | 3 | 4 | 1 | 2 | 1 | 1 | 2 | 14 |
| C11 | S19 and S20 | development/abiotic stress | 9 | 4 | 5 | 5 | 6 | 5 | 4 | 38 |
| C12 | S8 | secondary wall/lignin biosynthesis/abiotic stress | 6 | 5 | 6 | 6 | 6 | 6 | 0 | 35 |
| C13 | S16 | development/hypocotyl elongation | 3 | 2 | 2 | 2 | 2 | 2 | 0 | 13 |
| C14 | S32 | phenylpropanoid-derived secondary metabolites/secondary cell wall biosynthesis | 2 | 1 | 1 | 2 | 1 | 2 | 1 | 10 |
| C15 | S13 | lignin/triterpenoid/cellulose biosynthesis | 4 | 2 | 3 | 3 | 3 | 3 | 0 | 18 |
| C16 | S30 and S31 | starch biosynthesis/fructan synthesis/secondary cell wall biosynthesis | 3 | 6 | 8 | 8 | 7 | 8 | 0 | 40 |
| C17 | S3 | phenylpropanoid-derived secondary metabolites/flavonoid/lignin biosynthesis | 4 | 1 | 2 | 1 | 1 | 1 | 0 | 10 |
| C18 | S2 | abiotic stress | 3 | 5 | 5 | 5 | 4 | 5 | 1 | 28 |
| C19 | S4 | phenylpropanoid-derived secondary metabolites/proanthocyanin/lignin biosynthesis | 6 | 6 | 6 | 7 | 6 | 7 | 2 | 40 |
| C20 | S7 | phenylpropanoid-derived secondary metabolites/flavonol biosynthesis | 3 | 3 | 3 | 2 | 2 | 3 | 0 | 16 |
| C21 | NO | function unknown | 0 | 5 | 4 | 4 | 4 | 4 | 0 | 21 |
| C22 | S15 | development/root hair development | 4 | 1 | 1 | 1 | 1 | 1 | 0 | 9 |
| C23 | S44 | phenylpropanoid-derived secondary metabolites/proanthocyanin biosynthesis | 1 | 0 | 1 | 1 | 3 | 2 | 3 | 11 |
| C24 | S6 | phenylpropanoid-derived secondary metabolites/anthocyanin biosynthesis | 4 | 4 | 6 | 5 | 6 | 5 | 0 | 30 |
| C25 | S12 | glucosinolate biosynthesis | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 6 |
| C26 | NO | function unknown | 0 | 8 | 9 | 8 | 6 | 5 | 2 | 38 |
| C27 | S1 | abiotic stress/specialized metabolism | 5 | 5 | 4 | 4 | 4 | 4 | 0 | 26 |
| C28 | S33 and S46 | development/anther/pollen development | 2 | 1 | 2 | 1 | 2 | 2 | 0 | 10 |
| C29 | S14 | abiotic stress/specialized metabolism/development | 6 | 9 | 11 | 10 | 9 | 12 | 0 | 57 |
| C30 | NO | function unknown | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 10 |
| C31 | S11 | abiotic stress | 4 | 5 | 3 | 3 | 4 | 4 | 0 | 23 |
| C32 | S36 | development/inflorescence development/seed germination | 1 | 3 | 3 | 3 | 3 | 3 | 0 | 16 |
| C33 | S10 | suberin biosynthesis | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| C34 | S24 | development/lateral root development | 3 | 3 | 1 | 2 | 2 | 2 | 0 | 13 |
| C35 | S9 | development/trichome branching/cuticle formation | 2 | 2 | 2 | 2 | 1 | 2 | 0 | 11 |
| C36 | NO | function unknown | 0 | 11 | 8 | 9 | 5 | 0 | 0 | 33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhou, Y.; Li, K.; Guo, H. Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Six Ipomoea Species and the Identification of Anthocyanin-Related Members in Sweet Potatoes. Plants 2023, 12, 1731. https://doi.org/10.3390/plants12081731
Li M, Zhou Y, Li K, Guo H. Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Six Ipomoea Species and the Identification of Anthocyanin-Related Members in Sweet Potatoes. Plants. 2023; 12(8):1731. https://doi.org/10.3390/plants12081731
Chicago/Turabian StyleLi, Maoxing, Yuanping Zhou, Kaifeng Li, and Huachun Guo. 2023. "Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Six Ipomoea Species and the Identification of Anthocyanin-Related Members in Sweet Potatoes" Plants 12, no. 8: 1731. https://doi.org/10.3390/plants12081731
APA StyleLi, M., Zhou, Y., Li, K., & Guo, H. (2023). Genome-Wide Comparative Analysis of the R2R3-MYB Gene Family in Six Ipomoea Species and the Identification of Anthocyanin-Related Members in Sweet Potatoes. Plants, 12(8), 1731. https://doi.org/10.3390/plants12081731






