Automatic Clustering and Classification of Coffee Leaf Diseases Based on an Extended Kernel Density Estimation Approach
Abstract
1. Introduction
- The proposed framework is new in the domain of imbalanced data classification, which simultaneously treats major and minor classes by giving them the same priority.
- An effective and quick extraction operation finds symptoms to maintain only regions of infection; the number of classes is validated using the D.B.I.
- A new clustering strategy is adopted to investigate an existing region of infection and that categorises lesions as belonging to single or multiple symptoms.
- The proposed method does not need to predetermine any parameter, which makes it fully automated and flexible. Furthermore, there is no need for an influence dataset to categorise observations.
- The proposed model is simple. Unlike previous models, it allows the self-clustering of overlapped lesions to be classified individually, reducing or preventing misclassification.
2. Related Work
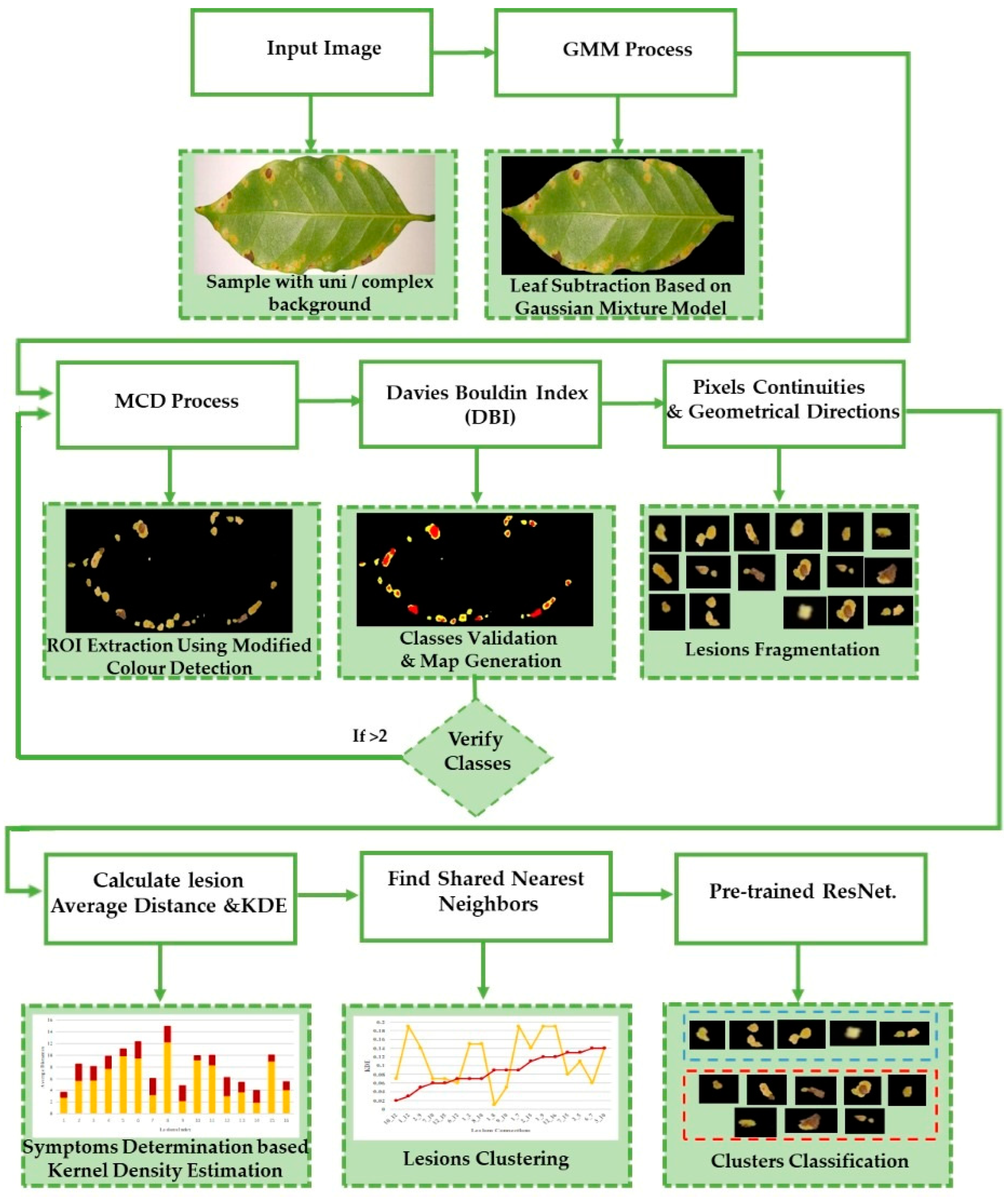
3. Methodology
3.1. Dataset
3.2. Proposed Framework
3.2.1. Stage1_ROI Extraction
| Algorithm 1: R.O.I. Extraction. | |
| Input: Coloured image of a leaf sample. | |
| Output: Two matrices of modified green pixels (M.G.P.) and modified red pixels (M.R.P.). | |
| Step 1: Process a modified colour-based detection method (M.C.D.) to check leaf pixels in an image. The red and green pixel values (R.P.V. and G.P.V.) are subtracted from the greyscale image value (G.I.V.): | |
| Step 2: Keep pixels with yellow and brown gradient only, which are responsible for determining symptoms regions. Equations (3) and (4) validate that: | |
3.2.2. Stage2_Class Determination and Validation
| Algorithm 2: Class Verification. | |
| Input: M.G.P. and M.R.P. matrices. | |
| Output: D.B.I. value to confirm the number of existing classes and ROI_img, the R.O.I. generated map. | |
| Step 1: is the mean of pixels obtained from Equation (4). | |
| Step 2: ) using the Euclidean distance function: | |
| Wi represents the average distance of all points in class Ci to their cluster centres, and Wj represents the average distance of all points in class Ci to the centre of class Cj. | |
| Step 3: Compute the D.B.I., where Cij represents the distance between the centres of classes Ci and Cj. | |
Step 4:
| |
| Step 5: Merge the M.G.P. and M.R.P. matrices to integrate both brown and yellow gradients in one R.O.I. image (ROI_img). | |
3.2.3. Stage3_Lesion Fragmentation
| Algorithm 3: Lesion Determination and Fragmentation. | |
| Input: updated ROI_img. | |
| Output: ROI_generated map, K lesions sub-images. | |
| Step 1: Unify colours by changing all yellow gradients to (R:255, G:255, B:0) and all brown gradients to (R:255, G:0, B:0). | |
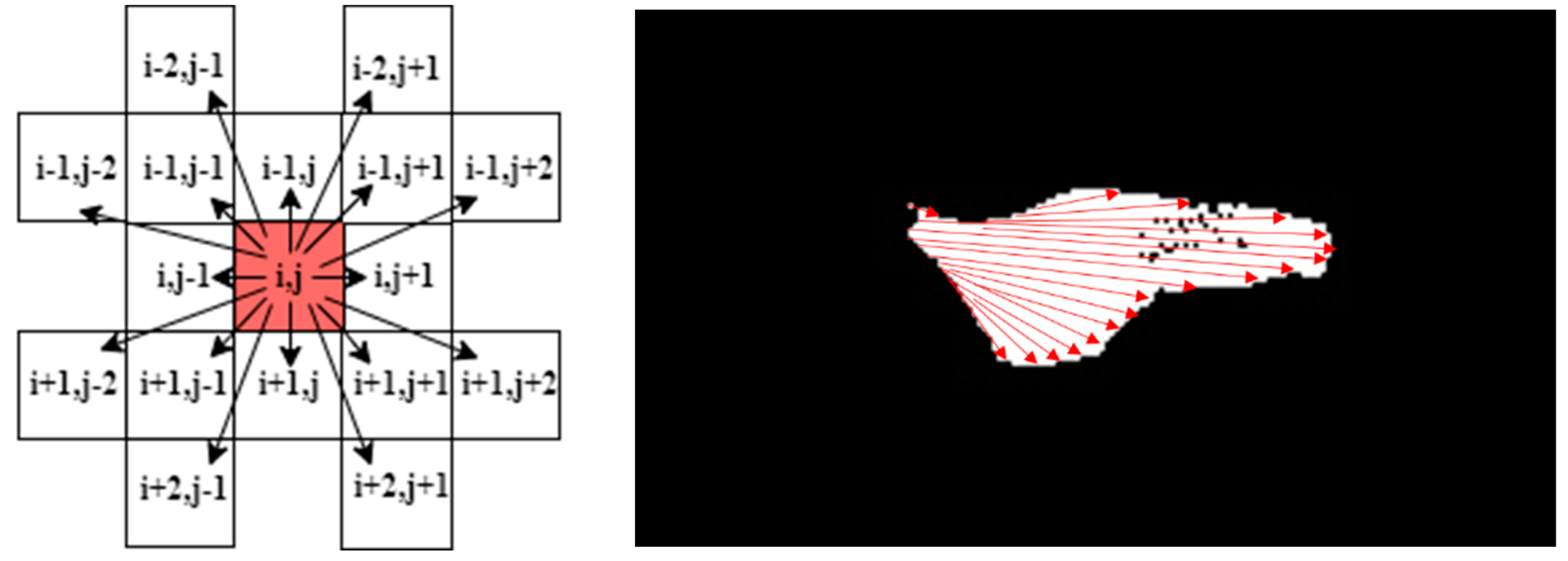
| Step 2: Detect points of ROI_img and trace continuities of successive pixels in the eight directions: | |
| While i < ROI_img (height) Do: | |
| While j < ROI_img (width) Do: | |
| If ROI_img (i, j) > 0: | |
| At each of the following directions (i, j + x), (i, j − x), (i − y, j − x), (i − y, j), (i − y, j + x), (i + y, j − x), (i + x, j), (i + y, j + x) assign the value of the detected point to its correspondent location in sub-image k. | |
| x and y are temporary counters initialised to the location of a current point; they increment by one to visit the next point, until they obtain a zero-pixel value, then jump to the next direction. | |
| Else continue searching for a new point until each point in this matrix is visited once. | |
| End | |
| End | |
| Step 2: Initialise K according to the number of extracted lesions. | |
| Step 3: For each lesion in the R.O.I., ensure that each point in that lesion is selected: | |
| where (i, j) is the location of a current point and (x, y) is the location of the farthest point in a lesion dimension. This equation supposes that each point in the map within these dimensions should have a corresponding point in the lesion sub-image. Otherwise, we assign the actual value of that pixel to the lesion sub-image. This step ensures that all the points in that lesion are integrated. Taking into account the noise and holes according to an acceptable spatial distance threshold ϵ compared to the surrounding points within the lesion: | |
| is a lesion with k index that belongs to an R.O.I. of a leaf sample. | |
3.2.4. Stage4_Symptom Determination and Classification
| Algorithm 4: Symptom Determination and Classification. | |||
| Input: Map_ image. | |||
| Output: Clusters of images. | |||
| Step 1: Initialise K according to the number of extracted lesions. | |||
| Step 2: For each lesion in the R.O.I.: | |||
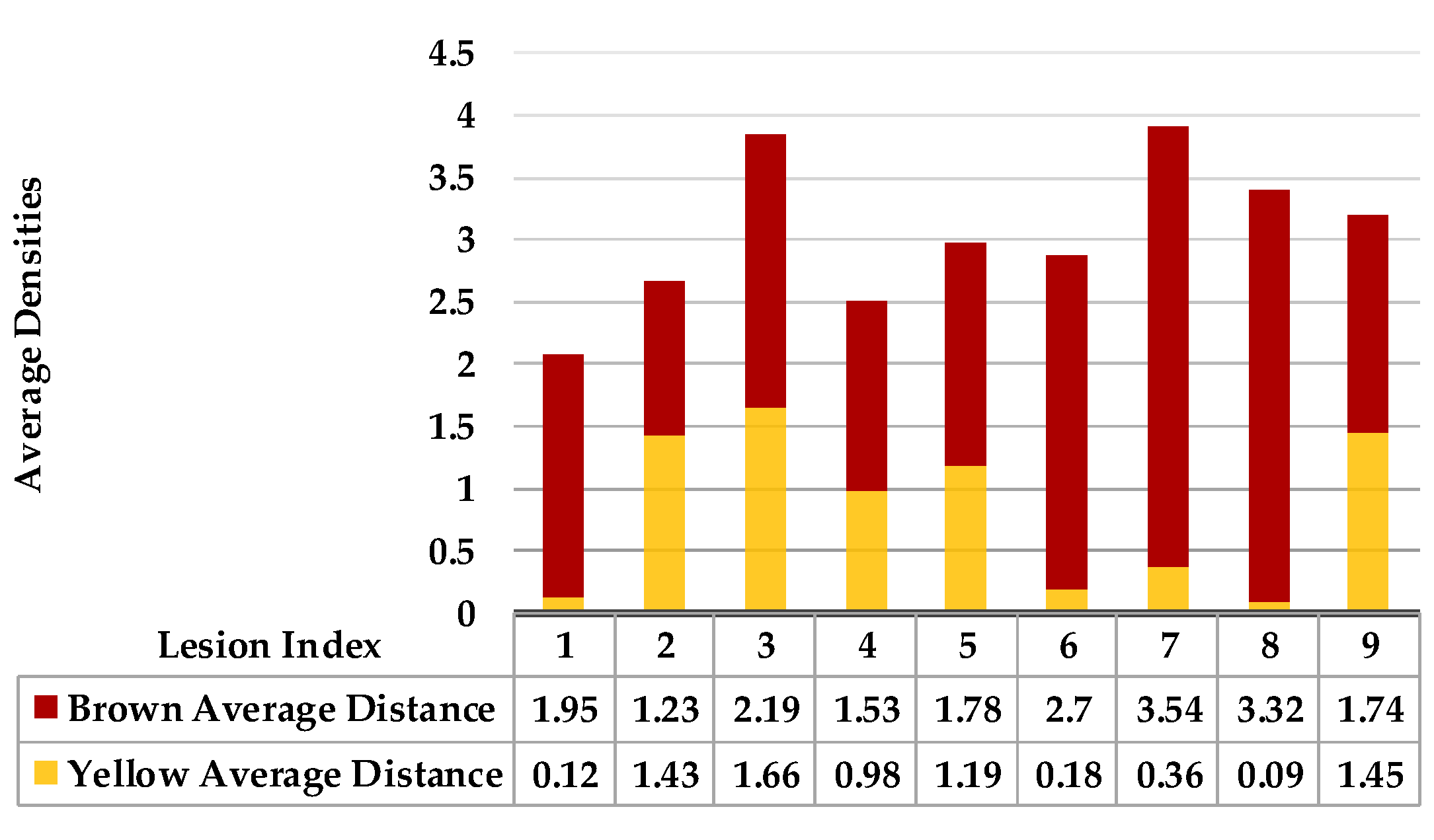
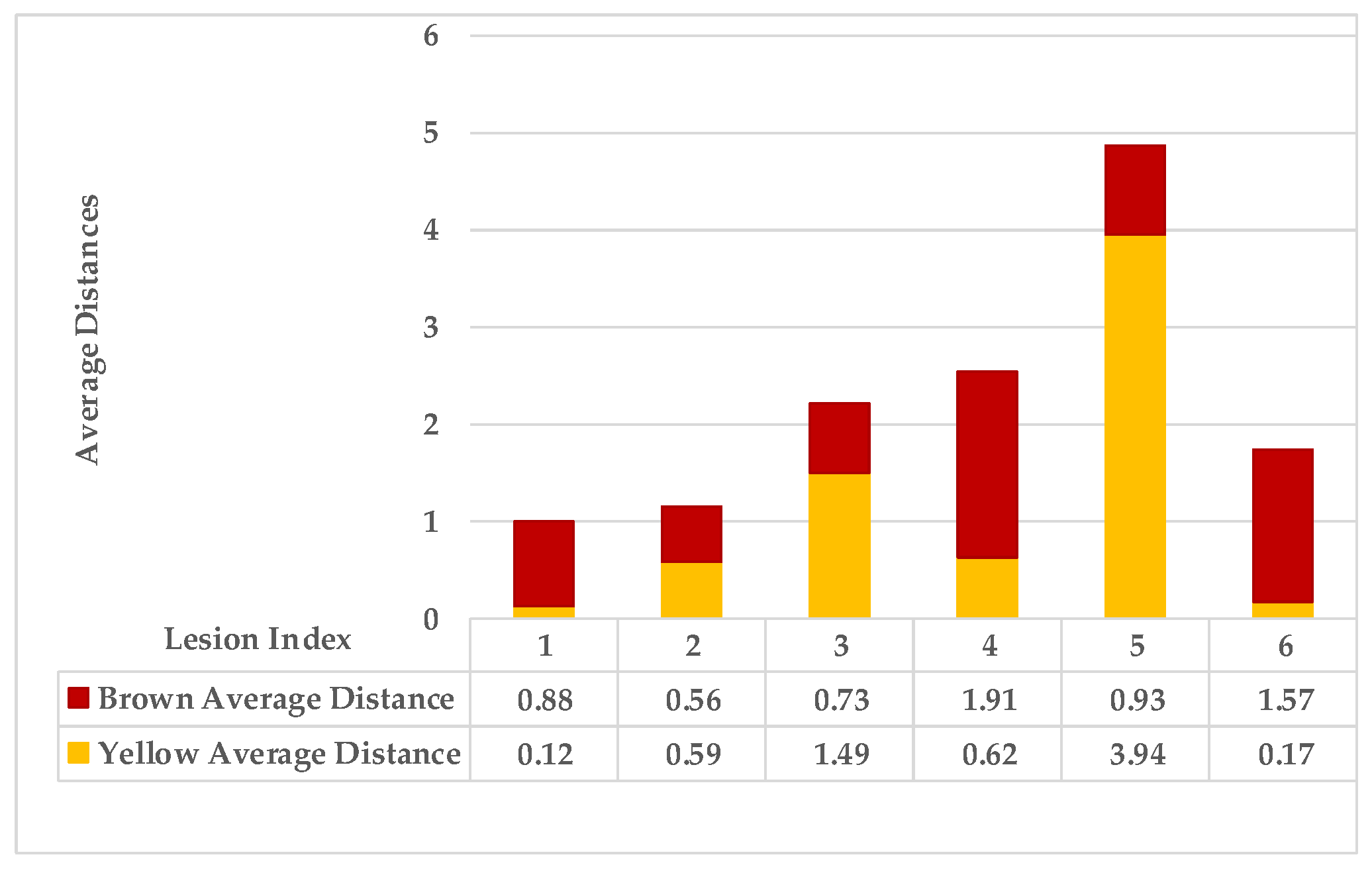
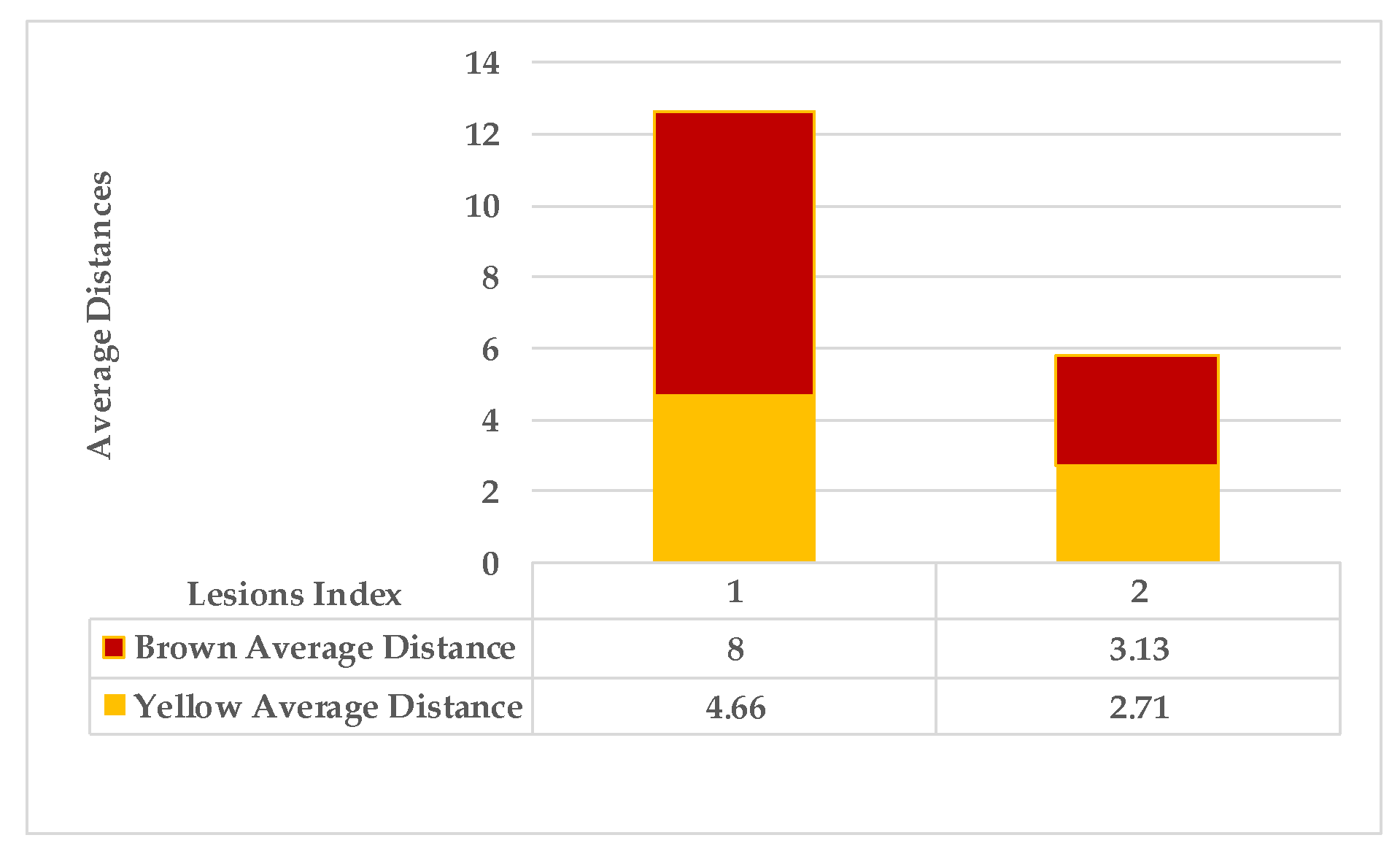
| Calculate the average distance for points characterised as yellow in the map separately from brown points using the Euclidean distance measure. | |||
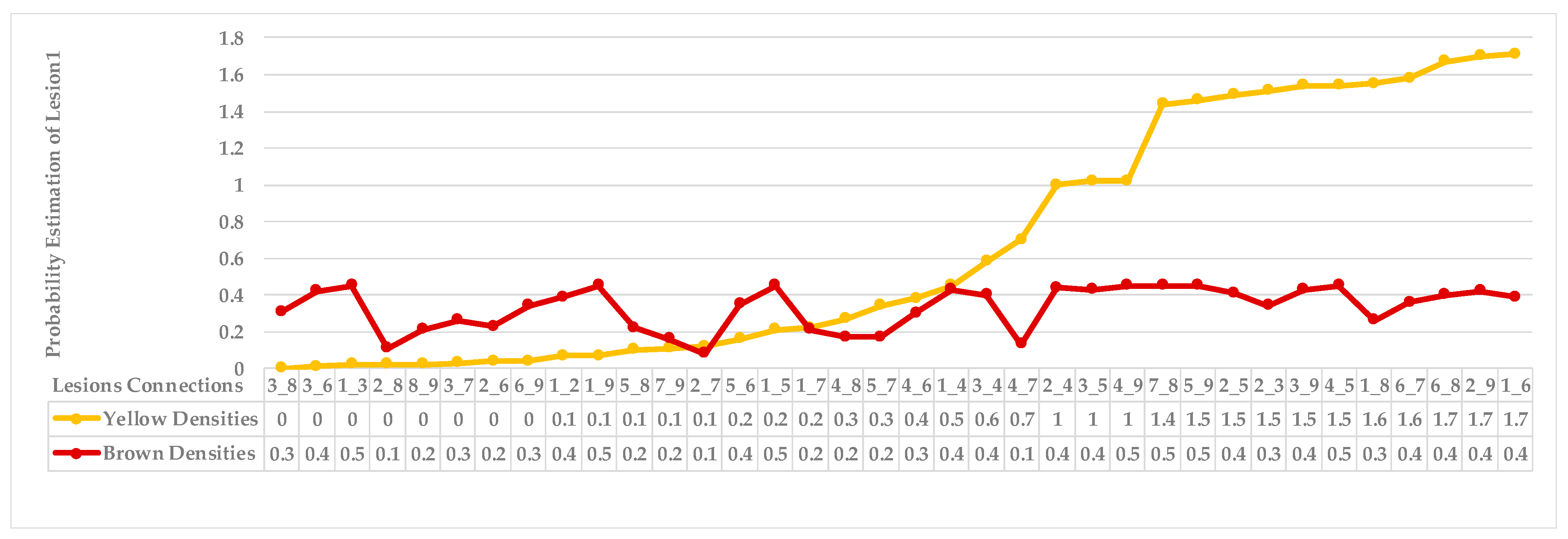
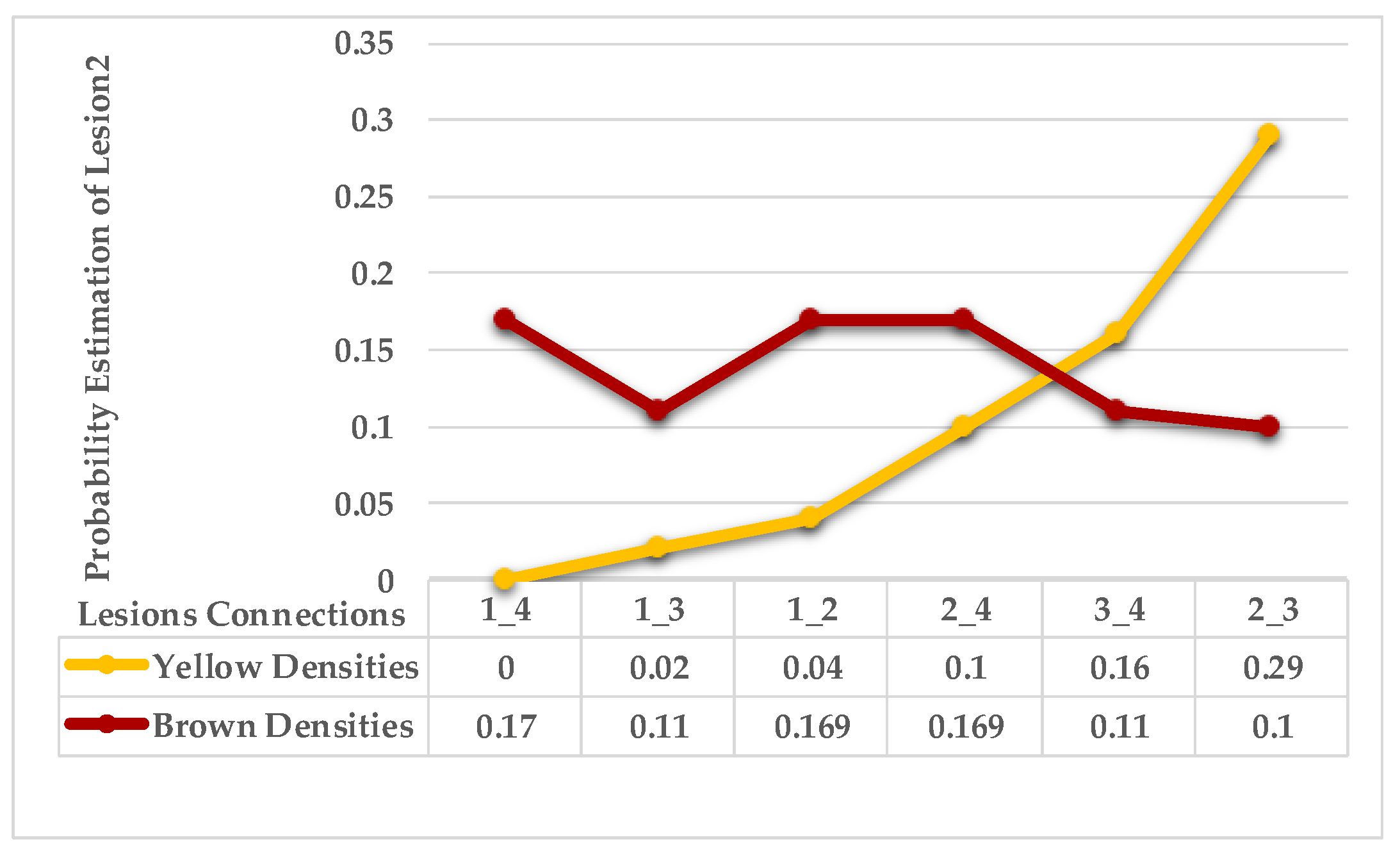
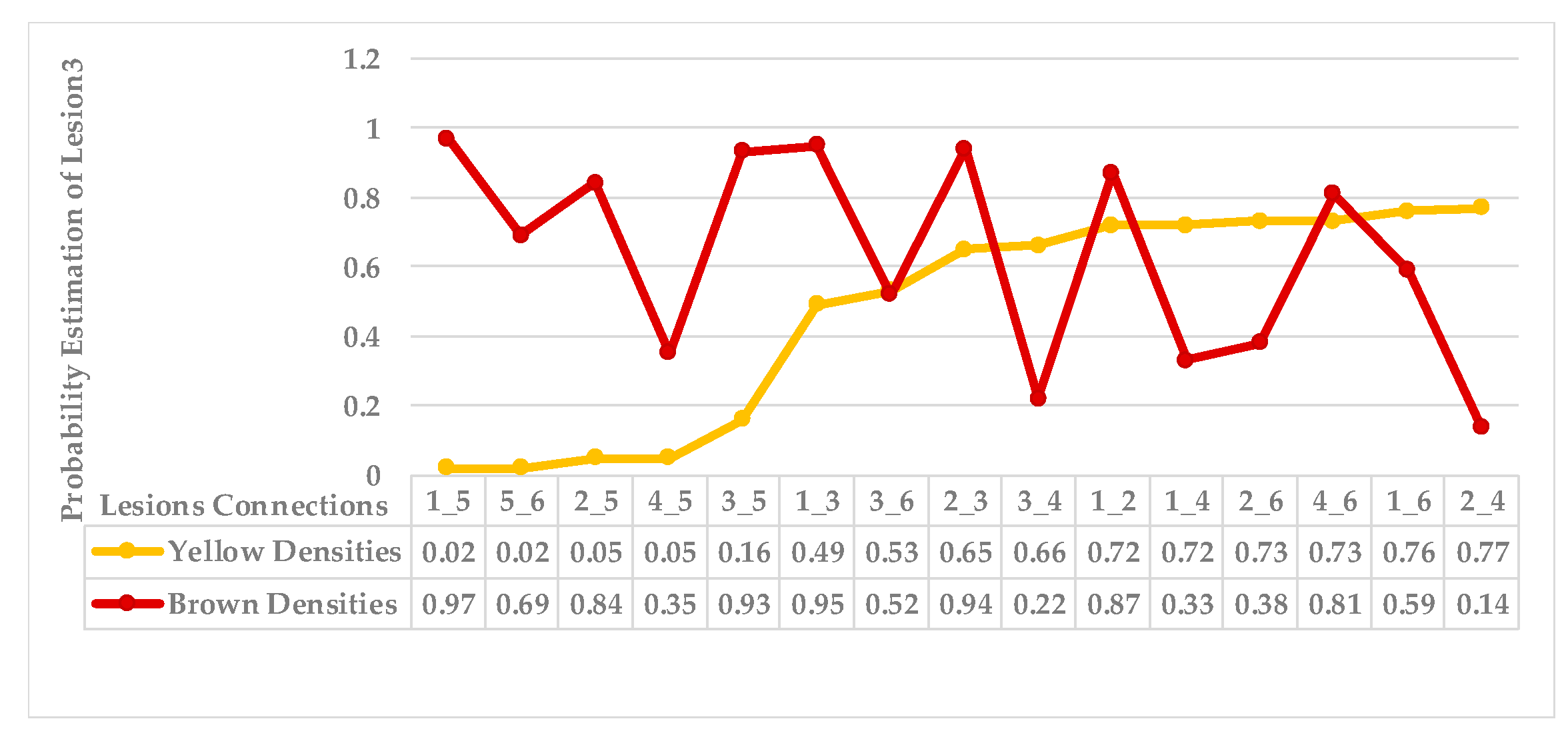
| Step 3: Find the nearest neighbours to a current lesion according to the similarity of characteristics based on the adaptive weighted Gaussian kernel function: | |||
| represents the weight computed by measuring the Euclidean distance between two lesions. | |||
| The h value is adapted to handle bandwidth estimation and accommodates each spot in a leaf. | |||
| are the maximum and minimum distances of yellow and then brown points for each lesion in a single leaf sample. | |||
| Step 4: Sort the obtained probabilities in ascending order, then arrange lesions into two main groups, where a group represents a symptom; points that share equal or similar probabilities are categorised as neighbours in one group. | |||
| Step 5: Classify each group independently using ResNet50 classifier. | |||
4. Results and Analyses
4.1. Parameter Settings
4.2. Experimental Results
4.3. Classification Results
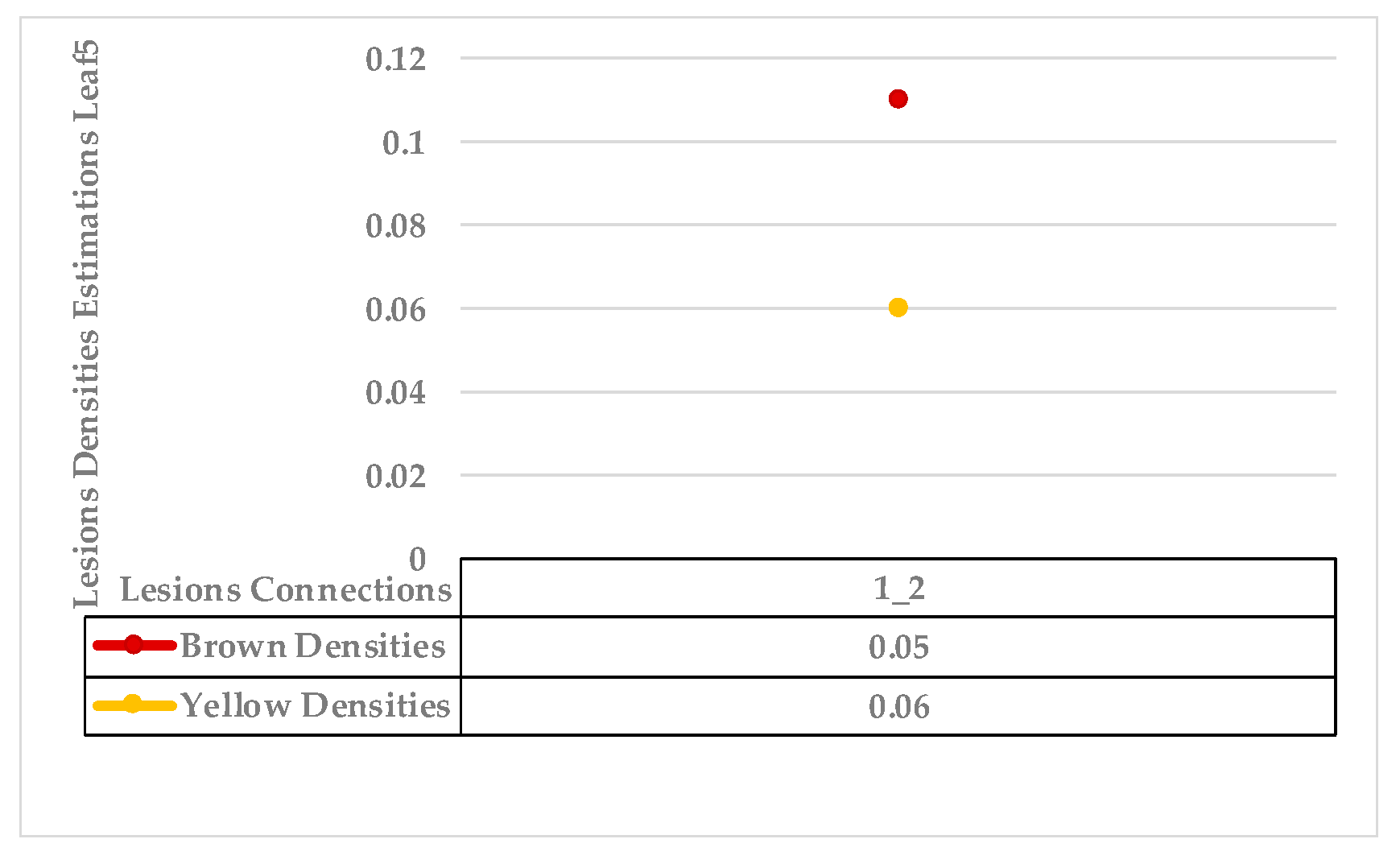
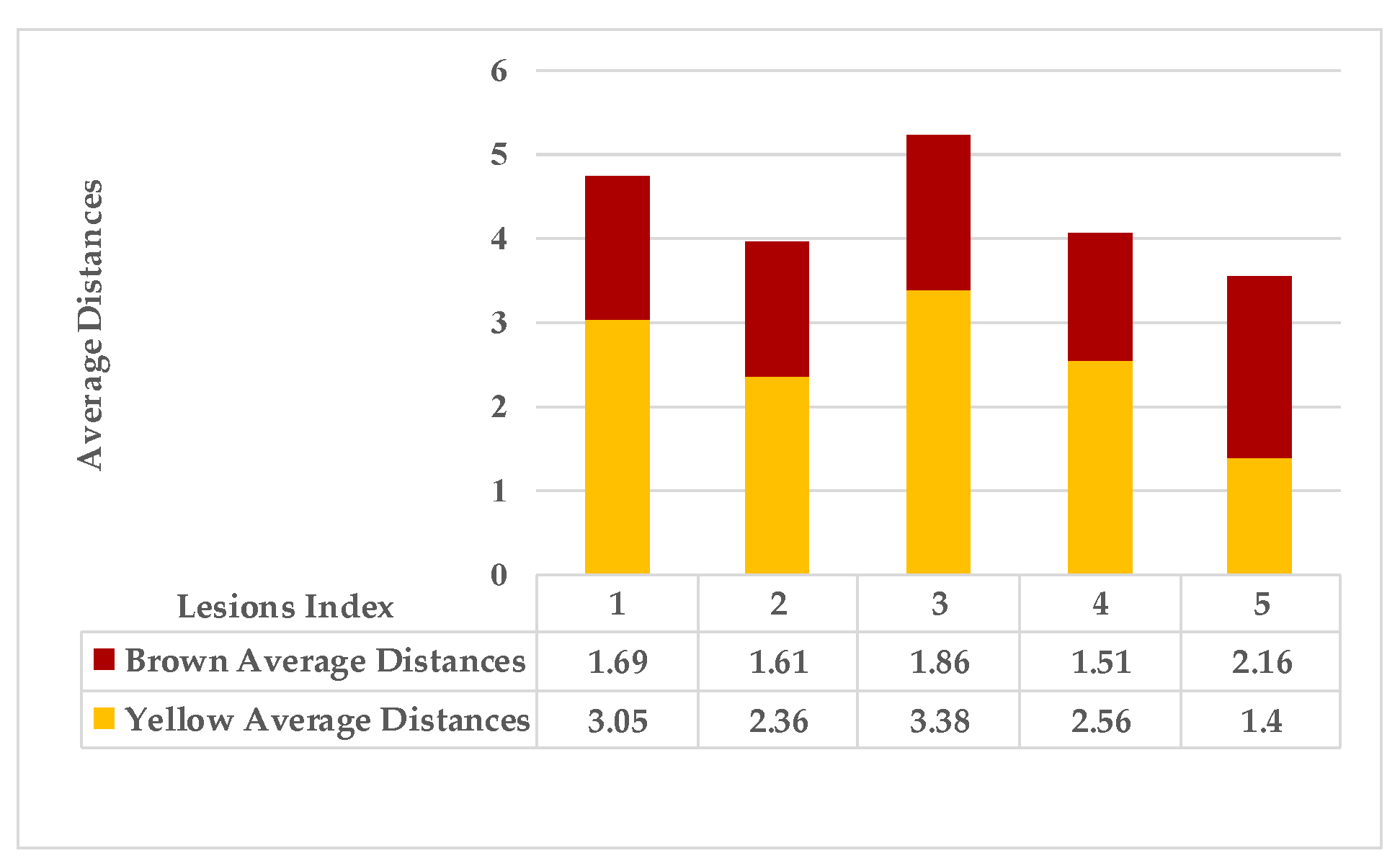
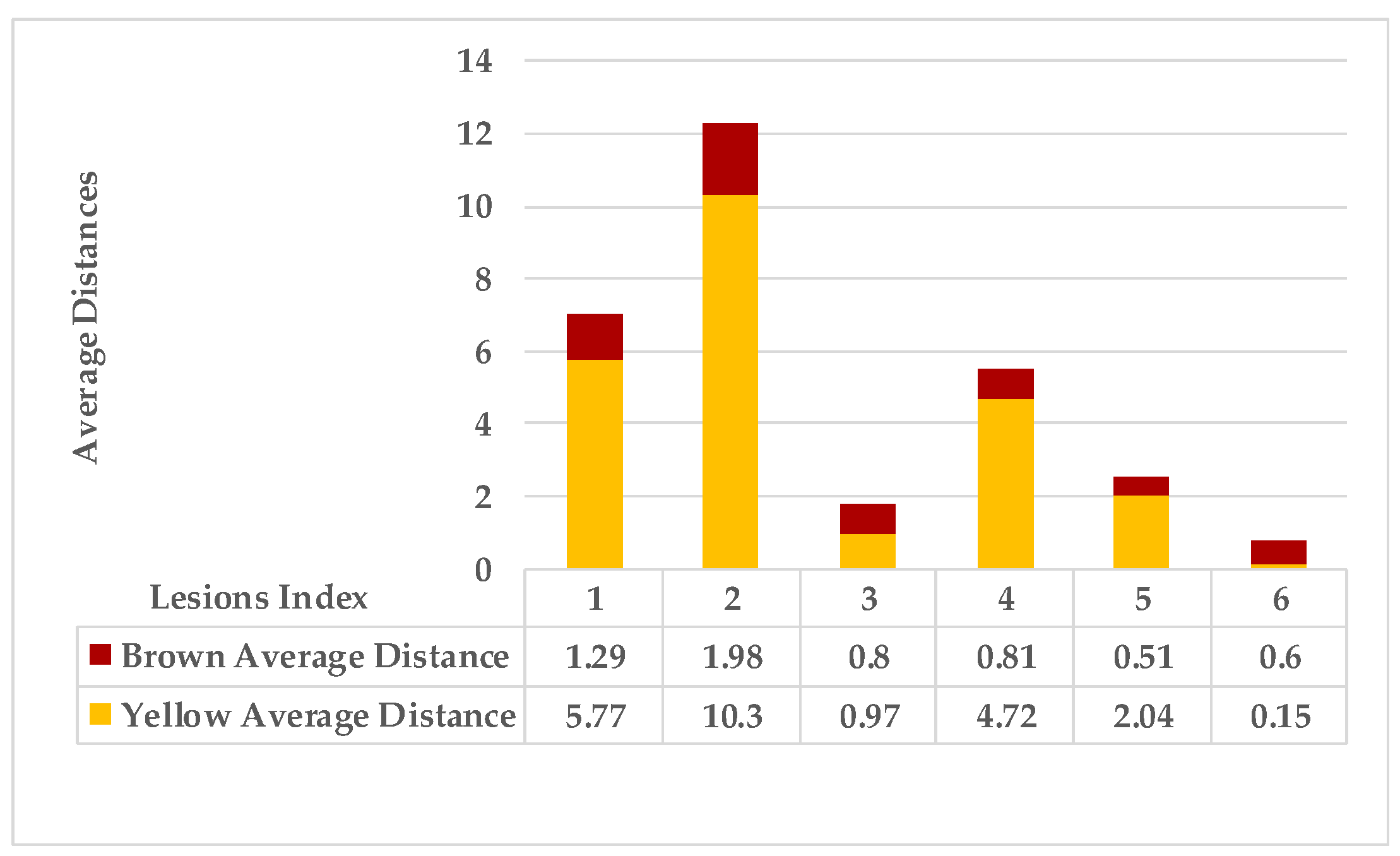
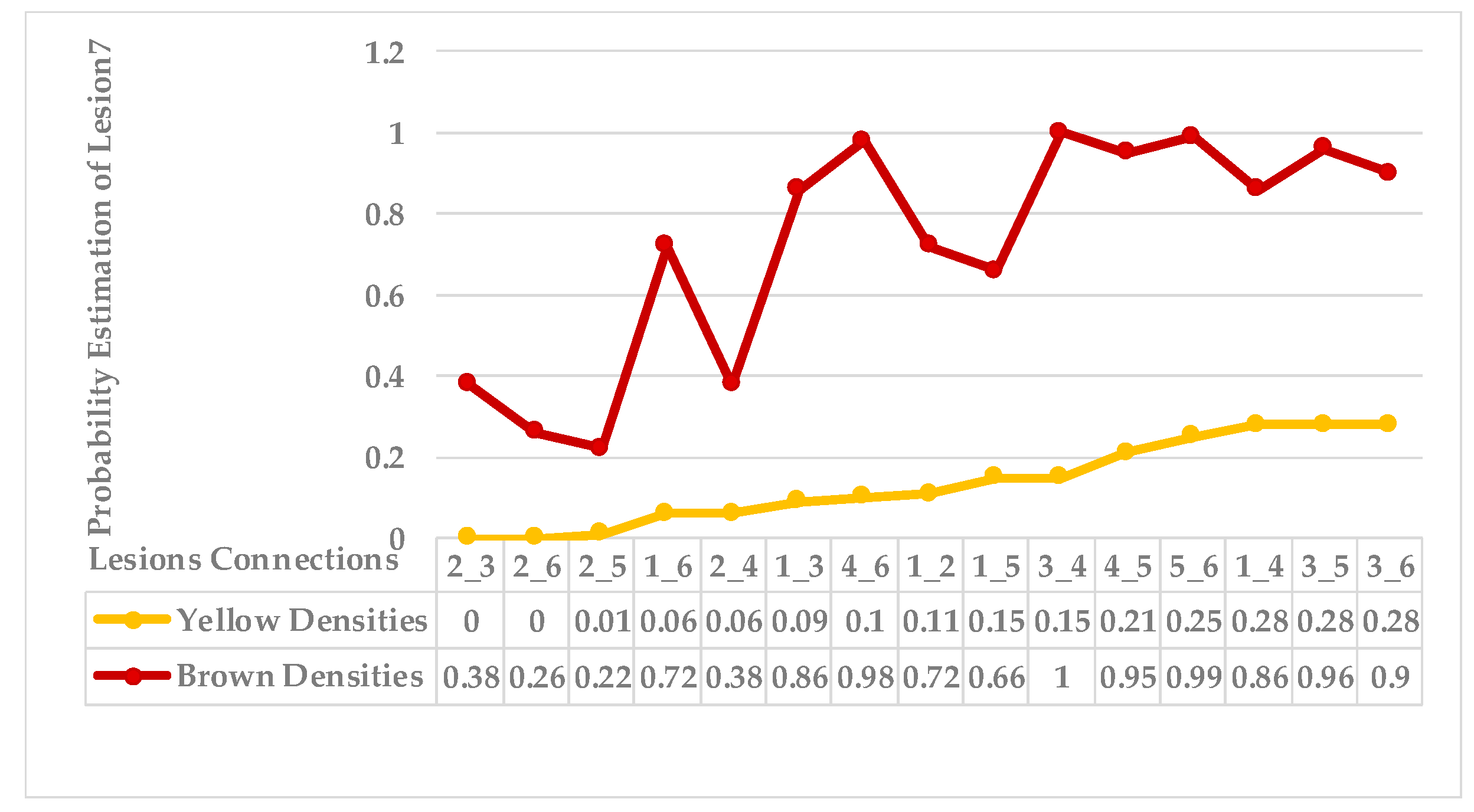
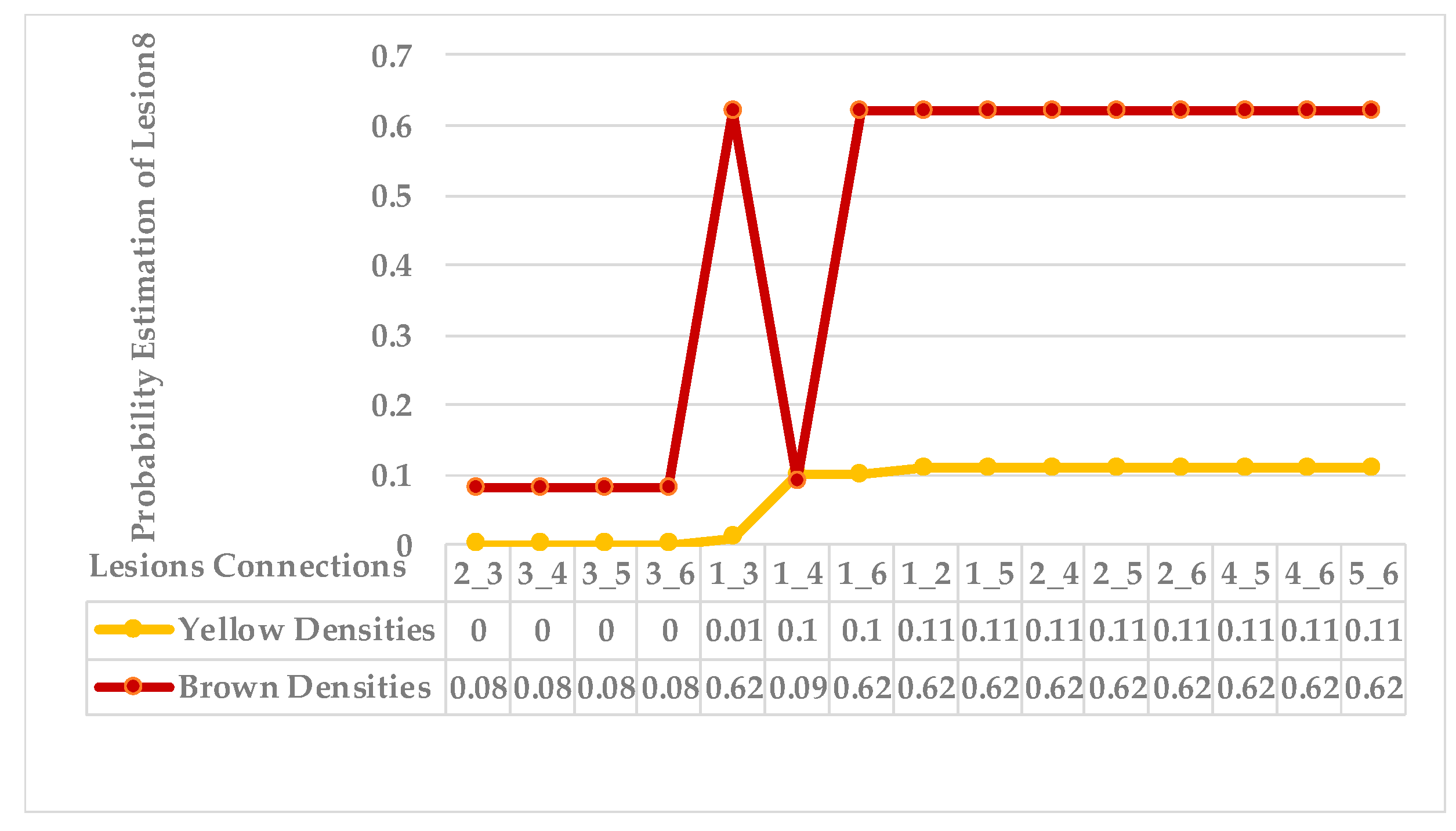
4.4. Extended KDE Analysis
4.5. Analytical Comparison with Other Kernels Methods
- RBK:
- Epanechnikov kernel:
5. Discussion
- There is a need to extract the R.O.I. method without losing region properties. The modified colour process is proposed to assume that the darkest gradients refer to the brown injured regions and the lightest gradients refer to the yellow injured regions.
- The best analytical technique to analyse variety in syndrome is self-clustering based on an extended Gaussian kernel density estimation method. This method avoids overfitting and over-generalising problems that result from resampling observations to provide a balanced dataset. Furthermore, it avoids over-smoothing that results from undesirable bandwidth selectors. Hence, the bandwidth is adaptive to the R.O.I. of each leaf.
- Most classification models are developed to detect prevalent diseases in a leaf. The solution is proposed to improve the classification of leaf disease diagnosis by making it able to characterise multiple infections in the same leaf by clustering symptoms and then training a classifier using a balanced symptoms dataset. So, each cluster is classified independently, reducing the classification error percentage.
6. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fujita, E.; Uga, H.; Kagiwada, S.; Iyatomi, H. A practical plant diagnosis system for field leaf images and feature visualization. Int. J. Eng. Technol. 2018, 7, 49–54. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Gao, L.; Lin, X. Fully automatic segmentation method for medicinal plant leaf images in complex background. Comput. Electron. Agric. 2019, 164, 104924. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Hari, D.P.R.K. Review on Fast Identification and Classification in Cultivation. Int. J. Adv. Sci. Technol. 2020, 29, 3498–3512. [Google Scholar]
- Amara, J.; Bouaziz, B.; Algergawy, A. A Deep Learning-Based Approach for Banana Leaf Diseases Classification. In Lecture Notes in Informatics (LNI); Gesellschaft für Informatik: Bonn, Germany, 2017; Volume 266, pp. 79–88. [Google Scholar]
- Zhang, S.; Huang, W.; Zhang, C. Three-channel convolutional neural networks for vegetable leaf disease recognition. Cogn. Syst. Res. 2019, 53, 31–41. [Google Scholar] [CrossRef]
- Ngugi, L.C.; Abdelwahab, M.M.; Abo-Zahhad, M. Recent advances in image processing techniques for automated leaf pest and disease recognition—A review. Inf. Process. Agric. 2020, 8, 27–51. [Google Scholar] [CrossRef]
- Sharif, M.; Khan, M.A.; Iqbal, Z.; Azam, M.F.; Lali, M.I.U.; Javed, M.Y. Detection and classification of citrus diseases in agriculture based on optimized weighted segmentation and feature selection. Comput. Electron. Agric. 2018, 150, 220–234. [Google Scholar] [CrossRef]
- Anjna; Sood, M.; Singh, P.K. Hybrid System for Detection and Classification of Plant Disease Using Qualitative Texture Features Analysis. Procedia Comput. Sci. 2020, 167, 1056–1065. [Google Scholar] [CrossRef]
- Haque, I.R.I.; Neubert, J. Deep learning approaches to biomedical image segmentation. Inform. Med. Unlocked 2020, 18, 100297. [Google Scholar] [CrossRef]
- Hasan, R.I.; Yusuf, S.M.; Alzubaidi, L. Review of the state of the art of deep learning for plant diseases: A broad analysis and discussion. Plants 2020, 9, 1302. [Google Scholar] [CrossRef]
- Sharif Razavian, A.; Azizpour, H.; Sullivan, J.; Carlsson, S. CNN features off-the-shelf: An astounding baseline for recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; pp. 806–813. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. Adv. Neural Inf. Prcoess. Syst. 2019, 32, 3347–3357. [Google Scholar]
- Sharma, P.; Berwal, Y.P.S.; Ghai, W. Performance analysis of deep learning CNN models for disease detection in plants using image segmentation. Inf. Process. Agric. 2019, in press. [Google Scholar] [CrossRef]
- Kamal, K.C.; Yin, Z.; Wu, M.; Wu, Z. Depthwise separable convolution architectures for plant disease classification. Comput. Electron. Agric. 2019, 165, 104948. [Google Scholar]
- Barbedo, J.G.A.; Koenigkan, L.V.; Halfeld-Vieira, B.A.; Costa, R.V.; Nechet, K.L.; Godoy, C.V.; Angelotti, F. Annotated plant pathology databases for image-based detection and recognition of diseases. IEEE Lat. Am. Trans. 2018, 16, 1749–1757. [Google Scholar] [CrossRef]
- Baso, C.D.; de la Cruz Rodriguez, J.; Danilovic, S. Solar image denoising with convolutional neural networks. Astron. Astrophys. 2019, 629, A99. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Liu, B.; He, D.; Liang, C. Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Ganesh, P.; Volle, K.; Burks, T.F.; Mehta, S.S. Deep orange: Mask R-CNN based orange detection and segmentation. IFAC-PapersOnLine 2019, 52, 70–75. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, J.; Fu, L.; Majeed, Y.; Feng, Y.; Li, R.; Cui, Y. Improved kiwifruit detection using pre-trained VGG16 with RGB and NIR information fusion. IEEE Access 2020, 8, 2327–2336. [Google Scholar] [CrossRef]
- Mao, S.; Li, Y.; Ma, Y.; Zhang, B.; Zhou, J.; Wang, K. Automatic cucumber recognition algorithm for harvesting robots in the natural environment using deep learning and multi-feature fusion. Comput. Electron. Agric. 2020, 170, 105254. [Google Scholar] [CrossRef]
- Juliano, P.G.; Francisco, A.C.P.; Daniel, M.Q.; Flora, M.M.V.; Jayme, G.A.B.; Emerson, M.D.P.; Ponte, D. Deep learning architectures for semantic segmentation and automatic estimation of severity of foliar symptoms caused by diseases or pests. Comput. Electron. Agric. 2021, 210, 129–142. [Google Scholar]
- Bhavsar, K.A.; Abugabah, A.; Singla, J.; AlZubi, A.A.; Bashir, A.K. A comprehensive review on medical diagnosis using machine learning. Comput. Mater. Contin. 2021, 67, 1997–2014. [Google Scholar] [CrossRef]
- Mirzaei, B.; Nikpour, B.; Nezamabadi-Pour, H. CDBH: A clustering and density-based hybrid approach for imbalanced data classification. Expert Syst. Appl. 2021, 164, 114035. [Google Scholar] [CrossRef]
- Nikpour, B.; Nezamabadi-pour, H. A memetic approach for training set selection in imbalanced data sets. Int. J. Mach. Learn. Cybern. 2019, 10, 3043–3070. [Google Scholar] [CrossRef]
- Liu, R.; Hall, L.O.; Bowyer, K.; Goldgof, D.B.; Gatenby, R.A.; Ahmed, K.B. Synthetic minority image over-sampling technique: How to improve A.U.C. for glioblastoma patient survival prediction. In Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics (S.M.C.), Banff, AB, Canada, 5–8 October 2017; pp. 1357–1362. [Google Scholar] [CrossRef]
- Desprez, M.; Zawada, K.; Ramp, D. Overcoming the ordinal imbalanced data problem by combining data processing and stacked generalizations. Mach. Learn. Appl. 2022, 7, 100241. [Google Scholar] [CrossRef]
- Ng, W.W.; Liu, Z.; Zhang, J.; Pedrycz, W. Maximizing minority accuracy for imbalanced pattern classification problems using cost-sensitive Localized Generalization Error Model. Appl. Soft Comput. 2021, 104, 107178. [Google Scholar] [CrossRef]
- Ren, J.; Wang, Y.; Mao, M.; Cheung, Y.M. Equalization ensemble for large scale highly imbalanced data classification. Knowl.-Based Syst. 2022, 242, 108295. [Google Scholar] [CrossRef]
- Shahabadi, M.S.E.; Tabrizchi, H.; Rafsanjani, M.K.; Gupta, B.B.; Palmieri, F. A combination of clustering-based under-sampling with ensemble methods for solving imbalanced class problem in intelligent systems. Technol. Forecast. Soc. Chang. 2021, 169, 120796. [Google Scholar] [CrossRef]
- Gulhane, V.A.; Kolekar, M.H. Diagnosis of diseases on cotton leaves using principal component analysis classifier. In Proceedings of the 2014 Annual IEEE India Conference (I.N.D.I.C.O.N.), Pune, India, 11–13 December 2014; pp. 1–5. [Google Scholar] [CrossRef]
- Xia, F.; Xie, X.; Wang, Z.; Jin, S.; Yan, K.; Ji, Z. A Novel Computational Framework for Precision Diagnosis and Subtype Discovery of Plant with Lesion. Front. Plant Sci. 2022, 12, 789630. [Google Scholar] [CrossRef]
- Saleem, R.; Shah, J.H.; Sharif, M.; Yasmin, M.; Yong, H.S.; Cha, J. Mango Leaf Disease Recognition and Classification Using Novel Segmentation and Vein Pattern Technique. Appl. Sci. 2021, 11, 11901. [Google Scholar] [CrossRef]
- Tsai, C.F.; Lin, W.C.; Hu, Y.H.; Yao, G.T. Under-sampling class imbalanced datasets by combining clustering analysis and instance selection. Inf. Sci. 2019, 477, 47–54. [Google Scholar] [CrossRef]
- Elyan, E.; Moreno-Garcia, C.F.; Jayne, C. CDSMOTE: Class decomposition and synthetic minority class oversampling technique for imbalanced-data classification. Neural Comput. Appl. 2021, 33, 2839–2851. [Google Scholar] [CrossRef]
- Hasan, R.I.; Yusuf, S.M.; Rahim, M.S.M.; Alzubaidi, L. Automated maps generation for coffee and apple leaf infected with single or multiple diseases-based color analysis approaches. Inform. Med. Unlocked 2022, 28, 100837. [Google Scholar] [CrossRef]
- Gurrala, K.K.; Yemineni, L.; Rayana, K.S.R.; Vajja, L.K. A New Segmentation method for Plant Disease Diagnosis. In Proceedings of the 2019 2nd International Conference on Intelligent Communication and Computational Techniques (I.C.C.T.), Jaipur, India, 28–29 September 2019; pp. 137–141. [Google Scholar] [CrossRef]
- Chowdhury, S.; Amorim, R.C.D. An efficient density-based clustering algorithm using reverse nearest neighbour. In Intelligent Computing-Proceedings of the Computing Conference; Springer: Cham, Switzerland; London, UK, 2019; pp. 29–42. [Google Scholar] [CrossRef]
- Fu, C.; Yang, J. Granular. classification for imbalanced datasets: A minkowski distance-based method. Algorithms 2021, 14, 54. [Google Scholar] [CrossRef]
- Deng, M.; Guo, Y.; Wang, C.; Wu, F. An oversampling method for multi-class imbalanced data based on composite weights. PLoS ONE 2021, 16, e0259227. [Google Scholar] [CrossRef]
- Tian, K.; Li, J.; Zeng, J.; Evans, A.; Zhang, L. Segmentation of tomato leaf images based on adaptive clustering number of K-means algorithm. Comput. Electron. Agric. 2019, 165, 104962. [Google Scholar] [CrossRef]
- Tang, B.; He, H. A local density-based approach for outlier detection. Neurocomputing 2017, 241, 171–180. [Google Scholar] [CrossRef]
- Li, K.; Gao, X.; Fu, S.; Diao, X.; Ye, P.; Xue, B.; Huang, Z. Robust outlier detection based on the changing rate of directed density ratio. Expert Syst. Appl. 2022, 207, 117988. [Google Scholar] [CrossRef]
- Yu, H.; Liu, J.; Chen, C.; Heidari, A.A.; Zhang, Q.; Chen, H.; Turabieh, H. Corn leaf diseases diagnosis based on K-means clustering and deep learning. IEEE Access 2021, 9, 143824–143835. [Google Scholar] [CrossRef]
- Zhang, L.; Lin, J.; Karim, R. Adaptive kernel density-based anomaly detection for nonlinear systems. Knowl.-Based Syst. 2018, 139, 50–63. [Google Scholar] [CrossRef]
- Fang, U.; Li, J.; Lu, X.; Gao, L.; Ali, M.; Xiang, Y. Self-supervised cross-iterative clustering for unlabeled plant disease images. Neurocomputing 2021, 456, 36–48. [Google Scholar] [CrossRef]
- Abdulghafoor, S.A.; Mohamed, L.A. Using Some Metric Distance in Local Density Based on Outlier Detection Methods. J. Posit. Psychol. Wellbeing 2022, 6, 189–202. [Google Scholar]
- Wahid, A.; Rao, A.C.S. Rkdos: A relative kernel density-based outlier score. IETE Tech. Rev. 2020, 37, 441–452. [Google Scholar] [CrossRef]
- Abdulghafoor, S.A.; Mohamed, L.A. A local density-based outlier detection method for high dimension data. Int. J. Nonlinear Anal. Appl. 2022, 13, 1683–1699. [Google Scholar]
- Parraga-Alava, J.; Cusme, K.; Loor, A.; Santander, E. RoCoLe: A robusta coffee leaf images dataset for evaluation of machine learning based methods in plant diseases recognition. Data Brief 2019, 25, 104414. [Google Scholar] [CrossRef]
- Esgario, J.G.; Krohling, R.A.; Ventura, J.A. Deep learning for classification and severity estimation of coffee leaf biotic stress. Comput. Electron. Agric. 2020, 169, 105162. [Google Scholar] [CrossRef]
- Tassis, L.M.; de Souza, J.E.T.; Krohling, R.A. A deep learning approach combining instance and semantic segmentation to identify diseases and pests of coffee leaves from in-field images. Comput. Electron. Agric. 2021, 186, 106191. [Google Scholar] [CrossRef]
- Tassis, L.M.; Krohling, R.A. Few-shot learning for biotic stress classification of coffee leaves. Artif. Intell. Agric. 2022, 6, 55–67. [Google Scholar] [CrossRef]
- Schubert, E.; Zimek, A.; Kriegel, H.P. Generalized outlier detection with flexible kernel density estimates. In Proceedings of the 2014 SIAM International Conference on Data Mining, Society for Industrial and Applied Mathematics. Philadelphia, PA, USA, 24 April 2014; pp. 542–550. [Google Scholar] [CrossRef]
- Cuturi, M. Fast global alignment kernels. In Proceedings of the 28th International Conference on Machine Learning (ICML-11), Bellevue, WA, USA, 28 June–2 July 2011; pp. 929–936. [Google Scholar]
- Oudjane, N.; Musso, C. L/sup 2/-density estimation with negative kernels. I.S.P.A. 2005. In Proceedings of the 4th International Symposium on Image and Signal Processing and Analysis, Zagreb, Croatia, 15–17 September 2005; pp. 34–39. [Google Scholar] [CrossRef]
- Scott, D.W. Multivariate Density Estimation: Theory, Practice and Visualization; John Wiley & Sons, Inc.: New York, NY, USA, 1992. [Google Scholar] [CrossRef]
- Vega, A.; Calderón, M.A.R.; Rey, J.C.; Lobo, D.; Gómez, J.A.; Landa, B.B. Identification of Soil Properties Associated with the Incidence of Banana Wilt Using Supervised Methods. Plants 2022, 11, 2070. [Google Scholar] [CrossRef]









| Dataset | Biotic Stress | No. R.O.I. Images |
|---|---|---|
| Coffee dataset | Miner | 593 |
| Rust | 991 | |
| Phoma | 504 | |
| Cercospora | 378 | |
| Healthy | 272 | |
| Miners and Phoma | 1 | |
| Rust and Phoma | 2 | |
| Brown spot and Cercospora | 7 | |
| Miners and Cercospora | 15 | |
| Miners and Rust | 112 | |
| Rust and Cercospora | 166 | |
| Total | 3041 | |
| RoCoLe dataset | Rust | 602 |
| Healthy | 300 | |
| Total | 902 |
| Symptoms | Leaf Image | D.B.I. | Generated Map | No. Detected Lesions | No. Groups |
|---|---|---|---|---|---|
| Rust and Cercospora |  | 2 |  | 9 |  |
| Rust and Phoma |  | 2 |  | 4 |  |
| Rust and Miner |  | 2 |  | 6 | 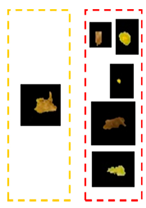 |
| Phoma and Cercospora |  | 2 |  | 4 |  |
| Miner and Cercospora |  | 2 |  | 2 |  |
| Miner |  | 2 |  | 5 | 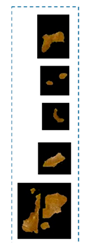 |
| Rust |  | 2 | 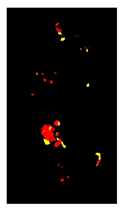 | 6 |  |
| Rust |  | 2 | 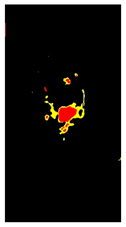 | 6 |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, R.I.; Yusuf, S.M.; Mohd Rahim, M.S.; Alzubaidi, L. Automatic Clustering and Classification of Coffee Leaf Diseases Based on an Extended Kernel Density Estimation Approach. Plants 2023, 12, 1603. https://doi.org/10.3390/plants12081603
Hasan RI, Yusuf SM, Mohd Rahim MS, Alzubaidi L. Automatic Clustering and Classification of Coffee Leaf Diseases Based on an Extended Kernel Density Estimation Approach. Plants. 2023; 12(8):1603. https://doi.org/10.3390/plants12081603
Chicago/Turabian StyleHasan, Reem Ibrahim, Suhaila Mohd Yusuf, Mohd Shafry Mohd Rahim, and Laith Alzubaidi. 2023. "Automatic Clustering and Classification of Coffee Leaf Diseases Based on an Extended Kernel Density Estimation Approach" Plants 12, no. 8: 1603. https://doi.org/10.3390/plants12081603
APA StyleHasan, R. I., Yusuf, S. M., Mohd Rahim, M. S., & Alzubaidi, L. (2023). Automatic Clustering and Classification of Coffee Leaf Diseases Based on an Extended Kernel Density Estimation Approach. Plants, 12(8), 1603. https://doi.org/10.3390/plants12081603











