Assessing Genetic Distinctness and Redundancy of Plant Germplasm Conserved Ex Situ Based on Published Genomic SNP Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of Published Genomic Data
2.2. Data Processing
2.3. APD Analysis
2.4. Analysis of APD Estimation with Large Genomic Data
3. Results
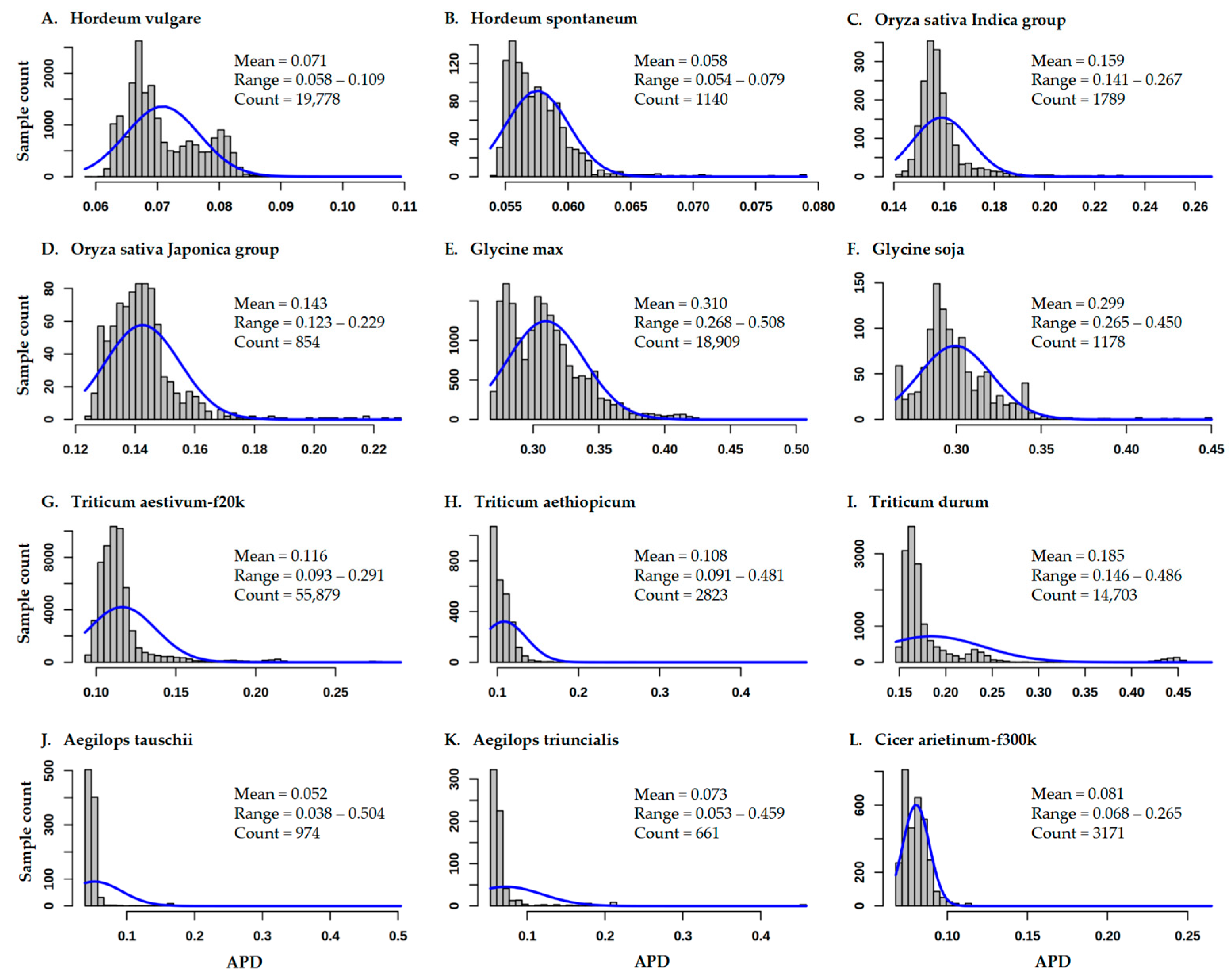
3.1. Variability of APD Estimates for 12 Germplasm Data Sets
3.2. Genetic Outliers, Genetically Distinctive and Redundant Sets
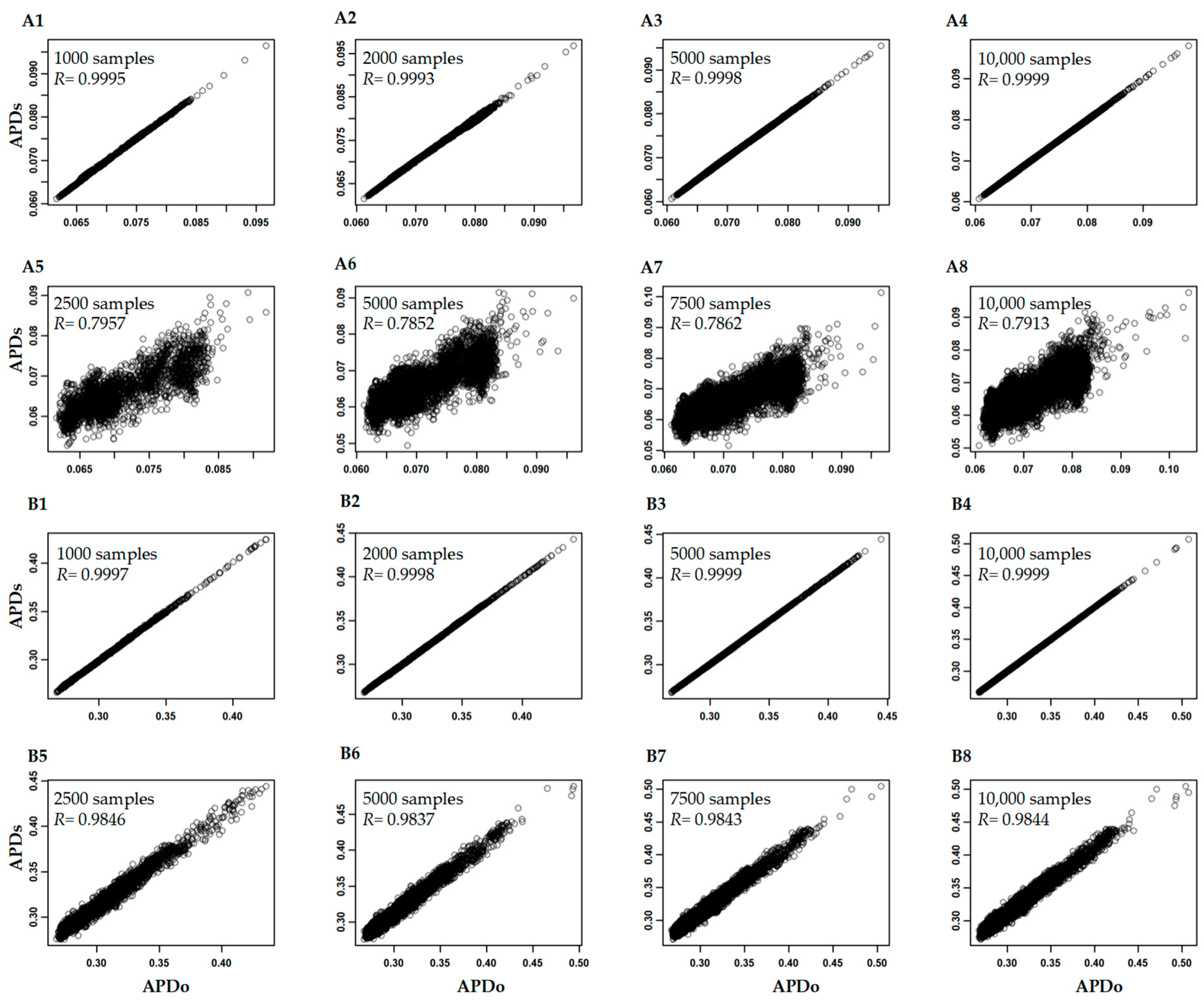
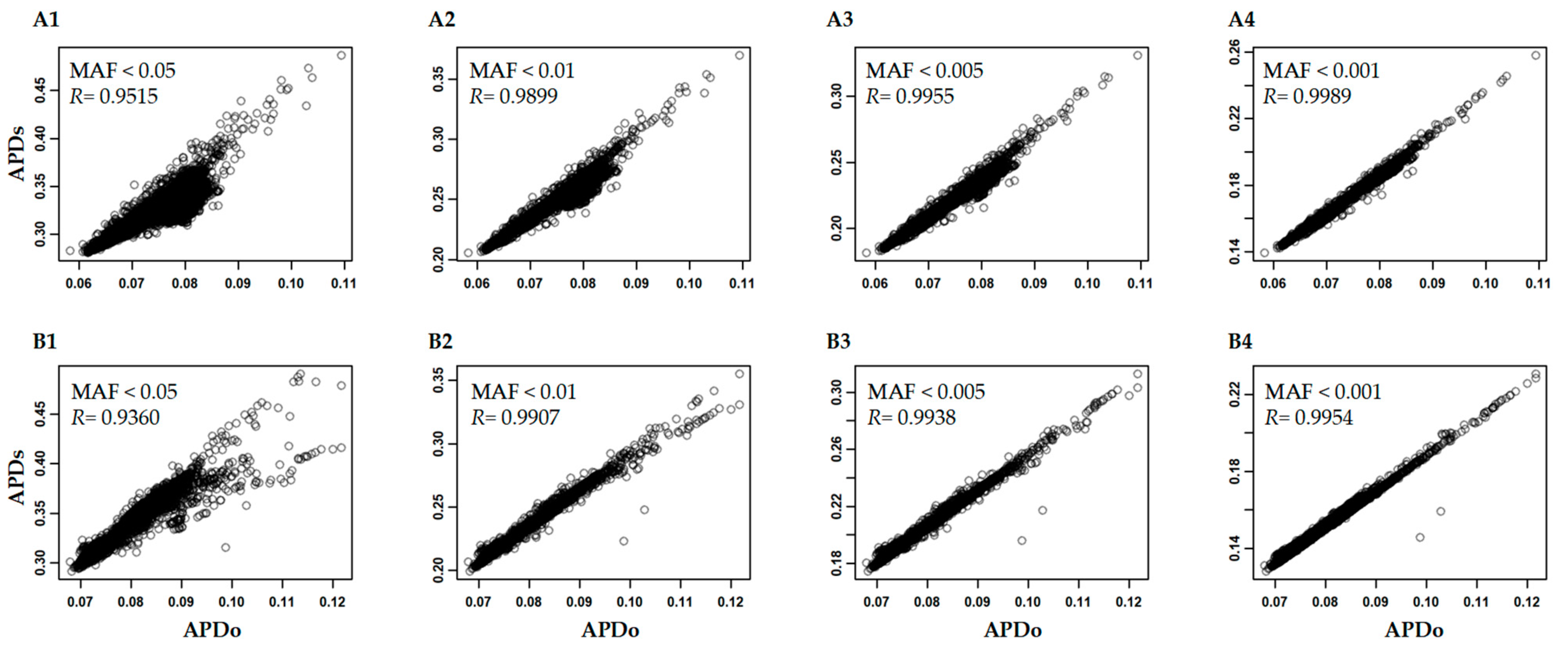
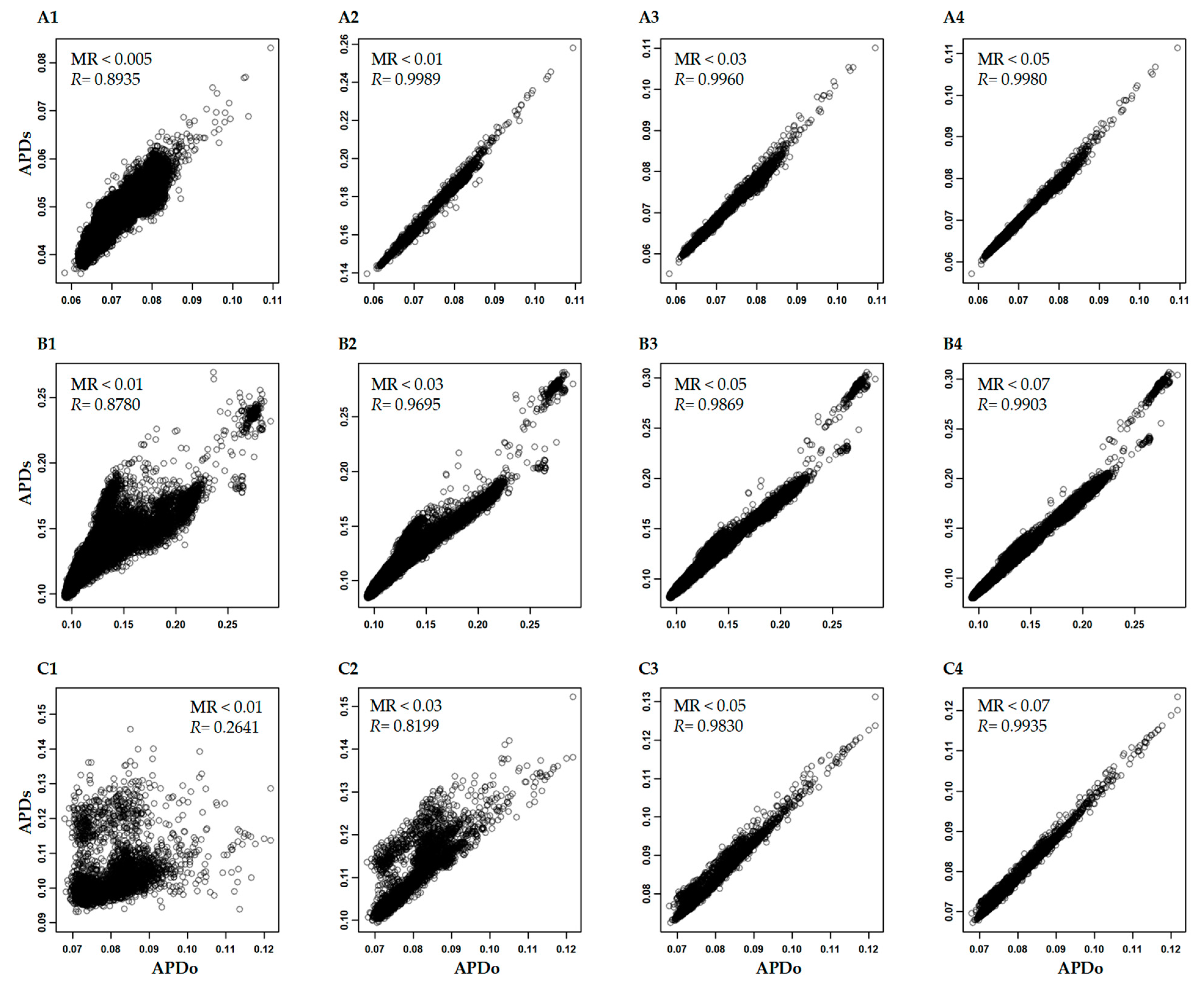
3.3. Variability of APD Estimation
4. Discussion
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Engels, J.M.M. Plant genetic resources management and conservation strategies: Problems and progress. Acta Hort. 2004, 634, 113–125. [Google Scholar] [CrossRef]
- FAO. The Second Report on the State of the World’s Plant; FAO: Rome, Italy, 2010. [Google Scholar]
- Fu, Y.B. The vulnerability of plant genetic resources conserved ex situ. Crop Sci. 2017, 57, 2314–2328. [Google Scholar] [CrossRef]
- Engels, J.M.M.; Ebert, A.W. A critical review of the current global ex situ conservation system for plant agrobiodiversity. II. strengths and weaknesses of the current system and recommendations for its improvement. Plants 2021, 10, 1904. [Google Scholar] [CrossRef] [PubMed]
- Hay, F.R.; Whitehouse, K.J.; Ellis, R.H.; Sackville Hamilton, N.R.; Lusty, C.; Ndjiondjop, M.N.; Tia, D.; Wenzel, P.; Santos, L.G.; Yazbek, M.; et al. CGIAR genebank viability data reveal inconsistencies in seed collection management. Glob. Food Sec. 2021, 30, 100557. [Google Scholar] [CrossRef]
- Lusty, C.; Sackville Hamilton, R.; Guarino, L.; Richards, C.; Jamora, N.; Hawtin, G. Envisaging an effective global long-term agrobiodiversity conservation system that promotes and facilitates use. Plants 2021, 10, 2764. [Google Scholar] [CrossRef]
- McCouch, S.; Navabi, Z.K.; Abberton, M.; Anglin, N.L.; Barbieri, R.L.; Baum, M.; Bett, K.; Booker, H.; Brown, G.L.; Bryan, G.J.; et al. Mobilizing crop biodiversity. Mol. Plant 2020, 13, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Díez, M.J.; De la Rosa, L.; Martín, I.; Guasch, L.; Cartea, M.E.; Mallor, C.; Casals, J.; Simó, J.; Rivera, A.; Anastasio, G.; et al. Plant genebanks: Present situation and proposals for their improvement. The case of the Spanish network. Front. Plant Sci. 2018, 9, 1794. [Google Scholar] [CrossRef]
- Waycott, W.; Fort, S.B. Differentiation of nearly identical germplasm accessions by a combination of molecular and morphological analyses. Genome 1994, 37, 577–583. [Google Scholar] [CrossRef]
- Virk, P.S.; Newbury, H.J.; Jackson, M.T.; Ford-Lloyd, B.V. The identification of duplicate accessions with a rice germplasm collection using RAPD analysis. Theor. Appl. Genet. 1995, 90, 1049–1055. [Google Scholar] [CrossRef]
- Chavarriaga-Aguirre, P.; Maya, M.M.; Tohme, J.; Duque, M.C.; Iglesias, C.; Bonierbale, M.W.; Kresovich, S.; Kochert, G. Using microsatellites, isozymes and AFLPs to evaluate genetic diversity and redundancy in the cassava core collection and to assess the usefulness of DNA-based markers to maintain germplasm collections. Mol. Breed. 1999, 5, 263–273. [Google Scholar] [CrossRef]
- Dean, R.E.; Dahlberg, J.A.; Hopkins, M.S.; Mitchell, S.E.; Kresovich, S. Genetic redundancy and diversity among ‘Orange’ accessions in the US national sorghum collection as assessed with simple sequence repeat (SSR) markers. Crop Sci. 1999, 39, 1215–1221. [Google Scholar] [CrossRef]
- Fu, Y.B. Redundancy and distinctness in flax germplasm as revealed by RAPD dissimilarity. Plant Gen. Res. 2006, 4, 117–124. [Google Scholar] [CrossRef]
- Kisha, T.J.; Cramer, C.S. Determining redundancy of short-day onion accessions in a germplasm collection using microsatellite and targeted region amplified polymorphic markers. J. Am. Soc. Hortic. 2011, 136, 129–134. [Google Scholar] [CrossRef]
- Motilal, L.A.; Zhang, D.; Mischke, S.; Meinhardt, L.W.; Umaharan, P. Microsatellite-aided detection of genetic redundancy improves management of the International Cocoa Genebank, Trinidad. Tree Genet. Genomes 2013, 9, 1395–1411. [Google Scholar] [CrossRef]
- Abbate, L.; Mercati, F.; Di Noto, G.; Heuertz, M.; Carimi, F.; Fatta del Bosco, S.; Schicchi, R. Genetic distinctiveness highlights the conservation value of a Sicilian manna ash germplasm collection assigned to Fraxinus angustifolia (Oleaceae). Plants 2020, 9, 1035. [Google Scholar] [CrossRef]
- Diederichsen, A.; Kusters, P.M.; Kessler, D.; Bainas, Z.; Gugel, R.K. Assembling a core collection from the flax world collection maintained by Plant Gene Resources of Canada. Genet. Resour. Crop Evol. 2013, 60, 1479–1485. [Google Scholar] [CrossRef]
- Fu, Y.B.; Cober, E.R.; Morrison, M.J.; Marsolais, F.; Zhou, R.; Xu, N.; Gahagan, A.C.; Horbach, C. Variability in maturity, oil and protein concentration, and genetic distinctness among soybean accessions conserved at Plant Gene Resources of Canada. Plants 2022, 11, 3525. [Google Scholar] [CrossRef]
- Yang, M.H.; Fu, Y.B. AveDissR: An R function for assessing genetic distinctness and genetic redundancy. Appl. Plant Sci. 2017, 5, apps.1700018. [Google Scholar] [CrossRef]
- Peterson, G.W.; Dong, Y.; Horbach, C.; Fu, Y.B. Genotyping-by-sequencing for plant genetic diversity analysis: A lab guide for SNP genotyping. Diversity 2014, 6, 665–680. [Google Scholar] [CrossRef]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics reveals the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef]
- Sansaloni, C.; Franco, J.; Santos, B.; Percival-Alwyn, L.; Singh, S.; Petroli, C.; Campos, J.; Dreher, K.; Payne, T.; Marshall, D.; et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 2020, 11, 4572. [Google Scholar] [PubMed]
- Varshney, R.K.; Roorkiwal, M.; Sun, S.; Bajaj, P.; Chitikineni, A.; Thudi, M.; Singh, N.P.; Du, X.; Upadhyaya, H.D.; Khan, A.W.; et al. A chickpea genetic variation map based on the sequencing of 3366 genomes. Nature 2021, 599, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Schreiber, M.; Scholz, U.; Graner, A.; Reif, J.C.; Stein, N. Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat. Genet. 2019, 51, 1076–1081. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Hyten, D.L.; Jia, G.; Quigley, C.V.; Fickus, E.W.; Nelson, R.L.; Cregan, P.B. Fingerprinting soybean germplasm and its utility in genomic research. G3 Genes Genomes Genet. 2015, 5, 1999–2006. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebortarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef]
- Danecek, P.; McCarthy, S.A.; HipSci Consortium; Durbin, R. A method for checking genomic integrity in cultured cell lines from SNP genotyping data. PLoS ONE 2016, 11, e0155014. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Mijangos, J.L.; Berry, O.F.; Pacioni, C.; Georges, A. dartR v2: An accessible genetic analysis platform for conservation, ecology and agriculture. Methods Ecol. Evol. 2022, 13, 2150–2158. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Anaconda Software Distribution. Computer Software. Anaconda. Web. 2021. Available online: https://anaconda.com (accessed on 17 February 2023).
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef]
- Weir, B.S.; Goudet, J. A unified characterization of population structure and relatedness. Genetics 2017, 206, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Dokan, K.; Kawamura, S.; Teshima, K.M. Effects of single nucleotide polymorphism ascertainment on population structure inferences. G3 Genes Genomes Genet. 2021, 11, jkab128. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.; Zhang, W.; Dubcovsky, J.; Sorrells, M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci. 2007, 47, 1018–1030. [Google Scholar] [CrossRef]
- Frankel, O.H. Genetic perspectives of germplasm conservation. In Genetic Manipulation: Impact on Man and Society; Arber, W.K., Llimensee, K., Peacock, W.J., Starlinger, P., Eds.; Cambridge University Press: Cambridge, UK, 1984; pp. 161–170. [Google Scholar]
- Brown, A.H.D. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- van Hintum, T.J.; Brown, A.; Spillane, C.; Hodgkin, T. Core Collections of Plant Genetic Resources; International Plant Genetic Resources Institute: Rome, Italy, 2000. [Google Scholar]

| Among-Group Variance | Group−Specific Fst and Group Size | ||||||
|---|---|---|---|---|---|---|---|
| Data Set | (%) and Sample Size | 1 (M + 3SD) | 2 (M + 2SD) | 3 (M + SD) | 4 (M) | 5 (M − SD) | 6 (M − 2SD) |
| Oryza sativa Indica group | 1.52 | −0.0499 | 0.0224 | −0.0606 | −0.0215 | 0.0499 | 0.2610 |
| 1789 | 38 | 29 | 87 | 430 | 1162 | 43 | |
| Oryza sativa Japonica group | 13.17 | −0.0551 | −0.0487 | 0.1555 | 0.0768 | 0.1547 | 0.6476 |
| 854 | 15 | 16 | 56 | 281 | 403 | 83 | |
| Glycine soja | 11.92 | −0.1049 | 0.0142 | 0.0479 | 0.0922 | 0.1677 | 0.7334 |
| 1178 | 11 | 16 | 133 | 318 | 589 | 111 | |
| Glycine max | 10.51 | 0.1773 | 0.0468 | 0.0723 | 0.1108 | 0.2325 | 0.4461 |
| 18,909 | 295 | 480 | 2087 | 5373 | 7574 | 3100 | |
| Hordeum spontaneum | 4.78 | 0.0619 | 0.0388 | −0.0126 | −0.0074 | 0.0528 | 0.2111 |
| 1140 | 20 | 16 | 89 | 354 | 599 | 62 | |
| Hordeum vulgare | 18.66 | 0.0677 | 0.1062 | 0.0850 | 0.0671 | 0.1938 | 0.5422 |
| 19,778 | 32 | 328 | 3783 | 3336 | 9566 | 2733 | |
| Triticum aestivum-f20k | 12.62 * | −0.2431 | 0.0243 | 0.0727 | 0.2105 | 0.3312 | 0.7615 |
| 24,847 * | 1657 | 682 | 2143 | 11,703 | 39,517 | 177 | |
| Triticum durum | 25.39 | 0.0129 | 0.0621 | 0.3753 | 0.2783 | 0.4692 | |
| 14,703 | 560 | 34 | 395 | 2265 | 11,449 | ||
| Triticum aethiopicum | 22.07 | −0.5228 | 0.1344 | 0.3092 | 0.4592 | 0.5490 | |
| 2822 | 35 | 13 | 46 | 912 | 1816 | ||
| Aegilops tauschii | 40.18 | −0.4481 | 0.5697 | 0.4207 | 0.6904 | 0.7273 | |
| 974 | 12 | 12 | 4 | 173 | 773 | ||
| Aegilops triuncialis | 53.37 | −0.1295 | 0.4138 | 0.4328 | 0.5576 | 0.6592 | |
| 661 | 11 | 18 | 10 | 47 | 575 | ||
| Cicer arietinum-f300k | 19.02 | 0.0579 | −0.0310 | 0.0522 | 0.1536 | 0.3037 | 0.5342 |
| 3171 | 31 | 63 | 273 | 1176 | 1279 | 349 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.-B. Assessing Genetic Distinctness and Redundancy of Plant Germplasm Conserved Ex Situ Based on Published Genomic SNP Data. Plants 2023, 12, 1476. https://doi.org/10.3390/plants12071476
Fu Y-B. Assessing Genetic Distinctness and Redundancy of Plant Germplasm Conserved Ex Situ Based on Published Genomic SNP Data. Plants. 2023; 12(7):1476. https://doi.org/10.3390/plants12071476
Chicago/Turabian StyleFu, Yong-Bi. 2023. "Assessing Genetic Distinctness and Redundancy of Plant Germplasm Conserved Ex Situ Based on Published Genomic SNP Data" Plants 12, no. 7: 1476. https://doi.org/10.3390/plants12071476
APA StyleFu, Y.-B. (2023). Assessing Genetic Distinctness and Redundancy of Plant Germplasm Conserved Ex Situ Based on Published Genomic SNP Data. Plants, 12(7), 1476. https://doi.org/10.3390/plants12071476






