Goss’s Wilt Resistance in Corn Is Mediated via Salicylic Acid and Programmed Cell Death but Not Jasmonic Acid Pathways
Abstract
1. Introduction
2. Results
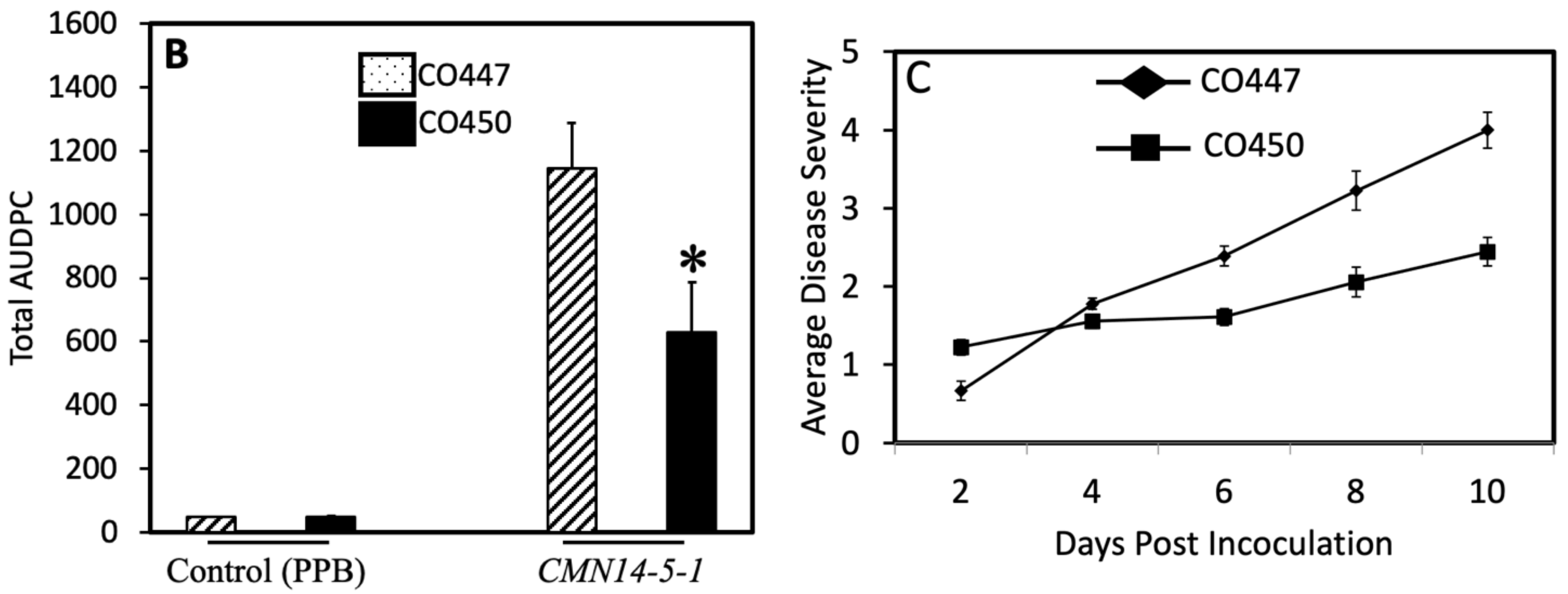
2.1. Pathogenicity
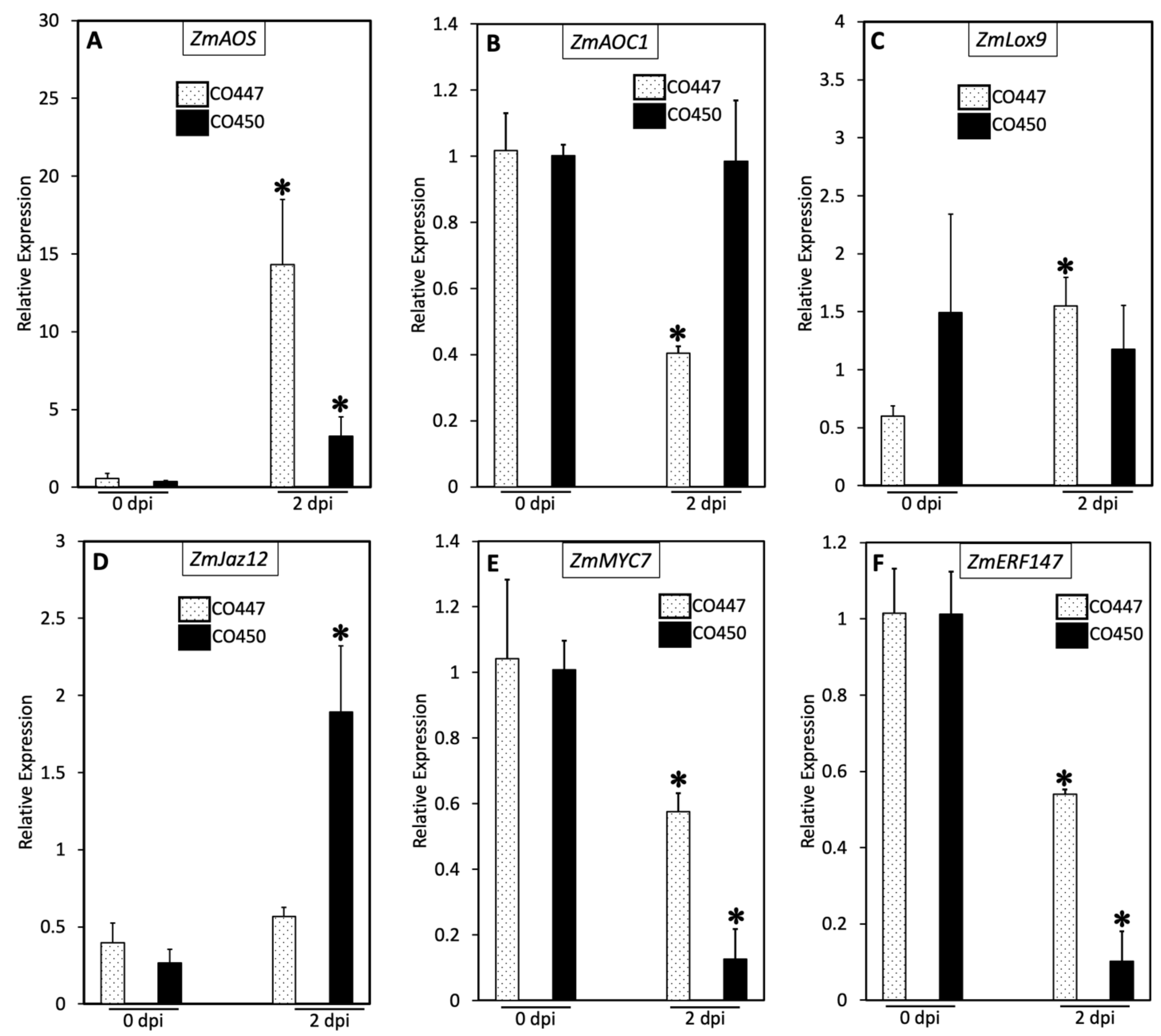
2.2. Plant Defense against Goss’s Wilt Is Not Enhanced via the Jasmonic Acid Pathway
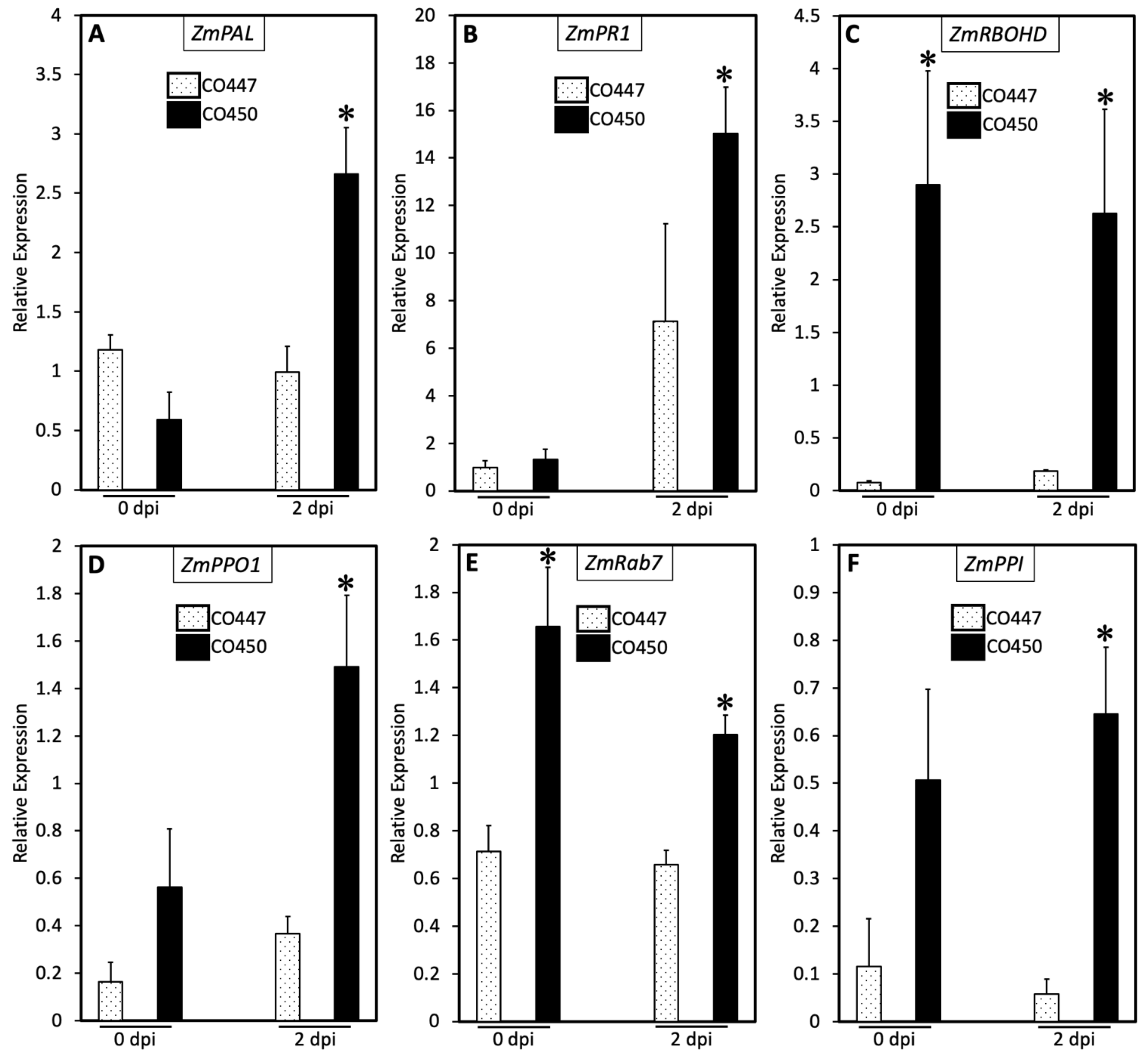
2.3. SA and PCD Regulate Goss’s Wilt Disease Resistance
2.4. Exogenous Application of SA and H2O2 Confers Partial Disease Resistance against CMN14-5-1
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Chemical Treatments
4.3. Bacterial Isolation, Preparation of Inoculum, and Leaf Inoculation
4.4. Measurement of Lesion Length, Disease Severity Rating, and Sampling
4.5. Measurement of Transcript Levels
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Staller, J.E. Maize Cobs and Cultures: History of Zea mays L.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Eggenberger, S.; Diaz-Arias, M.M.; Gougherty, A.V.; Nutter, F.W., Jr.; Sernett, J.; Robertson, A.E. Dissemination of Goss’s wilt of corn and epiphytic Clavibacter michiganensis subsp. nebraskensis from inoculum point sources. Plant Dis. 2016, 100, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Tambong, J.T. Comparative genomics of Clavibacter michiganensis subspecies, pathogens of important agricultural crops. PLoS ONE 2017, 12, e0172295. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ren, J.; Peng, Z.; Umana, A.A.; Le, H.; Danilova, T.; Fu, J.; Wang, H.; Robertson, A.; Hulbert, S.H. Analysis of Extreme Phenotype Bulk Copy Number Variation (XP-CNV) Identified the Association of rp1 with Resistance to Goss’s Wilt of Maize. Front. Plant Sci. 2018, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Eichenlaub, R.; Gartemann, K.-H. The Clavibacter michiganensis subspecies: Molecular investigation of gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 2011, 49, 445–464. [Google Scholar] [CrossRef]
- Gonzalez, A.J.; Trapiello, E. Clavibacter michiganensis subsp. phaseoli subsp. nov., pathogenic in bean. Int. J. Syst. Evol. Microbiol. 2014, 64, 1752–1755. [Google Scholar] [CrossRef]
- Li, X.; Tambong, J.; Yuan, K.X.; Chen, W.; Xu, H.; Lévesque, C.A.; De Boer, S.H. Re-classification of Clavibacter michiganensis subspecies on the basis of whole-genome and multi-locus sequence analyses. Int. J. Syst. Evol. Microbiol. 2018, 68, 234. [Google Scholar] [CrossRef]
- Wysong, D.; Doupnik, B., Jr.; Lane, L. Goss’s wilt and corn lethal necrosis: Can they become a major problem? [Corn Belt, in the United States, bacterial wilt and blight, virus disease]. In Proceedings of the Annual Corn and Sorghum Industry Research Conference American Seed Trade Association, Corn and Sorghum Division, Corn and Sorghum Research Conference, Chicago, IL, USA, 1982. [Google Scholar]
- Rocheford, T.; Gardner, C.; Vidaver, A. Genetic studies of resistance in maize (Zea mays L.) to Goss’s bacterial wilt and blight (Clavibacter michiganense ssp. nebraskense). J. Hered. 1989, 80, 351–356. [Google Scholar] [CrossRef]
- Jackson, T.A.; Harveson, R.M.; Vidaver, A.K. Reemergence of Goss’s wilt and blight of corn to the central high plains. Plant Health Prog. 2007, 8, 44. [Google Scholar] [CrossRef]
- McNally, R.R.; Ishimaru, C.A.; Malvick, D.K. PCR-mediated detection and quantification of the Goss’s wilt pathogen Clavibacter michiganensis subsp. nebraskensis via a novel gene target. Phytopathology 2016, 106, 1465–1472. [Google Scholar] [CrossRef]
- Mallowa, S.O.; Mbofung, G.Y.; Eggenberger, S.K.; Den Adel, R.L.; Scheiding, S.R.; Robertson, A.E. Infection of maize by Clavibacter michiganensis subsp. nebraskensis does not require severe wounding. Plant Dis. 2016, 100, 724–731. [Google Scholar] [CrossRef]
- Soliman, A.; Gulden, R.H.; Tambong, J.T.; Bajracharya, P.; Adam, L.R.; Xu, R.; Cott, M.; Daayf, F. Developed and validated inoculation and disease assessment methods for Goss’s bacterial wilt and leaf blight disease of corn. Crop Prot. 2018, 112, 159–167. [Google Scholar] [CrossRef]
- Pataky, J. Influence of host resistance and growth stage at the time of inoculation on Stewart’s wilt and Goss’s wilt development and sweet corn hybrid yield. Plant Dis. 1989, 73, 339–345. [Google Scholar]
- Agarkova, I.V.; Lambrecht, P.A.; Vidaver, A. Genetic diversity and population structure of Clavibacter michiganensis subsp. nebraskensis. Can. J. Microbiol. 2011, 57, 366–374. [Google Scholar] [CrossRef]
- Tambong, J.T.; Xu, R.; Adam, Z.; Cott, M.; Rose, K.; Reid, L.M.; Daayf, F.; Brière, S.; Bilodeau, G.J. Draft genome sequence of Clavibacter michiganensis subsp. nebraskensis Strain DOAB 397, isolated from an infected field corn plant in Manitoba, Canada. Genome Announc. 2015, 3, e00715–e00768. [Google Scholar] [CrossRef]
- Langemeier, C.B.; Jackson-Ziems, T.A.; Kruger, G.R. Four common Setaria species are alternative hosts for Clavibacter michiganensis subsp. nebraskensis, causal agent of Goss’s bacterial wilt and blight of corn. Plant Health Prog. 2014, 15, 57–60. [Google Scholar] [CrossRef]
- Mehl, K.M. Goss’s wilt and leaf blight of corn: Chemical control, residue management and resistance. Master’s Thesis, University of Illinois Urbana-Champaign, Champaign, IL, USA, 2015. Available online: http://hdl.handle.net/2142/88985 (accessed on 27 January 2023).
- Mehl, K.; Weems, J.; Ames, K.; Bradley, C. Evaluation of foliar-applied copper hydroxide and citric acid for control of Goss’s wilt and leaf blight of corn. Can. J. Plant Pathol. 2015, 37, 160–164. [Google Scholar] [CrossRef]
- Harding, M.W.; Jindal, K.; Tambong, J.T.; Daayf, F.; Howard, R.J.; Derksen, H.; Reid, L.M.; Tenuta, A.U.; Feng, J. Goss’s bacterial wilt and leaf blight of corn in Canada–disease update. Can. J. Plant Pathol. 2018, 40, 471–480. [Google Scholar] [CrossRef]
- Cooper, J.S.; Rice, B.R.; Shenstone, E.M.; Lipka, A.E.; Jamann, T.M. Genome-Wide Analysis and Prediction of Resistance to Goss’s Wilt in Maize. Plant Genome 2019, 12, 180045. [Google Scholar] [CrossRef]
- Ikley, J.T.; Wise, K.A.; Johnson, W.G. Annual Ryegrass (Lolium multiflorum), Johnsongrass (Sorghum halepense), and Large Crabgrass (Digitaria sanguinalis) are alternative hosts for Clavibacter michiganensis subsp. nebraskensis, causal agent of goss’s wilt of corn. Weed Sci. 2015, 63, 901–909. [Google Scholar] [CrossRef]
- Xu, X.; Miller, S.A.; Baysal-Gurel, F.; Gartemann, K.-H.; Eichenlaub, R.; Rajashekara, G. Bioluminescence imaging of Clavibacter michiganensis subsp. michiganensis infection of tomato seeds and plants. Appl. Environ. Microbiol. 2010, 76, 3978–3988. [Google Scholar] [CrossRef]
- Mbofung, G.C.; Sernett, J.; Horner, H.T.; Robertson, A.E. Comparison of susceptible and resistant maize hybrids to colonization by Clavibacter michiganensis subsp. nebraskensis. Plant Dis. 2016, 100, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.; Rampitsch, C.; Tambong, J.T.; Daayf, F. Secretome Analysis of Clavibacter nebraskensis Strains Treated with Natural Xylem Sap In Vitro Predicts Involvement of Glycosyl Hydrolases and Proteases in Bacterial Aggressiveness. Proteomes 2021, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Heitz, T.; Prasad, V.; Wiedemann-Merdinoglu, S.; Kauffmann, S.; Geoffroy, P.; Legrand, M.; Fritig, B. Plant ‘pathogenesis-related’proteins and their role in defense against pathogens. Biochimie 1993, 75, 687–706. [Google Scholar] [CrossRef] [PubMed]
- Omelichkina, Y.V.; Boyarkina, S.; Shafikova, T. Effector-activated immune responses in potato and tobacco cell cultures caused by phytopathogen Clavibacter michiganensis ssp. sepedonicus. Russ. J. Plant Physiol. 2017, 64, 423–430. [Google Scholar] [CrossRef]
- Owusu, V.; Mira, M.; Soliman, A.; Adam, L.; Daayf, F.; Hill, R.; Stasolla, C. Suppression of the maize phytoglobin ZmPgb1. 1 promotes plant tolerance against Clavibacter nebraskensis. Planta 2019, 250, 1803–1818. [Google Scholar] [CrossRef]
- Meyer, J.; Berger, D.K.; Christensen, S.A.; Murray, S.L. RNA-Seq analysis of resistant and susceptible sub-tropical maize lines reveals a role for kauralexins in resistance to grey leaf spot disease, caused by Cercospora zeina. BMC Plant Biol. 2017, 17, 197. [Google Scholar] [CrossRef]
- Christie, N.; Myburg, A.A.; Joubert, F.; Murray, S.L.; Carstens, M.; Lin, Y.C.; Meyer, J.; Crampton, B.G.; Christensen, S.A.; Ntuli, J.F. Systems genetics reveals a transcriptional network associated with susceptibility in the maize–grey leaf spot pathosystem. Plant J. 2017, 89, 746–763. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Cohu, C.M.; Amiard, V.; van Zadelhoff, G.; Veldink, G.A.; Muller, O.; Adams, W.W., III. Emerging trade-offs–impact of photoprotectants (PsbS, xanthophylls, and vitamin E) on oxylipins as regulators of development and defense. New Phytol. 2013, 197, 720–729. [Google Scholar] [CrossRef]
- Woldemariam, M.G.; Ahern, K.; Jander, G.; Tzin, V. A role for 9-lipoxygenases in maize defense against insect herbivory. Plant Signal. Behav. 2018, 13, 4709–4723. [Google Scholar] [CrossRef]
- Stenzel, I.; Otto, M.; Delker, C.; Kirmse, N.; Schmidt, D.; Miersch, O.; Hause, B.; Wasternack, C. ALLENE OXIDE CYCLASE (AOC) gene family members of Arabidopsis thaliana: Tissue-and organ-specific promoter activities and in vivo heteromerization. J. Exp. Bot. 2012, 63, 6125–6138. [Google Scholar] [CrossRef]
- Mao, L.; Ge, L.; Ye, X.; Xu, L.; Si, W.; Ding, T. ZmGLP1, a Germin-like Protein from Maize, Plays an Important Role in the Regulation of Pathogen Resistance. Int. J. Mol. Sci. 2022, 23, 14316. [Google Scholar] [CrossRef]
- Christensen, S.A.; Huffaker, A.; Kaplan, F.; Sims, J.; Ziemann, S.; Doehlemann, G.; Ji, L.; Schmitz, R.J.; Kolomiets, M.V.; Alborn, H.T. Maize death acids, 9-lipoxygenase–derived cyclopente (a) nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. USA 2015, 112, 11407–11412. [Google Scholar] [CrossRef]
- Murphree, C.; Kim, S.B.; Karre, S.; Samira, R.; Balint-Kurti, P. Use of virus-induced gene silencing to characterize genes involved in modulating hypersensitive cell death in maize. Mol. Plant Pathol. 2020, 21, 1662–1676. [Google Scholar] [CrossRef]
- Jiang, S.; Yao, J.; Ma, K.-W.; Zhou, H.; Song, J.; He, S.Y.; Ma, W. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef]
- Ishiga, Y.; Ishiga, T.; Uppalapati, S.R.; Mysore, K.S. Jasmonate ZIM-domain (JAZ) protein regulates host and nonhost pathogen-induced cell death in tomato and Nicotiana benthamiana. PLoS ONE 2013, 8, e75728. [Google Scholar] [CrossRef]
- Lyons, R.; Manners, J.M.; Kazan, K. Jasmonate biosynthesis and signaling in monocots: A comparative overview. Plant Cell Rep. 2013, 32, 815–827. [Google Scholar] [CrossRef]
- Sun, Q.; Wang, G.; Zhang, X.; Zhang, X.; Qiao, P.; Long, L.; Yuan, Y.; Cai, Y. Genome-wide identification of the TIFY gene family in three cultivated Gossypium species and the expression of JAZ genes. Sci. Rep. 2017, 7, srep42418. [Google Scholar] [CrossRef]
- Wang, Y.; Qiao, L.; Bai, J.; Wang, P.; Duan, W.; Yuan, S.; Yuan, G.; Zhang, F.; Zhang, L.; Zhao, C. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). Bmc Genom. 2017, 18, 1–19. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, K.; Li, W.; Pang, X.; Liu, P.; Si, H.; Zang, J.; Xing, J.; Dong, J. ZmMYC7 directly regulates ZmERF147 to increase maize resistance to Fusarium graminearum. Crop J. 2023, 11, 79–88. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Way, H.M.; Kazan, K.; Mitter, N.; Goulter, K.C.; Birch, R.G.; Manners, J.M. Constitutive expression of a phenylalanine ammonia-lyase gene from Stylosanthes humilis in transgenic tobacco leads to enhanced disease resistance but impaired plant growth. Physiol. Mol. Plant Pathol. 2002, 60, 275–282. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, X.; Zhang, F.; Dong, L.; Wu, J.; Cheng, Q.; Qi, D.; Yan, X.; Jiang, L.; Fan, S. Phenylalanine ammonia-lyase2. 1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z. Maize phenylalanine ammonia-lyases contribute to resistance to Sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol. Plant Pathol. 2019, 20, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, J.E.; Sanchez, J.P.; Zumstein, K.; Gilchrist, D.G. Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Mol. Plant Pathol. 2018, 19, 2111–2123. [Google Scholar] [CrossRef]
- Ghorbel, M.; Zribi, I.; Missaoui, K.; Drira-Fakhfekh, M.; Azzouzi, B.; Brini, F. Differential regulation of the durum wheat Pathogenesis-related protein (PR1) by Calmodulin TdCaM1. 3 protein. Mol. Biol. Rep. 2021, 48, 347–362. [Google Scholar] [CrossRef]
- Sagi, M.; Davydov, O.; Orazova, S.; Yesbergenova, Z.; Ophir, R.; Stratmann, J.W.; Fluhr, R. Plant respiratory burst oxidase homologs impinge on wound responsiveness and development in Lycopersicon esculentum. Plant Cell 2004, 16, 616–628. [Google Scholar] [CrossRef]
- Bhonwong, A.; Stout, M.J.; Attajarusit, J.; Tantasawat, P. Defensive role of tomato polyphenol oxidases against cotton bollworm (Helicoverpa armigera) and beet armyworm (Spodoptera exigua). J. Chem. Ecol. 2009, 35, 28–38. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef]
- Otulak-Kozieł, K.; Kozieł, E.; Valverde, R.A. The respiratory burst oxidase homolog D (RbohD) cell and tissue distribution in potato–potato virus Y (PVYNTN) hypersensitive and susceptible reactions. Int. J. Mol. Sci. 2019, 20, 2741. [Google Scholar] [CrossRef]
- Sagi, M.; Fluhr, R. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 2006, 141, 336–340. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 2005, 8, 397–403. [Google Scholar] [CrossRef]
- Kaur, G.; Pati, P.K. Analysis of cis-acting regulatory elements of Respiratory burst oxidase homolog (Rboh) gene families in Arabidopsis and rice provides clues for their diverse functions. Comput. Biol. Chem. 2016, 62, 104–118. [Google Scholar] [CrossRef]
- Wang, W.; Chen, D.; Zhang, X.; Liu, D.; Cheng, Y.; Shen, F. Role of plant respiratory burst oxidase homologs in stress responses. Free. Radic. Res. 2018, 52, 826–839. [Google Scholar] [CrossRef]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef]
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014, 164, 1191–1203. [Google Scholar] [CrossRef]
- Moshkov, I.; Novikova, G. Superfamily of plant monomeric GTP-binding proteins: 2. Rab proteins are the regulators of vesicles trafficking and plant responses to stresses. Russ. J. Plant Physiol. 2008, 55, 119–129. [Google Scholar] [CrossRef]
- Liu, F.; Guo, J.; Bai, P.; Duan, Y.; Wang, X.; Cheng, Y.; Feng, H.; Huang, L.; Kang, Z. Wheat TaRab7 GTPase is part of the signaling pathway in responses to stripe rust and abiotic stimuli. PLoS ONE 2012, 7, e37146. [Google Scholar] [CrossRef]
- Sui, J.; Li, G.; Chen, G.; Zhao, C.; Kong, X.; Hou, X.; Qiao, L.; Wang, J. RNA-seq analysis reveals the role of a small GTP-binding protein, Rab7, in regulating clathrin-mediated endocytosis and salinity-stress resistance in peanut. Plant Biotechnol. Rep. 2017, 11, 43–52. [Google Scholar] [CrossRef]
- Kaur, G.; Singh, S.; Singh, H.; Chawla, M.; Dutta, T.; Kaur, H.; Bender, K.; Snedden, W.; Kapoor, S.; Pareek, A. Characterization of peptidyl-prolyl cis-trans isomerase-and calmodulin-binding activity of a cytosolic Arabidopsis thaliana cyclophilin AtCyp19-3. PLoS ONE 2015, 10, e0136692. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ma, P.; Holtsmark, I.; Skaugen, M.; Eijsink, V.G.; Brurberg, M.B. New type of antimicrobial protein produced by the plant pathogen Clavibacter michiganensis subsp. michiganensis. Appl. Environ. Microbiol. 2013, 79, 5721–5727. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.-P.; Ryals, J.A. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef]
- Wang, C.; El-Shetehy, M.; Shine, M.; Yu, K.; Navarre, D.; Wendehenne, D.; Kachroo, A.; Kachroo, P. Free radicals mediate systemic acquired resistance. Cell Rep. 2014, 7, 348–355. [Google Scholar] [CrossRef]
- El-Shetehy, M.; Wang, C.; Shine, M.; Yu, K.; Kachroo, A.; Kachroo, P. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 2015, 10, e998544. [Google Scholar] [CrossRef] [PubMed]
- El-Shetehy, M.; Moradi, A.; Maceroni, M.; Reinhardt, D.; Petri-Fink, A.; Rothen-Rutishauser, B.; Mauch, F.; Schwab, F. Silica nanoparticles enhance disease resistance in Arabidopsis plants. Nat. Nanotechnol. 2021, 16, 344–353. [Google Scholar] [CrossRef]
- Morris, S.W.; Vernooij, B.; Titatarn, S.; Starrett, M.; Thomas, S.; Wiltse, C.C.; Frederiksen, R.A.; Bhandhufalck, A.; Hulbert, S.; Uknes, S. Induced resistance responses in maize. Mol. Plant-Microbe Interact. 1998, 11, 643–658. [Google Scholar] [CrossRef]
- Masuta, C.; Van den Bulcke, M.; Bauw, G.; Van Montagu, M.; Caplan, A.B. Differential effects of elicitors on the viability of rice suspension cells. Plant Physiol. 1991, 97, 619–629. [Google Scholar] [CrossRef]
- Vallélian-Bindschedler, L.; Métraux, J.-P.; Schweizer, P. Salicylic acid accumulation in barley is pathogen specific but not required for defense-gene activation. Mol. Plant-Microbe Interact. 1998, 11, 702–705. [Google Scholar] [CrossRef]
- Görlach, J.; Volrath, S.; Knauf-Beiter, G.; Hengy, G.; Beckhove, U.; Kogel, K.-H.; Oostendorp, M.; Staub, T.; Ward, E.; Kessmann, H. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 1996, 8, 629–643. [Google Scholar]
- Jindal, K.K.; Zhu, X.; Tenuta, A.; Javed, N.; Daayf, F.; Reid, L.R. Maize inbreds for multiple resistance breeding against major foliar, ear and stalk rot diseases. Maydica 2019, 64, 22. [Google Scholar]
- Gross, D.C.; Vidaver, A. A selective medium for isolation of Corynebacterium nebraskense from soil and plant parts. Phytopathology 1979, 69, 82–87. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shumilak, A.; El-Shetehy, M.; Soliman, A.; Tambong, J.T.; Daayf, F. Goss’s Wilt Resistance in Corn Is Mediated via Salicylic Acid and Programmed Cell Death but Not Jasmonic Acid Pathways. Plants 2023, 12, 1475. https://doi.org/10.3390/plants12071475
Shumilak A, El-Shetehy M, Soliman A, Tambong JT, Daayf F. Goss’s Wilt Resistance in Corn Is Mediated via Salicylic Acid and Programmed Cell Death but Not Jasmonic Acid Pathways. Plants. 2023; 12(7):1475. https://doi.org/10.3390/plants12071475
Chicago/Turabian StyleShumilak, Alexander, Mohamed El-Shetehy, Atta Soliman, James T. Tambong, and Fouad Daayf. 2023. "Goss’s Wilt Resistance in Corn Is Mediated via Salicylic Acid and Programmed Cell Death but Not Jasmonic Acid Pathways" Plants 12, no. 7: 1475. https://doi.org/10.3390/plants12071475
APA StyleShumilak, A., El-Shetehy, M., Soliman, A., Tambong, J. T., & Daayf, F. (2023). Goss’s Wilt Resistance in Corn Is Mediated via Salicylic Acid and Programmed Cell Death but Not Jasmonic Acid Pathways. Plants, 12(7), 1475. https://doi.org/10.3390/plants12071475






