Genetic Relationships of 118 Castanea Specific Germplasms and Construction of Their Molecular ID Based on Morphological Characteristics and SSR Markers
Abstract
1. Introduction
2. Results
2.1. Phenotypic Traits and Morphological Analysis of Appearance
2.2. Polymorphism and Genetic Diversity Analysis
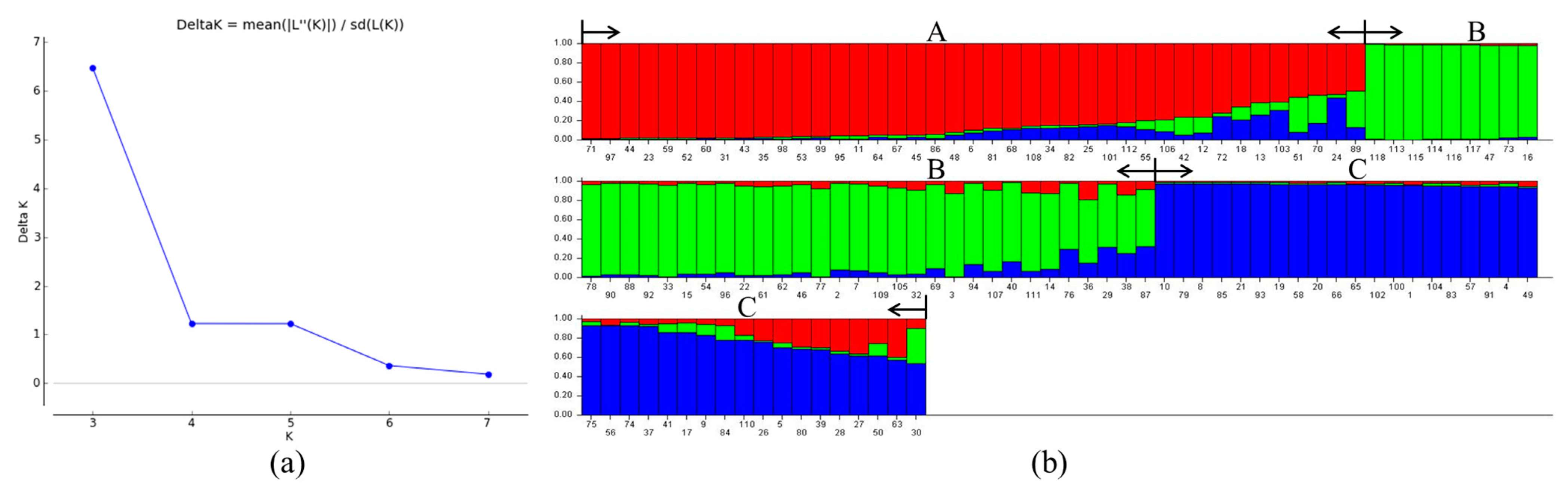
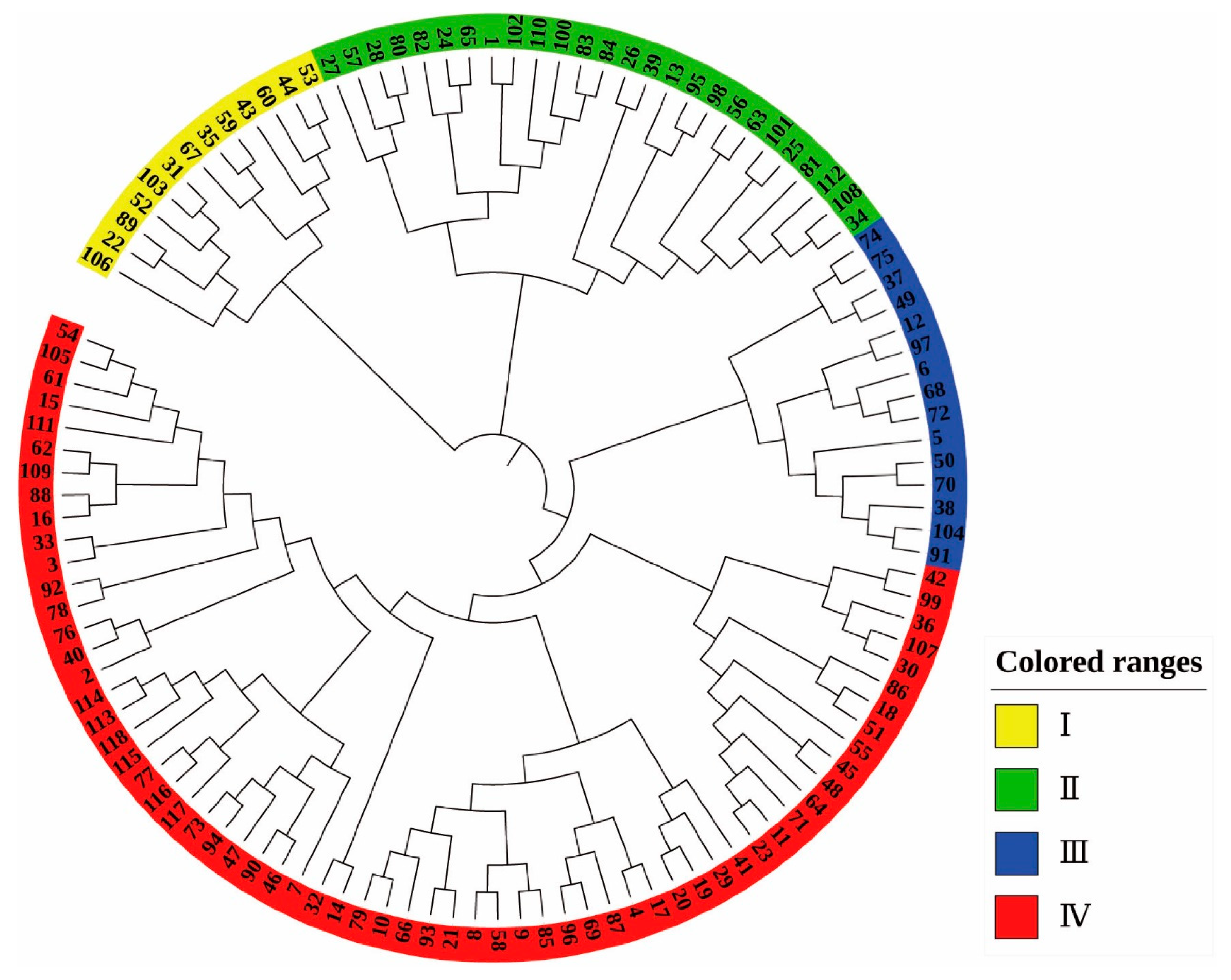
2.3. Population Structure Analysis
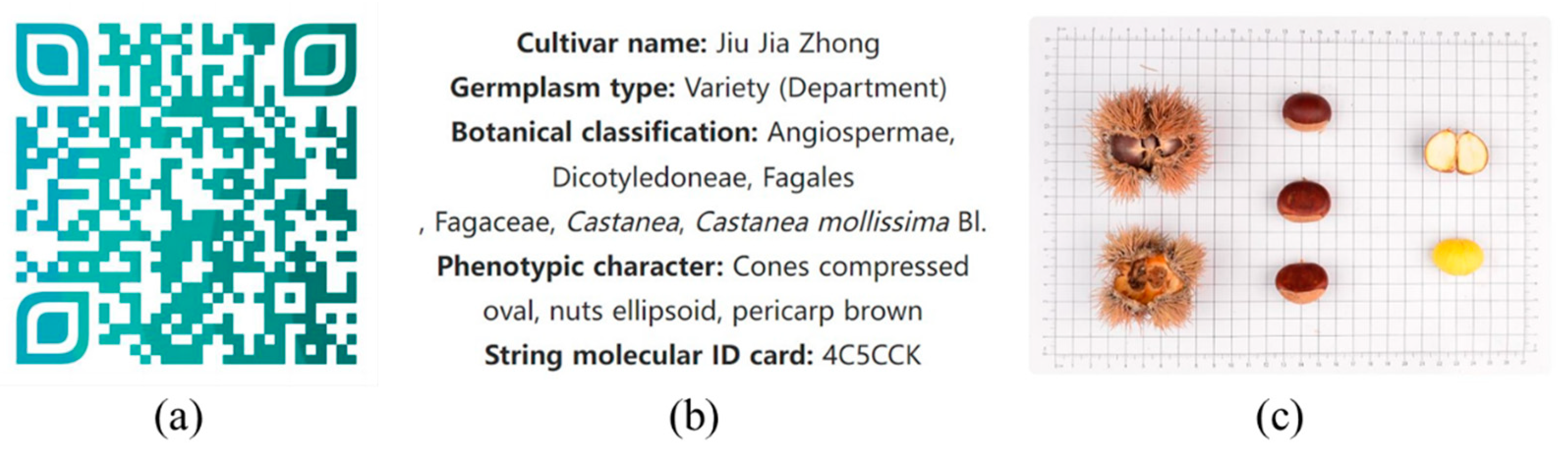
2.4. Establishment of DNA Molecular Identity
2.5. Combining Phenotypic Information with Molecular Information
3. Discussion
3.1. Phenotypic Trait Analysis
3.2. Genetic Diversity Analysis
3.3. Population Structure Analysis
3.4. Establishment of DNA Molecular Identity
3.5. Phenotype and Molecular Binding Analysis
4. Materials and Methods
4.1. Plant Materials
4.2. Phenotype Data Measurement
4.3. DNA Extraction and PCR Amplification
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anagnostakis, S.L. Chestnut Breeding in the United States for Disease and Insect Resistance. Plant Dis. 2012, 96, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Wang, J.; Liu, Y.; Ma, C.; Guo, S.; Lin, S.; Wang, J. Transcriptome analysis of genes involved in starch biosynthesis in developing Chinese chestnut (Castanea mollissima Blume) seed kernels. Sci. Rep. 2021, 11, 3570. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Zhang, P.; Guo, M.; Li, M.; Wang, L.; Adeel, M.; Shakoor, N.; Rui, Y. Effects of age on mineral elements, amino acids and fatty acids in Chinese chestnut fruits. Eur. Food Res. Technol. 2021, 247, 2079–2086. [Google Scholar] [CrossRef]
- Li, B.; Liu, C.; Tang, J.; Xin, Y.; Dong, Z.; Bai, B.; Xin, P. Genetic diversity analysis of Camellia fascicularis H. T. Chang based on SSR markers. J. Appl. Res. Med. Aromat. Plants 2022, 31, 100404. [Google Scholar] [CrossRef]
- Makhadmeh, I.M.; Thabet, S.G.; Ali, M.; Alabbadi, B.; Albalasmeh, A.; Alqudah, A.M. Exploring genetic variation among Jordanian Solanum lycopersicon L. landraces and their performance under salt stress using SSR markers. J. Genet. Eng. Biotechnol. 2022, 20, 45. [Google Scholar] [CrossRef]
- Ozkan, G.; Haliloglu, K.; Turkoglu, A.; Ozturk, H.I.; Elkoca, E.; Poczai, P. Determining Genetic Diversity and Population Structure of Common Bean (Phaseolus vulgaris L.) Landraces from Turkiye Using SSR Markers. Genes 2022, 13, 1418. [Google Scholar] [CrossRef]
- Santos, C.; Zhebentyayeva, T.; Serrazina, S.; Nelson, C.D.; Costa, R. Development and characterization of EST-SSR markers for mapping reaction to Phytophthora cinnamomi in Castanea spp. Sci. Hortic. 2015, 194, 181–187. [Google Scholar] [CrossRef]
- Shahnazari, N.; Noormohammadi, Z.; Sheidai, M.; Koohdar, F. A new insight on genetic diversity of sweet oranges: CAPs-SSR and SSR markers. J. Genet. Eng. Biotechnol. 2022, 20, 105. [Google Scholar] [CrossRef]
- Uddin, N.; Ali, N.; Nisar, M.; Liu, M.J.; Liu, Z.G.; Muhammad, N.; Rahman, I.U. SSR-based population structure, molecular diversity and identity cards of Ziziphus species from Pakistan and China. Genet. Resour. Crop Evol. 2021, 68, 2391–2409. [Google Scholar] [CrossRef]
- Zheng, X.W.; Cheng, T.; Yang, L.B.; Xu, J.X.; Tang, J.P.; Xie, K.Q.; Huang, X.F.; Bao, Z.Z.; Zheng, X.F.; Diao, Y.; et al. Genetic Diversity and DNA Fingerprints of Three Important Aquatic Vegetables by EST-SSR Markers. Sci. Rep. 2019, 9, 14074. [Google Scholar] [CrossRef]
- Parashuram, S.; Singh, N.V.; Gaikwad, N.N.; Corrado, G.; Sowjanya, P.R.; Basile, B.; Devaraja, N.S.; Chandra, R.; Babu, K.D.; Patil, P.G.; et al. Morphological, Biochemical, and Molecular Diversity of an Indian Ex Situ Collection of Pomegranate (Punica granatum L.). Plants 2022, 11, 3518. [Google Scholar] [CrossRef]
- Smith, A.C.; Shima, J.S. Variation in the effects of larval history on juvenile performance of a temperate reef fish. Austral Ecol. 2011, 36, 830–838. [Google Scholar] [CrossRef]
- Fu, Z.Y.; Sa, K.J.; Park, H.; Jang, S.J.; Kim, Y.J.; Lee, J.K. Utilization of Novel Perilla SSR Markers to Assess the Genetic Diversity of Native Perilla Germplasm Accessions Collected from South Korea. Plants 2022, 11, 2974. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, X.; Yu, W.; Xiang, P.; Zhang, S.; Wang, G. The Diversity of Melia azedarach L. from China Based on Transcriptome-Developed SSR Marker. Forests 2022, 13, 1011. [Google Scholar] [CrossRef]
- Inoue, E.; Ning, L.; Hara, H.; Ruan, S.; Anzai, H. Development of Simple Sequence Repeat Markers in Chinese Chestnut and Their Characterization in Diverse Chestnut Cultivars. J. Am. Soc. Hortic. Sci. 2009, 134, 610–617. [Google Scholar] [CrossRef]
- Nie, X.H.; Wang, Z.H.; Liu, N.W.; Song, L.; Yan, B.Q.; Xing, Y.; Zhang, Q.; Fang, K.F.; Zhao, Y.L.; Chen, X.; et al. Fingerprinting 146 Chinese chestnut (Castanea mollissima Blume) accessions and selecting a core collection using SSR markers. J. Integr. Agric. 2021, 20, 1277–1286. [Google Scholar] [CrossRef]
- Xiong, H.; Zou, F.; Guo, S.; Yuan, D.; Niu, G. Self-sterility May Be Due to Prezygotic Late-acting Self-incompatibility and Early-acting Inbreeding Depression in Chinese Chestnut. J. Am. Soc. Hortic. Sci. 2019, 144, 172–181. [Google Scholar] [CrossRef]
- Luan, M.B.; Yang, Z.M.; Zhu, J.J.; Deng, X.; Liu, C.C.; Wang, X.F.; Xu, Y.; Sun, Z.M.; Chen, J.H. Identification, evaluation, and application of the genomic-SSR loci in ramie. Acta Soc. Bot. Pol. 2016, 85. [Google Scholar] [CrossRef]
- Kongjaimun, A.; Takahashi, Y.; Yoshioka, Y.; Tomooka, N.; Mongkol, R.; Somta, P. Molecular Analysis of Genetic Diversity and Structure of the Lablab (Lablab purpureus (L.) Sweet) Gene Pool Reveals Two Independent Routes of Domestication. Plants 2023, 12, 57. [Google Scholar] [CrossRef]
- Chen, X.; Min, D.; Yasir, T.A.; Hu, Y.G. Genetic Diversity, Population Structure and Linkage Disequilibrium in Elite Chinese Winter Wheat Investigated with SSR Markers. PLoS ONE 2012, 7, e44510. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, F.; Li, S.; Tian, D.; Dong, L.; Chen, Y.; Su, Y. Genetic diversity of the wild Asian lotus (Nelumbo nucifera) from Northern China. Hortic. Plant J. 2021, 7, 488–500. [Google Scholar] [CrossRef]
- Guo, Q.; Cao, S.; Dong, L.; Li, X.; Zhang, J.; Zhang, Y.; Zhang, Z.; Sun, Y.; Long, C.; Fan, Y.; et al. Genetic diversity and population structure of Robinia pseudoacacia from six improved variety bases in China as revealed by simple sequence repeat markers. J. For. Res. 2022, 33, 611–621. [Google Scholar] [CrossRef]
- Laosatit, K.; Amkul, K.; Chankaew, S.; Somta, P. Molecular genetic diversity of winged bean gene pool in Thailand assessed by SSR markers. Hortic. Plant J. 2022, 8, 81–88. [Google Scholar] [CrossRef]
- Li, L.; Ou, W.; Wang, Y.; Peng, J.; Wang, D.; Xu, S. Comparison of genetic diversity between ancient and common populations of Docynia delavayi (Franch.) Schneid. Gene 2022, 829, 146498. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Tang, D.; Gong, B. Genetic diversity and association analysis of Chinese chestnut (Castanea mollissima Blume) cultivars based on SSR markers. Braz. J. Bot. 2017, 40, 235–246. [Google Scholar] [CrossRef]
- Nishio, S.; Iketani, H.; Fujii, H.; Yamamoto, T.; Terakami, S.; Takada, N.; Saito, T. Use of population structure and parentage analyses to elucidate the spread of native cultivars of Japanese chestnut. Tree Genet. Genomes 2014, 10, 1171–1180. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Maria Ramos-Cabrer, A.; Barreneche, T.; Mattioni, C.; Villani, F.; Belen Diaz-Hernandez, M.; Miguel Martin, L.; Martin, A. Database of European chestnut cultivars and definition of a core collection using simple sequence repeats. Tree Genet. Genomes 2017, 13, 114. [Google Scholar] [CrossRef]
- Chen, L.; Ma, Q.; Chen, Y.; Wang, B.; Pei, D. Identification of major walnut cultivars grown in China based on nut phenotypes and SSR markers. Sci. Hortic. 2014, 168, 240–248. [Google Scholar] [CrossRef]
- Kumar, P.P.; Janakiram, T.; Bhat, K.V. Microsatellite based DNA fingerprinting and assessment of genetic diversity in bougainvillea cultivars. Gene 2020, 753, 144794. [Google Scholar] [CrossRef]
- Li, C.; Wu, J.; Li, Q.; Yang, Y.; Zhang, K. Development of Simple Sequence Repeat Markers from Functional Genes and Establishment of Molecular Identity for Tree Peony. J. Plant Biochem. Biotechnol. 2022, 31, 22–36. [Google Scholar] [CrossRef]
- Li, L.; Xu, X.; Wu, P.; Zhang, G.; Zhang, X. Establishment of Molecular Identity Cards for Cucumis melo Cultivars Using SSR Markers. Hortscience 2018, 53, 138–143. [Google Scholar] [CrossRef]
- Luan, M.B.; Chen, B.F.; Zou, Z.Z.; Zhu, J.J.; Wang, X.F.; Xu, Y.; Sun, Z.M.; Chen, J.H. Molecular identity of ramie germplasms using simple sequence repeat markers. Genet. Mol. Res. 2015, 14, 2302–2311. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.B. Development and Integration of an SSR-Based Molecular Identity Database into Sugarcane Breeding Program. Agronomy 2016, 6, 28. [Google Scholar] [CrossRef]
- Liu, G.; Cao, J.; Lan, Y.; Wang, J. Construction of SSR fingerprint on 33 ancient chestnut trees. Acta Agric. Univ. Jiangxiensis 2017, 39, 134–139. [Google Scholar]
- Savoia, M.A.; Del Faro, L.; Venerito, P.; Gaeta, L.; Palasciano, M.; Montemurro, C.; Sabetta, W. The Relevance of Discovering and Recovering the Biodiversity of Apulian Almond Germplasm by Means of Molecular and Phenotypic Markers. Plants 2022, 11, 574. [Google Scholar] [CrossRef]
- Stavridou, E.; Lagiotis, G.; Kalaitzidou, P.; Grigoriadis, I.; Bosmali, I.; Tsaliki, E.; Tsiotsiou, S.; Kalivas, A.; Ganopoulos, I.; Madesis, P. Characterization of the genetic diversity present in a diverse sesame landrace collection based on phenotypic traits and est-ssr markers coupled with an hrm analysis. Plants 2021, 10, 656. [Google Scholar] [CrossRef] [PubMed]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Mashilo, J.; Zhang, X.; Zhang, Y.; Dossa, K.; Shayanowako, A.I.T. Genetic Variability and Population Structure of Ethiopian Sesame (Sesamum indicum L.) Germplasm Assessed through Phenotypic Traits and Simple Sequence Repeats Markers. Plants 2021, 10, 1129. [Google Scholar] [CrossRef] [PubMed]
- Villano, C.; Corrado, G.; Basile, B.; Di Serio, E.; Mataffo, A.; Ferrara, E.; Aversano, R. Morphological and Genetic Clonal Diversity within the ‘Greco Bianco’Grapevine (Vitis vinifera L.) Variety. Plants 2023, 12, 515. [Google Scholar] [CrossRef]
- Benbouza, H.; Baudoin, J.-P.; Mergeai, G. Improvement of the genomic DNA extraction method with CTAB for cotton leaves. Biotechnol. Agron. Soc. Environ. 2006, 10, 73–76. [Google Scholar]
- Wang, J.; Tian, S.; Sun, X.; Cheng, X.; Duan, N.; Tao, J.; Shen, G. Construction of Pseudomolecules for the Chinese Chestnut (Castanea mollissima) Genome. G3 Genes Genomes Genet. 2020, 10, 3565–3574. [Google Scholar] [CrossRef]
- Fan, W.; Gai, H.; Sun, X.; Yang, A.; Zhang, Z.; Ren, M. DataFormater, a software for SSR data formatting to develop population genetics analysis. Mol. Plant Breed 2016, 14, 265–270. [Google Scholar]
- Quardokus, E. PopGene. Science 2000, 288, 458. [Google Scholar] [CrossRef]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 129, 157. [Google Scholar]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; p. 512. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; VonHoldt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Mathiang, E.A.; Sa, K.J.; Park, H.; Kim, Y.J.; Lee, J.K. Genetic Diversity and Population Structure of Normal Maize Germplasm Collected in South Sudan Revealed by SSR Markers. Plants 2022, 11, 7878. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Diao, X.; Yu, K.; Dai, X.; Qu, P.; Crabbe, M.J.C.; Zhang, T.; Qiao, Q. Evaluation of genetic diversity and population structure of Fragaria nilgerrensis using EST-SSR markers. Gene 2021, 796, 145791. [Google Scholar] [CrossRef] [PubMed]
- Samarina, L.S.; Kulyan, R.V.; Koninskaya, N.G.; Gorshkov, V.M.; Ryndin, A.V.; Hanke, M.V.; Flachowsky, H.; Reim, S. Genetic diversity and phylogenetic relationships among citrus germplasm in the Western Caucasus assessed with SSR and organelle DNA markers. Sci. Hortic. 2021, 288, 110355. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]

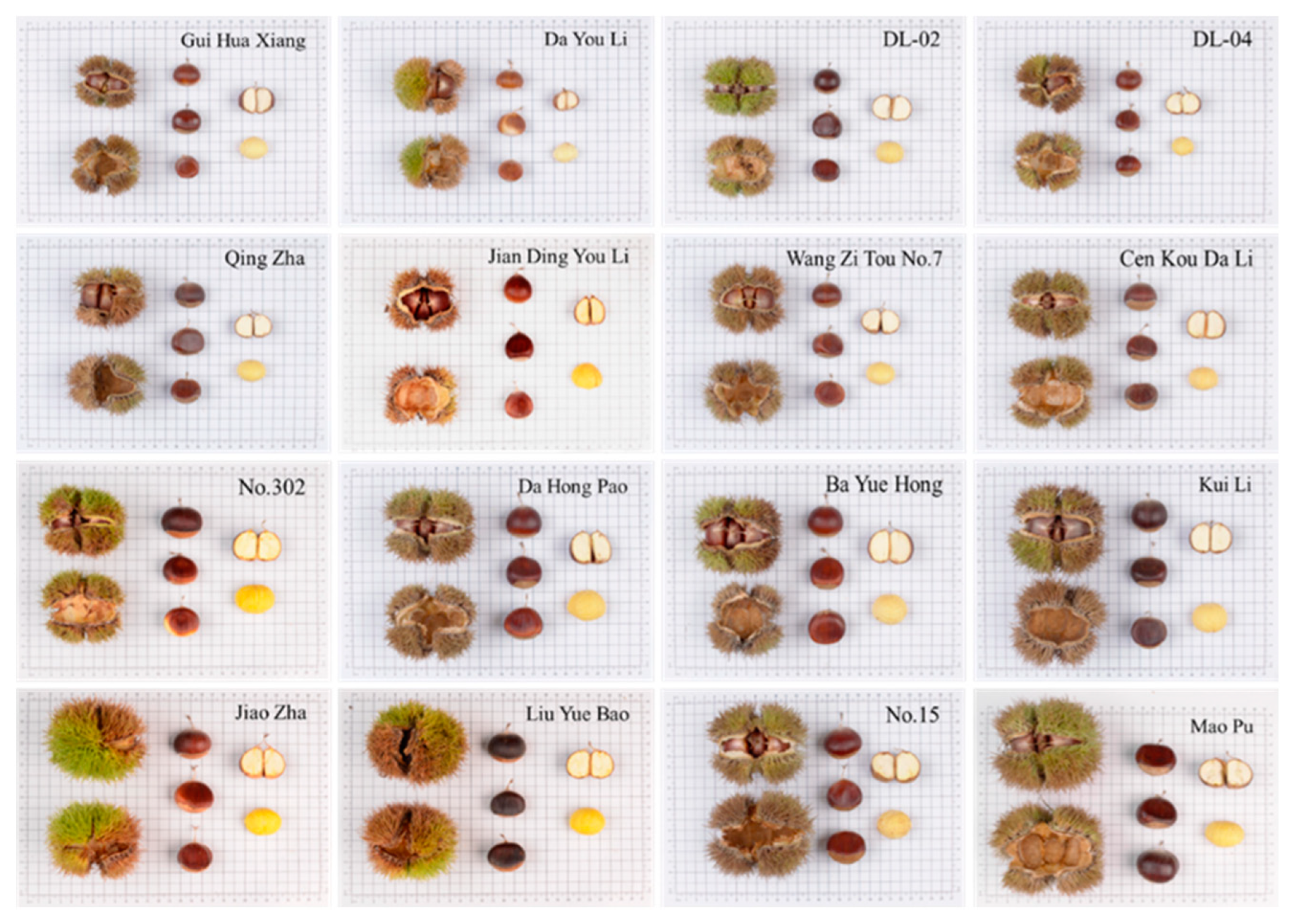

| Phenotypic Traits (Unit) | Mean | Max | Min | SD | CV (%) |
|---|---|---|---|---|---|
| Single cone weight (g) | 58.05 | 151.32 | 4.00 | 22.81 | 39.30% |
| Cone diameter (mm) | 71.84 | 140.02 | 31.67 | 17.76 | 24.72% |
| Cone length (mm) | 60.64 | 155.01 | 29.33 | 17.14 | 28.27% |
| Cone thickness (mm) | 56.60 | 115.38 | 26.67 | 15.06 | 26.61% |
| Spine length (mm) | 12.24 | 24.31 | 6.07 | 2.93 | 23.94% |
| Cone shell thickness (mm) | 2.40 | 6.37 | 0.79 | 0.89 | 37.08% |
| Single nut weight (g) | 10.45 | 37.67 | 2.00 | 5.19 | 49.67% |
| Nut diameter (mm) | 29.89 | 56.73 | 11.00 | 6.79 | 22.72% |
| Nut length (mm) | 24.06 | 46.42 | 11.33 | 5.26 | 21.86% |
| Nut thickness (mm) | 21.55 | 47.59 | 11.89 | 5.64 | 26.17% |
| Fruit shape index | 0.81 | 1.03 | 0.55 | 0.09 | 11.11% |
| Stigma length (mm) | 11.03 | 28.86 | 2.70 | 3.82 | 34.63% |
| Single kernel weight (g) | 8.33 | 36.17 | 1.70 | 5.03 | 60.38% |
| Kernel diameter (mm) | 27.82 | 52.07 | 9.87 | 6.61 | 23.76% |
| Kernel length (mm) | 21.93 | 42.49 | 10.10 | 4.86 | 22.16% |
| Kernel thickness (mm) | 18.22 | 151.32 | 8.02 | 5.19 | 28.49% |
| Primer Name | Na | NG | Ne | I | Ho | He | H | PIC | MAF |
|---|---|---|---|---|---|---|---|---|---|
| P4 | 5 | 8 | 2.7602 | 1.2316 | 0.1610 | 0.6404 | 0.6377 | 0.5909 | 0.5339 |
| P82 | 13 | 26 | 5.4042 | 2.0016 | 0.5254 | 0.8184 | 0.8150 | 0.7928 | 0.2966 |
| P106 | 12 | 23 | 4.2003 | 1.7045 | 0.8898 | 0.7652 | 0.7619 | 0.7265 | 0.3517 |
| P108 | 9 | 14 | 3.6012 | 1.5635 | 0.2373 | 0.7254 | 0.7223 | 0.6894 | 0.4534 |
| P127 | 7 | 13 | 3.6792 | 1.4494 | 0.5763 | 0.7313 | 0.7282 | 0.6790 | 0.3305 |
| P138 | 12 | 21 | 4.0063 | 1.6584 | 0.6017 | 0.7536 | 0.7504 | 0.7124 | 0.3602 |
| Mean | 9.6667 | 17.5 | 3.9419 | 1.6015 | 0.4986 | 0.7390 | 0.7359 | 0.6985 | 0.3877 |
| Primer Combination | Materials Can Be Distinguished | Subtotal | Total |
|---|---|---|---|
| P4 | 115, 117, 118 | 3 | 3 |
| P4 + P82 | 2, 3, 7, 11, 14, 22, 26, 30, 32, 33, 38, 39, 40, 45, 55, 61, 64, 66, 73, 76, 77, 78, 83, 84, 85, 95, 97, 100, 103, 113, 114, 116 | 32 | 35 |
| P4 + P82 + P106 | 4, 5, 6, 8, 12, 15, 17, 18, 19, 20, 24, 29, 31, 41, 42, 43, 48, 51, 58, 67, 70, 71, 81, 82, 86, 87, 89, 92, 93, 98, 99, 106, 107, 109, 110 | 35 | 70 |
| P4 + P82 + P106 + P108 | 9, 10, 13, 21, 23, 25, 36, 46, 47, 49, 50, 52, 56, 57, 65, 69, 74, 75, 79, 88, 90, 94, 96, 111, 112 | 25 | 95 |
| P4 + P82 + P106 + P108 + P127 | 1, 27,28, 35, 37, 44, 53, 54, 59, 60, 63, 80, 91, 101, 102, 104, 105 | 17 | 112 |
| P4 + P82 + P106 + P108 + P127 + P138 | 16, 34, 62, 68, 72, 108 | 6 | 118 |
| Code | Name of Variety | Molecular Identity Code | Code | Name of Variety | Molecular Identity Code |
|---|---|---|---|---|---|
| 1 | Qing Zha | 2CHCCK | 60 | Shen Ci Da Ban Li | 2E784H |
| 2 | Shu He No.1 | 6O9A3A | 61 | Xiao Jing Tie Li | 65J92D |
| 3 | Shu He No.7 | 6Q983A | 62 | Zhong Chi Li | 6C942K |
| 4 | Shu He No.10 | 37574D | 63 | Yue You No.9 | 27793G |
| 5 | Da Di Qing | 2P5A8G | 64 | Te Zao | 21783F |
| 6 | Da Hong Pao | 2P788G | 65 | Wu Mao Tie Li | 2CHD4D |
| 7 | Da Gong Shu No.4 | 1C844D | 66 | Duan Zhi Li | 495C4B |
| 8 | You Zao No.1 | 474ACH | 67 | Ban Li Zi | 2HM888 |
| 9 | Zao Li Zi | 4C5B6D | 68 | Hong Ming Jian | 23788G |
| 10 | Jiu Jia Zhong | 4C5CCK | 69 | Hu Bei You Li | 3B513B |
| 11 | Jiao Zha | 3G783K | 70 | Qing Mao Ruan Zha | 23584A |
| 12 | Jian Ding You Li | 3Q58CC | 71 | CKD | 2HD83F |
| 13 | Dong Wang Ming Li | 2772DC | 72 | DL-01 | 23788C |
| 14 | Hong Li | 1G4C8G | 73 | DL-02 | 136C5E |
| 15 | Xiao Luan Shi | 6CJCB2 | 74 | DL-03 | 3O7D7C |
| 16 | Huang Qian Zhong Wan | 6C942B | 75 | DL-04 | 3O7C8C |
| 17 | Lian Hua Li | 225727 | 76 | MJH | 83AA4C |
| 18 | Yue You No.8 | 3AH87H | 77 | W4 | 1EF82I |
| 19 | Mi Feng Qiu | 47ID7G | 78 | W5 | 6PJ844 |
| 20 | Wang Zi Tou No.7 | 475A7H | 79 | XHC | 425CCD |
| 21 | Gao Yuan No.1 | 42564D | 80 | XBC | 2O79CH |
| 22 | Shi Men Zao Shuo | 5EE963 | 81 | YBH | 22742H |
| 23 | Er Shui Zao | 2G783F | 82 | YML | 2QMA2K |
| 24 | Xin Zhuang No.2 | 2GKD4D | 83 | Y46 | 287C4B |
| 25 | Wei Hai Zao Shu | 27784H | 84 | Y47 | 2JDC4B |
| 26 | Chu Shu Hong | 2A7ACH | 85 | ZMZ | 4O4D8D |
| 27 | Yan Hong | 27798K | 86 | ZA | 3QH83H |
| 28 | Xiao Xue | 2O794K | 87 | No.6 | 3A534D |
| 29 | Duan Zha | 62JB7H | 88 | No.9 | 6C983B |
| 30 | Tai Shan Hong Li | 57588D | 89 | No.15 | 3EB75D |
| 31 | Gui Hua Xiang | 2EM86F | 90 | No.17 | 1O523C |
| 32 | Yan Shan Zao Feng | 6E9C7H | 91 | No.18 | 2O5C2G |
| 33 | Yang Guang No.2 | 6M982L | 92 | No.101 | 7CF83A |
| 34 | Bo Ke Chi Li | 2C782K | 93 | 102B | 428D4K |
| 35 | Huang Li Pu | 2HH87H | 94 | No.105 | 1O6C4G |
| 36 | Mao Pu | 3B588D | 95 | No.108 | 24D27H |
| 37 | Xin Yi You Li | 2CHC7H | 96 | No.203 | 6C533C |
| 38 | Chui Zhi Li | 3P5948 | 97 | No.207 | 26789G |
| 39 | Nian Di Ban | 2N7A3H | 98 | No.213 | 2Q727G |
| 40 | Yue Xi No.2 | 8LAA3L | 99 | No.302 | 2E588K |
| 41 | Wu Ke Li | 37FA3H | 100 | No.1059 | 297C4B |
| 42 | Jie Jie Hong | 3E588D | 101 | No.1061 | 2C784G |
| 43 | Cen Kou Da Li | 2GD4CH | 102 | No.1504 | 2CHC4D |
| 44 | Liu Yue Bao | 2E78AH | 103 | 8017 | 2FMC5K |
| 45 | Ba Yue Hong | 2D783F | 104 | Liu He Hong Li | 2O5C3C |
| 46 | Hua Gai | 1O588C | 105 | - | 67J83K |
| 47 | Huang Qian Wu Hua | 1O648I | 106 | - | 2EKA8I |
| 48 | Su Cheng Da Li | 27H83F | 107 | - | 62587D |
| 49 | Da You Li | 2C7C8G | 108 | - | 2C782H |
| 50 | Gui Xuan 72-1 | 2O584B | 109 | - | 67924H |
| 51 | Long An No.1 | 37N47K | 110 | Gan Yu No.1 | 22H54C |
| 52 | He Bei Zun Yu | 2G745K | 111 | - | 6C582K |
| 53 | Kui Li | 2E78CJ | 112 | - | 2O742H |
| 54 | Jiu Yue Han | 67J82D | 113 | Yin Ji | 1I2879 |
| 55 | Mei Gui Hong | 3C783K | 114 | - | 1J3879 |
| 56 | Wang Jie Gen | 277B3G | 115 | - | 6I1871 |
| 57 | Chen Guo You Li | 2C76CK | 116 | - | 1KG429 |
| 58 | Shuang He Da Hong Pao | 4C4A2K | 117 | - | 1JCEC5 |
| 59 | Er Xin Zao | 2HH89H | 118 | - | 1IL516 |
| Primer | Primer Sequence (5′−3′) | Chromosome No. | Allele Size | Fluorescent Dyes |
|---|---|---|---|---|
| P4 | F-GATTGTGCAACAACACCTGC R-CAACCCTGCCAAGAGATTGT | 1 | 173–181 | TET |
| P82 | F-CTCTGGGTTTACCTTGGGCT R-CGGGCTGAGTTTGGTTAAAA | 5 | 155–189 | TAMRA |
| P106 | F-GAGCAAGCTGCTACCCTGAC R-CGGTCTCAGATTTCAGGCTC | 7 | 163–181 | Fam |
| P108 | F-TCGCATGTTGTCCTTTACGA R-TCTCCGAGTTCTCCCTCTGA | 8 | 176–189 | Hex |
| P127 | F-TTCCAATGGACCAATACCGT R-CCCACATGGCCTCATTCTAT | 9 | 184–250 | ROX |
| P138 | F-CGAGGGAATTATGAGGGTTTT R-CAAAATGCTCAAGGGGGTAA | 11 | 174–201 | Fam |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, X.; Zhang, S.; Wang, W.; Chen, Y.; Zhao, Y.; Shi, F.; Zhu, C. Genetic Relationships of 118 Castanea Specific Germplasms and Construction of Their Molecular ID Based on Morphological Characteristics and SSR Markers. Plants 2023, 12, 1438. https://doi.org/10.3390/plants12071438
Bai X, Zhang S, Wang W, Chen Y, Zhao Y, Shi F, Zhu C. Genetic Relationships of 118 Castanea Specific Germplasms and Construction of Their Molecular ID Based on Morphological Characteristics and SSR Markers. Plants. 2023; 12(7):1438. https://doi.org/10.3390/plants12071438
Chicago/Turabian StyleBai, Xiaoqian, Shijie Zhang, Wu Wang, Yu Chen, Yuqiang Zhao, Fenghou Shi, and Cancan Zhu. 2023. "Genetic Relationships of 118 Castanea Specific Germplasms and Construction of Their Molecular ID Based on Morphological Characteristics and SSR Markers" Plants 12, no. 7: 1438. https://doi.org/10.3390/plants12071438
APA StyleBai, X., Zhang, S., Wang, W., Chen, Y., Zhao, Y., Shi, F., & Zhu, C. (2023). Genetic Relationships of 118 Castanea Specific Germplasms and Construction of Their Molecular ID Based on Morphological Characteristics and SSR Markers. Plants, 12(7), 1438. https://doi.org/10.3390/plants12071438




