Self-DNA Early Exposure in Cultivated and Weedy Setaria Triggers ROS Degradation Signaling Pathways and Root Growth Inhibition
Abstract
1. Introduction
2. Results
2.1. Root Elongation in Response to DNA Exposure
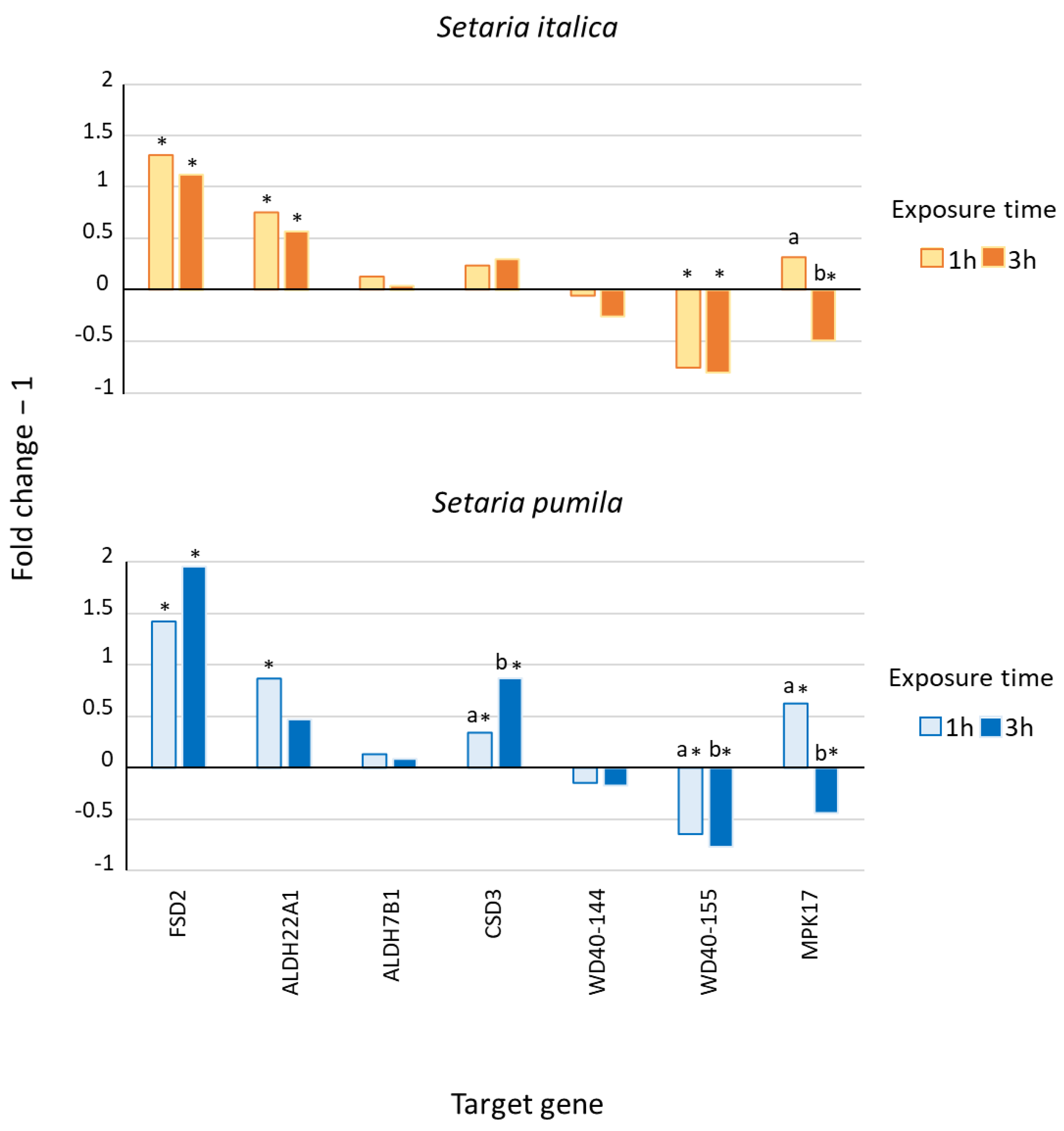
2.2. Expression of Abiotic Stress Responsive Genes in Response to Self-DNA
3. Discussion
3.1. Species-Specificity of Self-DNA Inhibitory Effect
3.2. Expression of Abiotic Stress Responsive Genes after Self-DNA Exposure
3.2.1. Drought and Dehydration Stress
3.2.2. Osmotic and Oxidative Stress
3.2.3. Thermic Stress
4. Materials and Methods
4.1. Root Elongation Experiments
4.1.1. Leaf Biomass Production for DNA Extraction
4.1.2. DNA Extraction and RNase Treatment
4.1.3. DNA Treatment Solution Preparation
4.1.4. Experimental Setup
4.2. qPCR Experiment for Abiotic Stress Responsive Genes
4.2.1. Gene Selection and Primer Design
4.2.2. Self-DNA Exposure
4.2.3. RNA Extraction, Purification and cDNA Synthesis
4.2.4. Real Time qPCR
4.2.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mazzoleni, S.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Mingo, A.; Senatore, M.; Giannino, F.; Carteni, F.; Rietkerk, M.; et al. Inhibitory and toxic effects of extracellular self-DNA in litter: A mechanism for negative plant-soil feedbacks? New Phytol. 2015, 205, 1195–1210. [Google Scholar] [CrossRef]
- Bonanomi, G.; Zotti, M.; Idbella, M.; Termolino, P.; De Micco, V.; Mazzoleni, S. Field evidence for litter and self-DNA inhibitory effects on Alnus glutinosa roots. New Phytol. 2022, 236, 399–412. [Google Scholar] [CrossRef]
- Rietkerk, M. Spatial pattern formation, community assembly and resilience: Comment on “Belowground feedbacks as drivers of spatial self-organization and community assembly” by Inderjit, Ragan M. Callaway, Ehud Meron. Phys. Life Rev. 2022, 40, 51–53. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Carteni, F.; Bonanomi, G.; Senatore, M.; Termolino, P.; Giannino, F.; Incerti, G.; Rietkerk, M.; Lanzotti, V.; Chiusano, M.L. Inhibitory effects of extracellular self-DNA: A general biological process? New Phytol. 2015, 206, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Palomba, E.; Chiaiese, P.; Termolino, P.; Paparo, R.; Filippone, E.; Mazzoleni, S.; Chiusano, M.L. Effects of Extracellular Self- and Nonself-DNA on the Freshwater Microalga Chlamydomonas reinhardtii and on the Marine Microalga Nannochloropsis gaditana. Plants 2022, 11, 1436. [Google Scholar] [CrossRef] [PubMed]
- Carteni, F.; Bonanomi, G.; Giannino, F.; Incerti, G.; Vincenot, C.E.; Chiusano, M.L.; Mazzoleni, S. Self-DNA inhibitory effects: Underlying mechanisms and ecological implications. Plant Signal Behav. 2016, 11, e1158381. [Google Scholar] [CrossRef] [PubMed]
- Duran-Flores, D.; Heil, M. Growth inhibition by self-DNA: A phenomenon and its multiple explanations. New Phytol. 2015, 207, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Barbero, F.; Guglielmotto, M.; Islam, M.; Maffei, M.E. Extracellular Fragmented Self-DNA Is Involved in Plant Responses to Biotic Stress. Front. Plant Sci. 2021, 12, 686121. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Cartenì, F.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Migliozzi, A.; Senatore, M.; Giannino, F.; Rietkerk, M.; et al. New perspectives on the use of nucleic acids in pharmacological applications: Inhibitory action of extracellular self-DNA in biological systems. Phytochem. Rev. 2014, 13, 937–946. [Google Scholar] [CrossRef]
- Ferrusquía-Jiménez, N.I.; Chandrakasan, G.; Torres-Pacheco, I.; Rico-Garcia, E.; Feregrino-Perez, A.A.; Guevara-González, R.G. Extracellular DNA: A Relevant Plant Damage-Associated Molecular Pattern (DAMP) for Crop Protection Against Pests—A Review. J. Plant Growth Regul. 2021, 40, 451–463. [Google Scholar] [CrossRef]
- Morrone, O.; Aliscioni, S.S.; Veldkamp, J.F.; Pensiero, J.F.; Zuloaga, F.O.; Kellogg, E.A. Revision of the Old World Species of Setaria (Poaceae: Panicoideae: Paniceae). In Systematic Botany Monographs; American Society of Plant Taxonomists: Lansin, MI, USA, 2014; Volume 96. [Google Scholar]
- Doust, A.; Diao, X. Genetics and Genomics of Setaria, 1st ed.; Springer International Publishing AG: New York, NY, USA, 2017; p. 377. [Google Scholar]
- Tozer, K.N.; Cameron, C.A.; Matthews, L. Grazing defoliation and nutritive value of Setaria pumila and Digitaria sanguinalis in Lolium perenne-based swards. Crop Pasture Sci. 2015, 66, 184–191. [Google Scholar] [CrossRef]
- Orlandi, S.; Nucera, E.; Mosimann, E.; D’Adda, G.; Garzoli, D.; Bertossa, M.; Lonati, M.; Lombardi, G. Drivers of Setaria pumila (Poir.) Roem. et Schult growth and impact on forage quality in lowland Switzerland meadows. Grass Forage Sci. 2015, 72, 154–162. [Google Scholar] [CrossRef]
- Satchivi, N.M.; deBoer, G.J.; Bell, J.L. Understanding the Differential Response of Setaria viridis L. (green foxtail) and Setaria pumila Poir (yellow foxtail) to Pyroxsulam. J. Agric. Food Chem. 2017, 65, 7328–7336. [Google Scholar] [CrossRef] [PubMed]
- Mazzoleni, S. Composition Comprising Nucleic Acids of Parasitic, Pathogenic or Weed Biological Systems for Inhibiting and/or Controlling the Growth of Said Systems. International Patent WO 2014020624 A9, 6 February 2014. [Google Scholar]
- Bennetzen, J.L.; Schmutz, J.; Wang, H.; Percifield, R.; Hawkins, J.; Pontaroli, A.C.; Estep, M.; Feng, L.; Vaughn, J.N.; Grimwood, J.; et al. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012, 30, 555–561. [Google Scholar] [CrossRef]
- Duran-Flores, D.; Heil, M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 2018, 72, 78–88. [Google Scholar] [CrossRef]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef]
- Heil, M.; Vega-Muñoz, I. Nucleic Acid Sensing in Mammals and Plants: Facts and Caveats. Int. Rev. Cell Mol. Biol. 2019, 345, 225–285. [Google Scholar]
- Barbero, F.; Guglielmotto, M.; Capuzzo, A.; Maffei, M.E. Extracellular Self-DNA (esDNA), but Not Heterologous Plant or Insect DNA (etDNA), Induces Plasma Membrane Depolarization and Calcium Signaling in Lima Bean (Phaseolus lunatus) and Maize (Zea mays). Int. J. Mol. Sci. 2016, 17, 1659. [Google Scholar] [CrossRef]
- Lanzotti, V.; Grauso, L.; Mangoni, A.; Termolino, P.; Palomba, E.; Anzano, A.; Incerti, G.; Mazzoleni, S. Metabolomics and molecular networking analyses in Arabidopsis thaliana show that extracellular self-DNA affects nucleoside/nucleotide cycles with accumulation of cAMP, cGMP and N6-methyl-AMP. Phytochemistry 2022, 204, 113453. [Google Scholar] [CrossRef]
- Chiusano, M.L.; Incerti, G.; Colantuono, C.; Termolino, P.; Palomba, E.; Monticolo, F.; Benvenuto, G.; Foscari, A.; Esposito, A.; Marti, L.; et al. Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure. Plants 2021, 10, 1744. [Google Scholar] [CrossRef]
- Zerebecki, R.A.; Sorte, C.J.B. Temperature Tolerance and Stress Proteins as Mechanisms of Invasive Species Success. PLoS ONE 2011, 6, e14806. [Google Scholar] [CrossRef]
- Godoy, O.; de Lemos-Filho, J.P.; Valladares, F. Invasive species can handle higher leaf temperature under water stress than Mediterranean natives. Environ. Exp. Bot. 2011, 71, 207–214. [Google Scholar] [CrossRef]
- Podda, L.; Santo, A.; Leone, C.; Mayoral, O.; Bacchetta, G. Seed germination, salt stress tolerance and seedling growth of Opuntia ficus-indica (Cactaceae), invasive species in the Mediterranean Basin. Flora 2017, 229, 50–57. [Google Scholar] [CrossRef]
- Clements, D.R.; Jones, V.L. Rapid Evolution of Invasive Weeds Under Climate Change: Present Evidence and Future Research Needs. Front. Agron. 2021, 3, 664034. [Google Scholar] [CrossRef]
- Leal, R.P.; Silveira, M.J.; Petsch, D.K.; Mormul, R.P.; Thomaz, S.M. The success of an invasive Poaceae explained by drought resilience but not by higher competitive ability. Environ. Exp. Bot. 2022, 194, 104717. [Google Scholar] [CrossRef]
- Serrano-Jamaica, L.M.; Villordo-Pineda, E.; Gonzalez-Chavira, M.M.; Guevara-Gonzalez, R.G.; Medina-Ramos, G. Effect of Fragmented DNA From Plant Pathogens on the Protection Against Wilt and Root Rot of Capsicum annuum L. Plants. Front. Plant Sci. 2020, 11, 581891. [Google Scholar] [CrossRef]
- Germoglio, M.; Adamo, A.; Incerti, G.; Carteni, F.; Gigliotti, S.; Storlazzi, A.; Mazzoleni, S. Self-DNA Exposure Induces Developmental Defects and Germline DNA Damage Response in Caenorhabditis elegans. Biology 2022, 11, 262. [Google Scholar] [CrossRef] [PubMed]
- Ferrusquía-Jiménez, N.I.; Serrano-Jamaica, L.M.; Martínez-Camacho, J.E.; Sáenz de la O, D.; Villagomez-Aranda, A.L.; González-Chavira, M.M.; Guerrero-Aguilar, B.Z.; Torres-Pacheco, I.; Feregrino-Pérez, A.A.; Medina-Ramos, G.; et al. Extracellular self-DNA plays a role as a damage-associated molecular pattern (DAMP) delaying zoospore germination rate and inducing stress-related responses in Phytophthora capsici. Plant Pathol. 2022, 71, 1066–1075. [Google Scholar] [CrossRef]
- Zhang, Q.; Bartels, D. Molecular responses to dehydration and desiccation in desiccation-tolerant angiosperm plants. J. Exp. Bot. 2018, 69, 3211–3222. [Google Scholar] [CrossRef] [PubMed]
- Martignago, D.; Rico-Medina, A.; Blasco-Escamez, D.; Fontanet-Manzaneque, J.B.; Cano-Delgado, A.I. Drought Resistance by Engineering Plant Tissue-Specific Responses. Front. Plant Sci. 2019, 10, 1676. [Google Scholar] [CrossRef]
- Shao, H.B.; Chu, L.Y.; Jaleel, C.A.; Zhao, C.X. Water-deficit stress-induced anatomical changes in higher plants. C. R. Biol. 2008, 331, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef]
- Abobatta, W.F. Drought adaptive mechanisms of plants—A review. Adv. Agric. Environ. Sci. 2019, 2, 42–45. [Google Scholar] [CrossRef]
- Anjum, S.A.; Xie, X.Y.; Wang, L.C.; Saleem, M.F.; Man, C.; Lei, W. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agric. Res. 2011, 6, 2026–2032. [Google Scholar] [CrossRef]
- Wang, T.; Song, H.; Zhang, B.; Lu, Q.; Liu, Z.; Zhang, S.; Guo, R.; Wang, C.; Zhao, Z.; Liu, J.; et al. Genome-wide identification, characterization, and expression analysis of superoxide dismutase (SOD) genes in foxtail millet (Setaria italica L.). 3 Biotech 2018, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Muthamilarasan, M.; Khan, Y.; Parida, S.K.; Prasad, M. Genome-wide investigation and expression analyses of WD40 protein family in the model plant foxtail millet (Setaria italica L.). PLoS ONE 2014, 9, e86852. [Google Scholar] [CrossRef] [PubMed]
- Lata, C.; Sahu, P.P.; Prasad, M. Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem. Biophys. Res. Commun. 2010, 393, 720–727. [Google Scholar] [CrossRef]
- Scandalios, J.G. Oxygen stress and superoxide dismutases. Plant Physiol. 1993, 101, 7–12. [Google Scholar] [CrossRef]
- Tjia, T.O.S.; Meitha, K.; Septiani, P.; Awaludin, R.; Sumardi, D. Extracellular self-DNA induces local inhibition of growth, regulates production of reactive oxygen species, and gene expression in rice roots. Biol. Plant. 2023, 67, 9–18. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The formation and function of plant cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef]
- Lee, J.H.; Yoon, H.J.; Terzaghi, W.; Martinez, C.; Dai, M.; Li, J.; Byun, M.O.; Deng, X.W. DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 2010, 22, 1716–1732. [Google Scholar] [CrossRef] [PubMed]
- Dar, N.A.; Amin, I.; Wani, W.; Wani, S.A.; Shikari, A.B.; Wani, S.H.; Masoodi, K.Z. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 2017, 11, 106–111. [Google Scholar] [CrossRef]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef]
- Son, Y.; Cheong, Y.K.; Kim, N.H.; Chung, H.T.; Kang, D.G.; Pae, H.O. Mitogen-Activated Protein Kinases and Reactive Oxygen Species: How Can ROS Activate MAPK Pathways? J. Signal Transduct. 2011, 2011, 792639. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, T.; Lan, J.; Ma, J.; Xu, H.; Yang, Z.; Guo, Y.; Chen, Y.; Zhang, J.; Dou, S.; et al. Rice MPK17 Plays a Negative Role in the Xa21-Mediated Resistance Against Xanthomonas oryzae pv. oryzae. Rice 2022, 15, 41. [Google Scholar] [CrossRef]
- Lu, Y.Q.; Ye, Z.H.; Wu, W.R. Analysis of the phylogenetic relationships among several species of Gramineae using ACGM markers. Yi Chuan Xue Bao 2006, 33, 1127–1131. [Google Scholar] [CrossRef]
- Fraire-Velázquez, S.; Balderas-Hernández, V.E. Abiotic Stress in Plants and Metabolic Responses. In Abiotic Stress—Plant Responses and Applications in Agriculture; Vahdati, K., Leslie, C., Eds.; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef]
- Qamer, Z.; Muhammad Tanees, C.; Xiongming, D.; Lori, H.; Muhammad Tehseen, A. Review of oxidative stress and antioxidative defense mechanisms in Gossypium hirsutum L. in response to extreme abiotic conditions. J. Cotton Res. 2021, 4, 9. [Google Scholar] [CrossRef]
- Zhu, C.; Ming, C.; Zhao-shi, X.; Lian-cheng, L.; Xue-ping, C.; You-zhi, M. Characteristics and Expression Patterns of the Aldehyde Dehydrogenase (ALDH) Gene Superfamily of Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e0101136. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Filippou, P.; Bouchagier, P.; Skotti, E.; Fotopoulos, V. Proline and reactive oxygen/nitrogen species metabolism is involved in the tolerant response of the invasive plant species Ailanthus altissima to drought and salinity. Environ. Exp. Bot. 2014, 97, 1–10. [Google Scholar] [CrossRef]
- Moustafa, K.; Vos, D.L.; Leprince, A.; Savouré, A.; Laurière, C. Analysis of the Arabidopsis Mitogen-activated protein kinase families: Organ specificity and transcriptional regulation upon water stresses. Sch. Res. Exch. 2008, 2008, 143656. [Google Scholar] [CrossRef]
- Frick, E.M.; Strader, L.C. Kinase MPK17 and the Peroxisome Division Factor PMD1 Influence Salt-induced Peroxisome Proliferation. Plant Physiol. 2018, 176, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, M.; Kong, X.; Xing, X.; Liu, Y.; Zhou, Y.; Liu, Y.; Sun, L.; Li, D. ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 2012, 235, 661–676. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Ashraf, M.A.; Cheng, Y.; Wang, B.; Qin, Y. Plant Low-Temperature Stress: Signaling and Response. Agronomy 2022, 12, 702. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Rudi, K.; Kroken, M.; Dahlberg, O.J.; Deggerdal, A.; Jakobsen, K.S.; Larsen, F. Rapid, Universal Method to Isolate PCR-Ready DNA Using Magnetic Beads. Biotechniques 1997, 22, 506–511. [Google Scholar] [CrossRef]
- Beckman Coulter. Agencourt AMPure XP Information for Use Guide PCR Purification; Beckman Coulter, Inc.: Brea, CA, USA, 2016; p. 36. [Google Scholar]
- Kumar, K.; Muthamilarasan, M.; Prasad, M. Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tissue Organ Cult. 2013, 115, 13–22. [Google Scholar] [CrossRef]
- Mareri, L.; Parrotta, L.; Cai, G. Environmental Stress and Plants. Int. J. Mol. Sci. 2022, 23, 5416. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
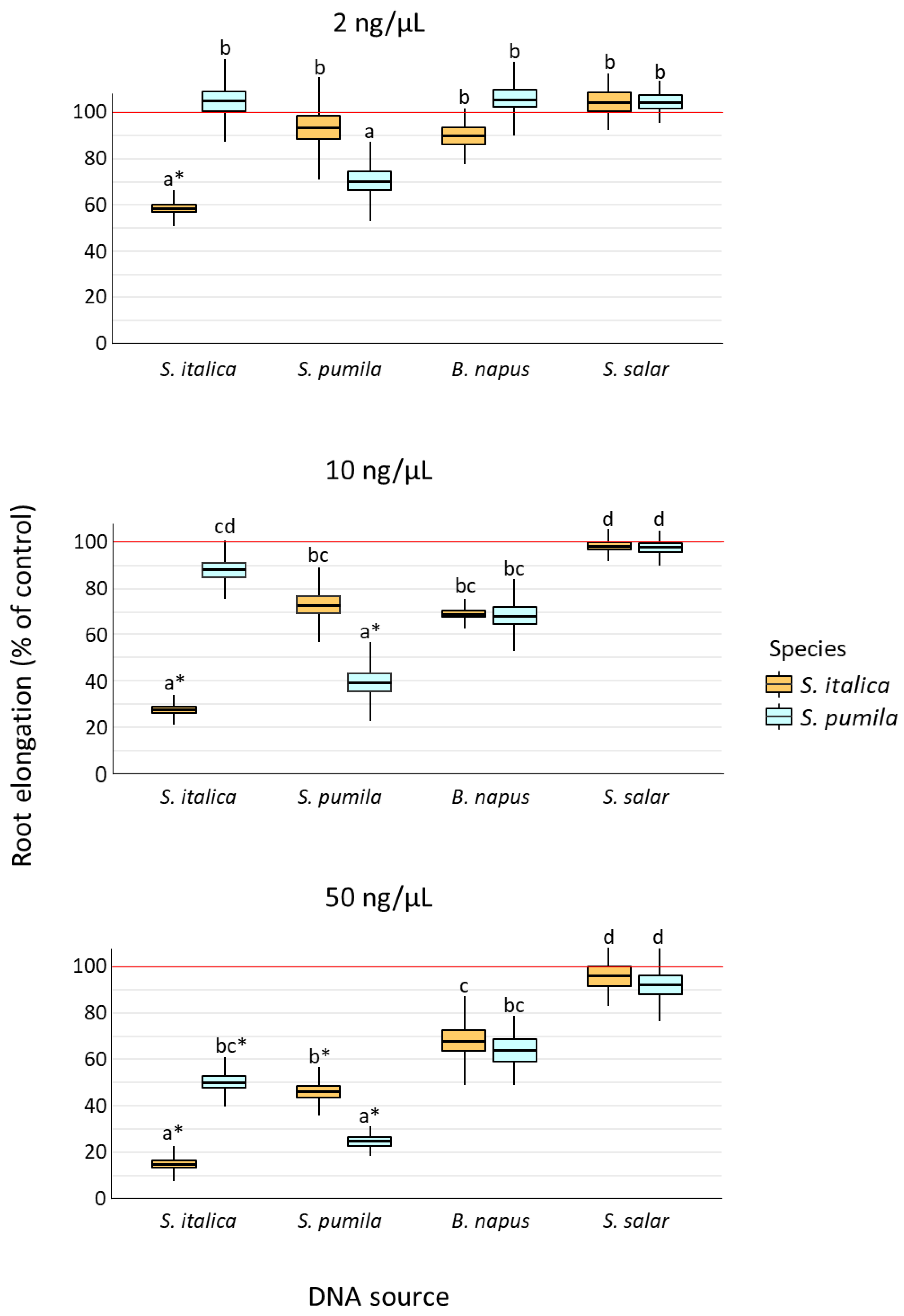

| Effect | Df | SS | MS | F | p |
|---|---|---|---|---|---|
| Target species (S) | 1 | 178.8 | 178.8 | 4.66 | 0.0353 |
| DNA source (D) | 3 | 24,862.5 | 8287.5 | 216.13 | <0.0001 |
| Concentration (C) | 2 | 15,268.6 | 7634.3 | 199.09 | <0.0001 |
| S × D | 3 | 13,069.7 | 4356.6 | 113.61 | <0.0001 |
| S × C | 2 | 279.5 | 139.7 | 3.64 | 0.0328 |
| D × C | 6 | 3641.8 | 607.0 | 15.83 | <0.0001 |
| Error | 54 | 2070.7 | 38.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ronchi, A.; Foscari, A.; Zaina, G.; De Paoli, E.; Incerti, G. Self-DNA Early Exposure in Cultivated and Weedy Setaria Triggers ROS Degradation Signaling Pathways and Root Growth Inhibition. Plants 2023, 12, 1288. https://doi.org/10.3390/plants12061288
Ronchi A, Foscari A, Zaina G, De Paoli E, Incerti G. Self-DNA Early Exposure in Cultivated and Weedy Setaria Triggers ROS Degradation Signaling Pathways and Root Growth Inhibition. Plants. 2023; 12(6):1288. https://doi.org/10.3390/plants12061288
Chicago/Turabian StyleRonchi, Alessia, Alessandro Foscari, Giusi Zaina, Emanuele De Paoli, and Guido Incerti. 2023. "Self-DNA Early Exposure in Cultivated and Weedy Setaria Triggers ROS Degradation Signaling Pathways and Root Growth Inhibition" Plants 12, no. 6: 1288. https://doi.org/10.3390/plants12061288
APA StyleRonchi, A., Foscari, A., Zaina, G., De Paoli, E., & Incerti, G. (2023). Self-DNA Early Exposure in Cultivated and Weedy Setaria Triggers ROS Degradation Signaling Pathways and Root Growth Inhibition. Plants, 12(6), 1288. https://doi.org/10.3390/plants12061288







