Nanopore Technology Applied to Targeted Detection of Tomato Brown Rugose Fruit Virus Allows Sequencing of Related Viruses and the Diagnosis of Mixed Infections
Abstract
1. Introduction
2. Results
2.1. ToBRFV RT-PCR and RT-qPCR Detection
2.2. Nanopore Sequencing of Virus-Infected Tomato Samples
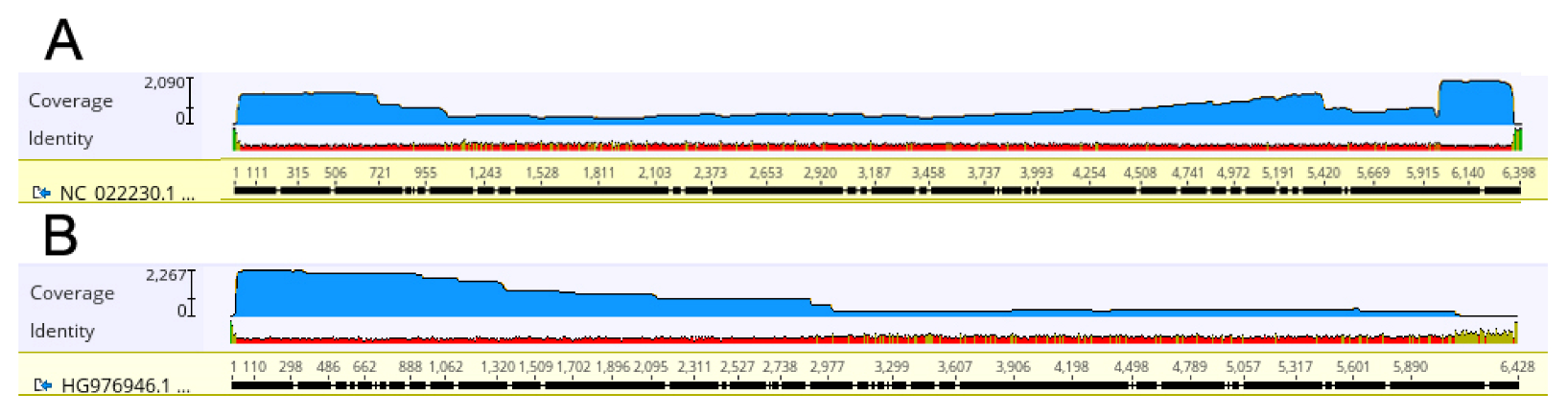
2.3. ToBRFV Genome Reconstruction by Mapping of ONT Reads
2.4. ToBRFV Full-Length Genome Characterization and Phylogenetic Analysis
2.5. Nanopore Sequencing with ToBRFV Specific Primers Detects the ToMMV Genome
2.6. PepMV Sequences Identification and Full-Length Genome Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Description
4.2. Nucleic Acid Extraction and ToBRFV Molecular Detection
4.3. Primer Design Strategy and Oxford Nanopore Technologies (ONT) Sequencing
4.4. Sanger Sequencing of the Viral Genomes Terminal Regions
4.5. ONT Datasets Analysis
4.6. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capobianco-Uriarte, M.d.l.M.; Aparicio, J.; De Pablo-Valenciano, J.; Casado-Belmonte, M.d.P. The European tomato market. An approach by export competitiveness maps. PLoS ONE 2021, 16, e0250867. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M. Major tomato viruses in the Mediterranean basin. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2012; Volume 84, pp. 31–66. [Google Scholar]
- Hanssen, I.M.; Mumford, R.; Blystad, D.-R.; Cortez, I.; Hasiów-Jaroszewska, B.; Hristova, D.; Pagán, I.; Pereira, A.-M.; Peters, J.; Pospieszny, H. Seed transmission of Pepino mosaic virus in tomato. Eur. J. Plant Pathol. 2010, 126, 145–152. [Google Scholar] [CrossRef]
- Li, R.; Baysal-Gurel, F.; Abdo, Z.; Miller, S.A.; Ling, K.-S. Evaluation of disinfectants to prevent mechanical transmission of viruses and a viroid in greenhouse tomato production. Virol. J. 2015, 12, 5. [Google Scholar] [CrossRef]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A. A new Israeli Tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef]
- Zhang, S.; Griffiths, J.S.; Marchand, G.; Bernards, M.A.; Wang, A. Tomato brown rugose fruit virus: An emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 2022, 23, 1262–1277. [Google Scholar] [CrossRef]
- Nagai, A.; Duarte, L.M.L.; Chaves, A.L.R.; Alexandre, M.A.V.; Ramos-González, P.L.; Chabi-Jesus, C.; Harakava, R.; Dos Santos, D.Y.A.C. First complete genome sequence of an isolate of tomato mottle mosaic virus infecting plants of Solanum lycopersicum in South America. Genome Announc. 2018, 6, e00427-18. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Hu, J.; Xiao, L.; Tan, G.; Lan, P.; Liu, Y.; Li, F. The complete genome sequence, occurrence and host range of Tomato mottle mosaic virus Chinese isolate. Virol. J. 2017, 14, 15. [Google Scholar] [CrossRef]
- Li, R.; Gao, S.; Fei, Z.; Ling, K.-S. Complete Genome Sequence of a New Tobamovirus Naturally Infecting Tomatoes in Mexico. Genome Announc. 2013, 1, e00794-13. [Google Scholar] [CrossRef]
- Van der Vlugt, R.; Stijger, C.; Verhoeven, J.T.J.; Lesemann, D.E. First report of Pepino mosaic virus on tomato. Plant Dis. 2000, 84, 103. [Google Scholar] [CrossRef]
- EPPO—European and Mediterranean Plant Protection Organization. PM 7/146 (1) Tomato brown rugose fruit virus. EPPO Bull. 2021, 51, 178–197. [Google Scholar] [CrossRef]
- Alkowni, R.; Alabdallah, O.; Fadda, Z. Molecular identification of tomato brown rugose fruit virus in tomato in Palestine. J. Plant Pathol. 2019, 101, 719–723. [Google Scholar] [CrossRef]
- Klap, C.; Luria, N.; Smith, E.; Hadad, L.; Bakelman, E.; Sela, N.; Belausov, E.; Lachman, O.; Leibman, D.; Dombrovsky, A. Tomato brown rugose fruit virus contributes to enhanced pepino mosaic virus titers in tomato plants. Viruses 2020, 12, 879. [Google Scholar] [CrossRef]
- European Commission. Commission Implementing Regulation (EU) 2020/1191 of August 11, 2020 Establishing Measures to Prevent the Introduction into and the Spread within the Union of Tomato Brown Rugose Fruit Virus (ToBRFV) and Repealing Implementing Decision (EU) 2019/1615. (Document 32020R1191). Available online: https://eur-lex.europa.eu/eli/reg_impl/2020/1191/oj2020 (accessed on 10 January 2023).
- USDA Federal Order. Tomato Brown Rugose Fruit Virus. Available online: https://www.aphis.usda.gov/aphis/ourfocus/planthealth/import-information/federal-import-orders/tobrfv/tomato-brown-rugose-fruit-virus2019,2020 (accessed on 10 January 2023).
- China. Phytosanitary Requirements of Import Solanum lycopersicum L. and Capsicum spp. Seeds on Tomato Brown Rugose Fruit Virus. WTO Code: G/SPS/N/CHN/1223. Available online: https://epingalert.org/en/Search?viewData%3DG%2fSPS%2fN%2fCHN%2f12232021 (accessed on 10 January 2023).
- Salem, N.M.; Sulaiman, A.; Samarah, N.; Turina, M.; Vallino, M. Localization and mechanical transmission of tomato brown rugose fruit virus in tomato seeds. Plant Dis. 2022, 106, 275–281. [Google Scholar] [CrossRef]
- Kabas, A.; Fidan, H.; Kucukaydin, H.; Atan, H.N. Screening of wild tomato species and interspecific hybrids for resistance/tolerance to Tomato brown rugose fruit virus (ToBRFV). Chil. J. Agric. Res. 2022, 82, 189–196. [Google Scholar] [CrossRef]
- Jewehan, A.; Salem, N.; Tóth, Z.; Salamon, P.; Szabó, Z. Screening of Solanum (sections Lycopersicon and Juglandifolia) germplasm for reactions to the tomato brown rugose fruit virus (ToBRFV). J. Plant Dis. Prot. 2022, 129, 117–123. [Google Scholar] [CrossRef]
- Zinger, A.; Lapidot, M.; Harel, A.; Doron-Faigenboim, A.; Gelbart, D.; Levin, I. Identification and mapping of tomato genome loci controlling tolerance and resistance to tomato brown rugose fruit virus. Plants 2021, 10, 179. [Google Scholar] [CrossRef]
- Kutnjak, D.; Tamisier, L.; Adams, I.; Boonham, N.; Candresse, T.; Chiumenti, M.; De Jonghe, K.; Kreuze, J.F.; Lefebvre, M.; Silva, G.; et al. A Primer on the Analysis of High-Throughput Sequencing Data for Detection of Plant Viruses. Microorganisms 2021, 9, 841. [Google Scholar] [CrossRef]
- Sun, K.; Liu, Y.; Zhou, X.; Yin, C.; Zhang, P.; Yang, Q.; Mao, L.; Shentu, X.; Yu, X. Nanopore sequencing technology and its application in plant virus diagnostics. Front. Microbiol. 2022, 13, 2811. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Nanopore technology and its applications in gene sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Chanda, B.; Rivera, Y.; Nunziata, S.O.; Galvez, M.E.; Gilliard, A.; Ling, K.-S. Complete genome sequence of a tomato brown rugose fruit virus isolated in the United States. Microbiol. Resour. Announc. 2020, 9, e00630-20. [Google Scholar] [CrossRef]
- Chalupowicz, L.; Dombrovsky, A.; Gaba, V.; Luria, N.; Reuven, M.; Beerman, A.; Lachman, O.; Dror, O.; Nissan, G.; Manulis-Sasson, S. Diagnosis of plant diseases using the Nanopore sequencing platform. Plant Pathol. 2019, 68, 229–238. [Google Scholar] [CrossRef]
- Amoia, S.S.; Minafra, A.; Nicoloso, V.; Loconsole, G.; Chiumenti, M. A New Jasmine Virus C Isolate Identified by Nanopore Sequencing Is Associated to Yellow Mosaic Symptoms of Jasminum officinale in Italy. Plants 2022, 11, 309. [Google Scholar] [CrossRef]
- Çelik, A.; Coşkan, S.; Morca, A.F.; Santosa, A.I.; Koolivand, D. Insight into Population Structure and Evolutionary Analysis of the Emerging Tomato Brown Rugose Fruit Virus. Plants 2022, 11, 3279. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Thomma, B.P.H.J. Pepino mosaic virus: A successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol. Plant Pathol. 2010, 11, 179–189. [Google Scholar] [CrossRef]
- Mackie, J.; Kinoti, W.M.; Chahal, S.I.; Lovelock, D.A.; Campbell, P.R.; Tran-Nguyen, L.T.T.; Rodoni, B.C.; Constable, F.E. Targeted Whole Genome Sequencing (TWG-Seq) of Cucumber Green Mottle Mosaic Virus Using Tiled Amplicon Multiplex PCR and Nanopore Sequencing. Plants 2022, 11, 2716. [Google Scholar] [CrossRef]
- Della Bartola, M.; Byrne, S.; Mullins, E. Characterization of potato virus Y isolates and assessment of nanopore sequencing to detect and genotype potato viruses. Viruses 2020, 12, 478. [Google Scholar] [CrossRef]
- Fellers, J.P.; Webb, C.; Fellers, M.C.; Shoup Rupp, J.; De Wolf, E. Wheat virus identification within infected tissue using nanopore sequencing technology. Plant Dis. 2019, 103, 2199–2203. [Google Scholar] [CrossRef]
- Grädel, C.; Terrazos Miani, M.A.; Baumann, C.; Barbani, M.T.; Neuenschwander, S.; Leib, S.L.; Suter-Riniker, F.; Ramette, A. Whole-Genome Sequencing of Human Enteroviruses from Clinical Samples by Nanopore Direct RNA Sequencing. Viruses 2020, 12, 841. [Google Scholar] [CrossRef]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J. Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. J. Clin. Microbiol. 2019, 58, e00963-19. [Google Scholar] [CrossRef]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Andino, R.; Domingo, E. Viral quasispecies. Virology 2015, 479, 46–51. [Google Scholar] [CrossRef]
- Rietveld, K.; Linschooten, K.; Pleij, C.W.A.; Bosch, L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984, 3, 2613–2619. [Google Scholar] [CrossRef]
- Alcaide, C.; Rabadán, M.P.; Moreno-Pérez, M.G.; Gómez, P. Chapter Five—Implications of mixed viral infections on plant disease ecology and evolution. In Advances in Virus Research; Kielian, M., Mettenleiter, T.C., Roossinck, M.J., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 106, pp. 145–169. [Google Scholar]
- Zhan, B.-H.; Ning, C.A.O.; Wang, K.-N.; Zhou, X.-P. Detection and characterization of an isolate of Tomato mottle mosaic virus infecting tomato in China. J. Integr. Agric. 2018, 17, 1207–1212. [Google Scholar] [CrossRef]
- Moreira, S.R.; Eiras, M.; Chaves, A.L.; Galleti, S.R.; Colariccio, A. Characterization of a new Tomato mosaic virus strain isolated from tomato in the State of São Paulo, Brazil. Fitopatol. Bras. 2003, 28, 602–607. [Google Scholar] [CrossRef]
- Leger, A.; Leonardi, T. PycoQC, interactive quality control for Oxford Nanopore Sequencing. J. Open Source Softw. 2019, 4, 1236. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T.-A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]




| Flye | Canu | |||
|---|---|---|---|---|
| Blastn Annotation | Contig Length (bp) | Blastn Annotation | Contig Length (bp) | |
| ToMMV (Bar03) | Unknown | 19,102 | ToMMV | 5941 |
| Unknown | 9070 | S. lycopersicum | 2765 | |
| ToBRFV (Bar04) | ToBRFV | 615 | ToBRFV | 5932 |
| ToBRFV | 699 | |||
| S. lycopersicum | 12,778 | |||
| PepMV | 8482 | |||
| Library | Ref. Sequence (Acc. No.) | Nr of Mapping Reads | % out of Total Quality Filtered Reads |
|---|---|---|---|
| ToBRFV (Bar04) | S. lycopersicum genome (assembly SL3.0) | 74,432 | 57.24 |
| S. lycopersicum chloroplast (NC_007898.3) | 145 | 0.11 | |
| S. lycopersicum mitochondrion (NC_035963.1) | 58 | 0.04 | |
| ToBRFV (MN882023) | 40,114 | 30.85 | |
| PepMV (HG976946) | 3281 | 2.52 | |
| TOTAL | 118,030 | 90.77 | |
| ToMMV (Bar03) | S. lycopersicum genome (assembly SL3.0) | 114,042 | 76.83 |
| S. lycopersicum chloroplast (NC_007898.3) | 2542 | 1.71 | |
| S. lycopersicum mitochondrion (NC_035963.1) | 68 | 0.04 | |
| ToMMV (NC_022230) | 7264 | 4.89 | |
| TOTAL | 123,916 | 83.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abou Kubaa, R.; Amoia, S.S.; Altamura, G.; Minafra, A.; Chiumenti, M.; Cillo, F. Nanopore Technology Applied to Targeted Detection of Tomato Brown Rugose Fruit Virus Allows Sequencing of Related Viruses and the Diagnosis of Mixed Infections. Plants 2023, 12, 999. https://doi.org/10.3390/plants12050999
Abou Kubaa R, Amoia SS, Altamura G, Minafra A, Chiumenti M, Cillo F. Nanopore Technology Applied to Targeted Detection of Tomato Brown Rugose Fruit Virus Allows Sequencing of Related Viruses and the Diagnosis of Mixed Infections. Plants. 2023; 12(5):999. https://doi.org/10.3390/plants12050999
Chicago/Turabian StyleAbou Kubaa, Raied, Serafina Serena Amoia, Giuseppe Altamura, Angelantonio Minafra, Michela Chiumenti, and Fabrizio Cillo. 2023. "Nanopore Technology Applied to Targeted Detection of Tomato Brown Rugose Fruit Virus Allows Sequencing of Related Viruses and the Diagnosis of Mixed Infections" Plants 12, no. 5: 999. https://doi.org/10.3390/plants12050999
APA StyleAbou Kubaa, R., Amoia, S. S., Altamura, G., Minafra, A., Chiumenti, M., & Cillo, F. (2023). Nanopore Technology Applied to Targeted Detection of Tomato Brown Rugose Fruit Virus Allows Sequencing of Related Viruses and the Diagnosis of Mixed Infections. Plants, 12(5), 999. https://doi.org/10.3390/plants12050999






