Salinity Alleviation and Reduction in Oxidative Stress by Endophytic and Rhizospheric Microbes in Two Rice Cultivars
Abstract
1. Introduction
2. Results
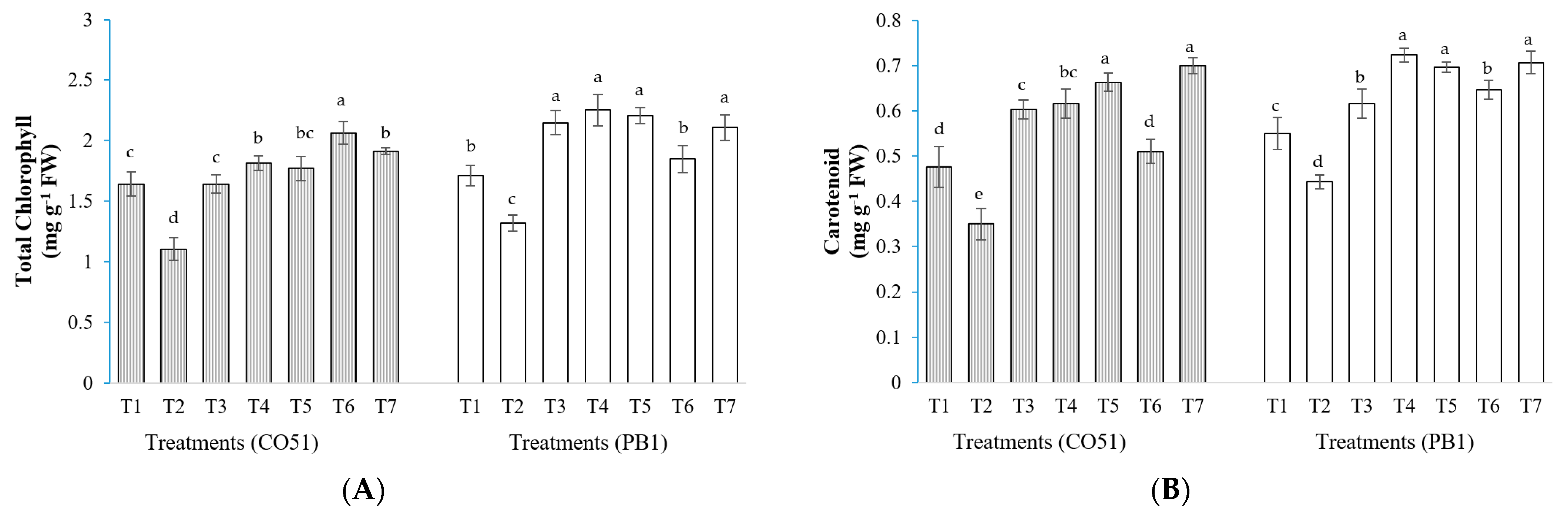
2.1. Chlorophyll and Carotenoids Content
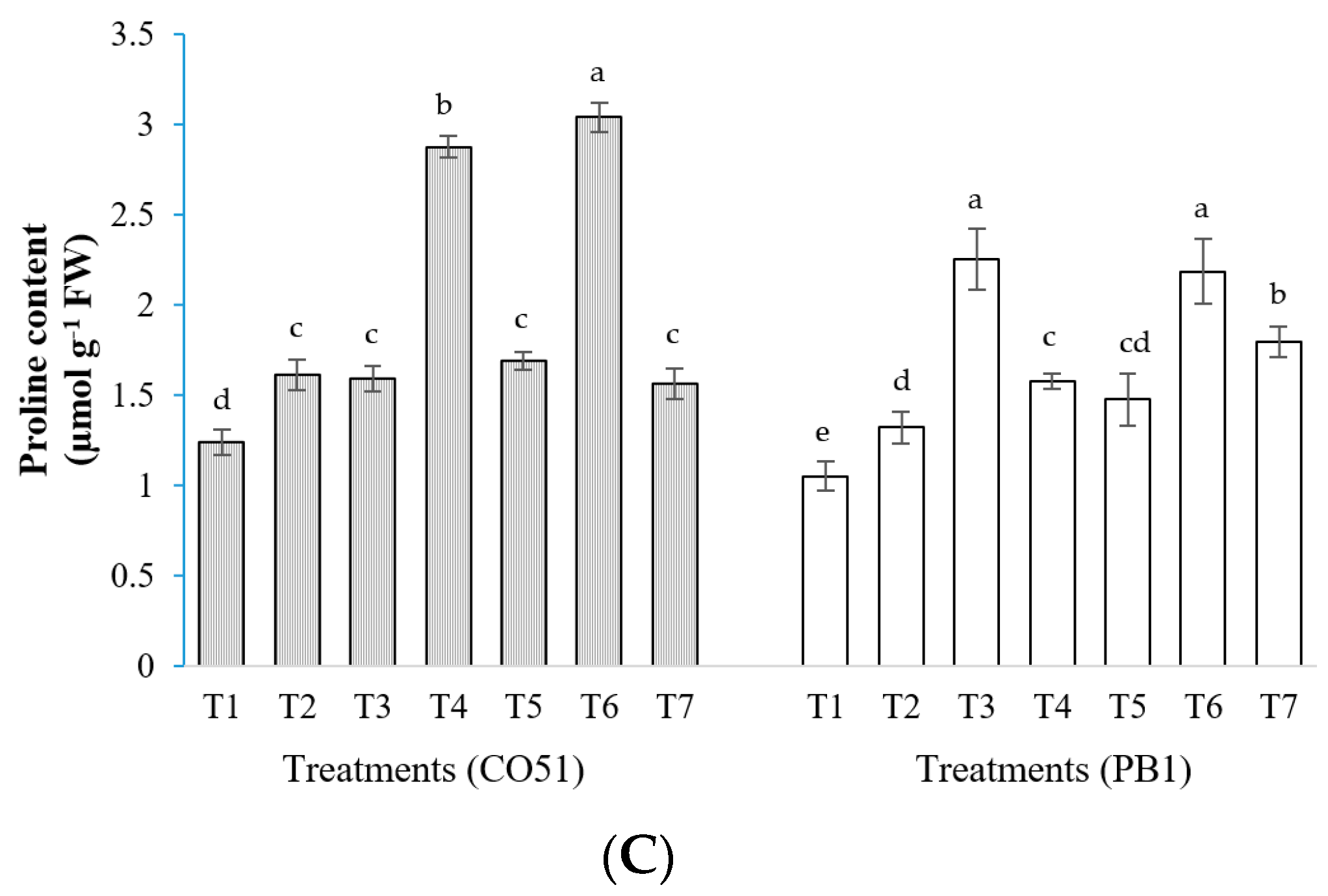
2.2. Proline Content
2.3. Antioxidant Enzymes
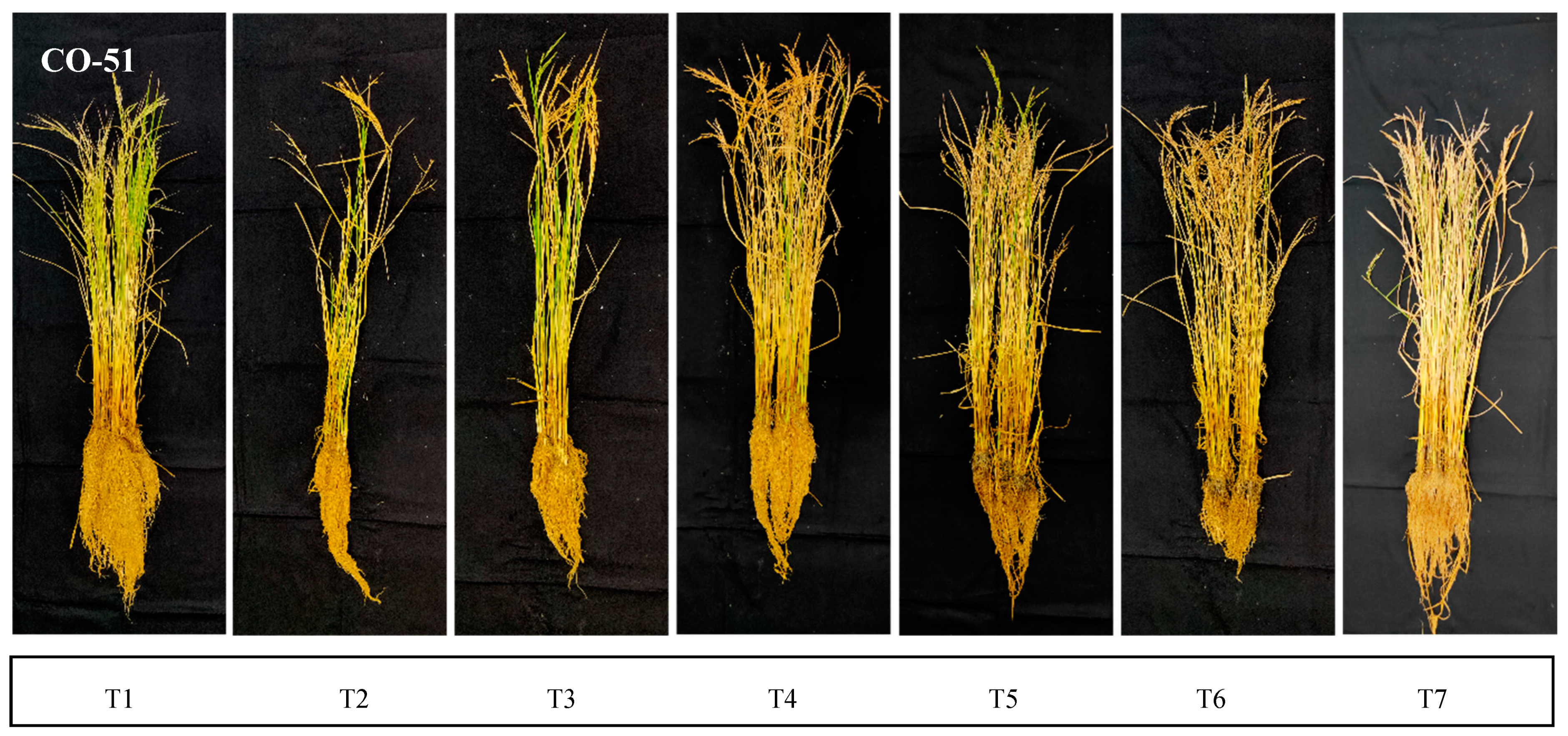
2.4. Shoot and Root Dry Weight
2.5. Shoot and Root Length
2.6. Number of Tillers
2.7. Root Parameters
2.8. Gene Expression Study
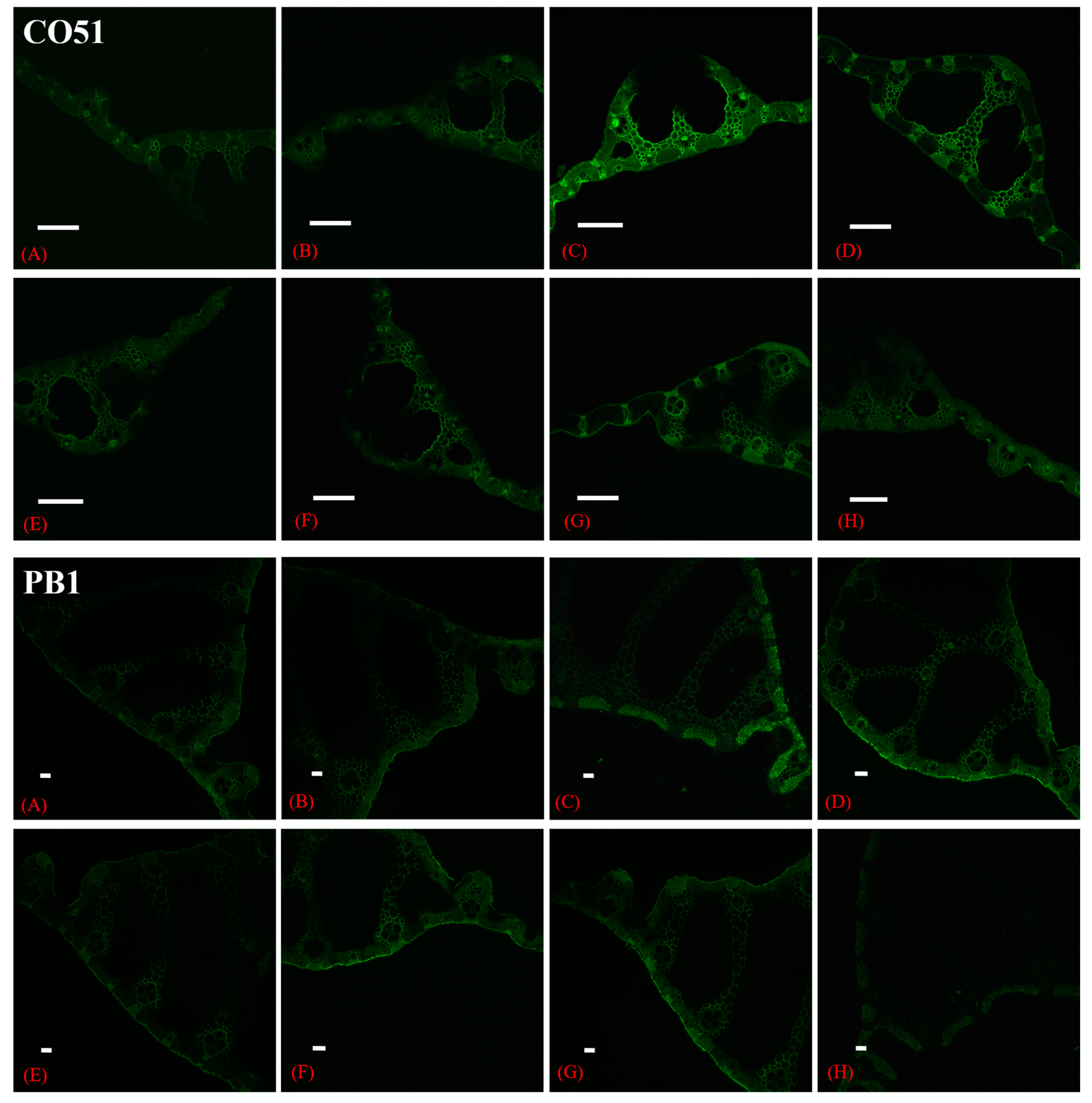
2.9. CSLM Study for Na+ Accumulation in Leaf Tissues
3. Discussion
4. Materials and Methods
4.1. Preparation of Pots, Inoculation and Transplanting
4.2. Chlorophyll and Carotenoids Content
4.3. Proline Content
4.4. Electrolyte Leakage
4.5. Antioxidant Enzymes
4.5.1. Peroxidase (PO)
4.5.2. Catalase (CAT)
4.5.3. Superoxide Dismutase (SOD)
4.5.4. Phenylalanine Ammonia Lyase (PAL)
4.5.5. Polyphenol Oxidase (PPO)
4.5.6. Ascorbate Peroxidase (APx)
4.6. Biomass Accumulation
4.7. Shoot and Root Length and Number of Tillers
4.8. Root Scanning
4.9. Gene Expression Study
4.10. CSLM Study for Na+ Accumulation in Leaf Tissues
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fujimori, S.; Hasegawa, T.; Krey, V.; Riahi, K.; Bertram, C.; Bodirsky, B.L.; Bosetti, V.; Callen, J.; Després, J.; Doelman, J.; et al. A multi-model assessment of food security implications of climate change mitigation. Nat. Sustain. 2019, 2, 386–396. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic stress responses and microbe-mediated mitigation in plants: The omics strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Sahu, P.K.; Kumari, N.; Gupta, A.; Manzar, N. Rhizospheric and endophytic microorganisms and their role in alleviation of salinity stress in plants. In Plant, Soil and Microbes in Tropical Ecosystems; Springer: Singapore, 2021; pp. 19–37. [Google Scholar]
- Sahu, P.K.; Singh, S.; Singh, U.B.; Chakdar, H.; Sharma, P.K.; Sarma, B.K.; Teli, B.; Bajpai, R.; Bhowmik, A.; Singh, H.V.; et al. Inter-genera colonization of Ocimum tenuiflorum endophytes in tomato and their complementary effects on na+/k+ balance, oxidative stress regulation, and root architecture under elevated soil salinity. Front. Microbiol. 2021, 12, 744733. [Google Scholar] [CrossRef]
- Vimal, S.R.; Patel, V.K.; Singh, J.S. Plant growth promoting Curtobacterium albidum strain SRV4: An agriculturally important microbe to alleviate salinity stress in paddy plants. Ecol. Indic. 2019, 105, 553–562. [Google Scholar] [CrossRef]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D.; et al. ROS generated from biotic stress: Effects on plants and alleviation by endophytic microbes. Front. Plant Sci. 2022, 13, 1042936. [Google Scholar] [CrossRef]
- Sahu, P.K.; Thomas, P.; Singh, S.; Gupta, A. Taxonomic and functional diversity of cultivable endophytes with respect to the fitness of cultivars against Ralstonia solanacearum. J. Plant Dis. Prot. 2020, 127, 667–676. [Google Scholar] [CrossRef]
- Robin, S.; Mohanasundaram, K.; Manonmani, S.; Rajeswari, S.; Jeyaprakash, P.; Pushpam, R.; Thiagarajan, K.; Rabindran, R.; Suresh, S.; Ravichandran, V.; et al. TNAU Rice CO 51 (IET 21605)—A high yielding short duration fine grain rice variety for Tamil Nadu. Electron. J. Plant Breed. 2019, 10, 324–333. [Google Scholar] [CrossRef]
- Siddiq, E.A.; Vemireddy, L.R.; Nagaraju, J. Basmati rices: Genetics, breeding and trade. Agric. Res. 2012, 1, 25–36. [Google Scholar] [CrossRef]
- Wegner, L.H.; Stefano, G.; Shabala, L.; Rossi, M.; Mancuso, S.; Shabala, S. Sequential depolarization of root cortical and stelar cells induced by an acute salt shock–implications for Na+ and K+ transport into xylem vessels. Plant Cell Environ. 2011, 34, 859–869. [Google Scholar] [CrossRef]
- Minta, A.; Tsien, R.Y. Fluorescent indicators for cytosolic sodium. J. Biol. Chem. 1989, 264, 19449–19457. [Google Scholar] [CrossRef]
- Schmidt, R.; Mieulet, D.; Hubberten, H.M.; Obata, T.; Hoefgen, R.; Fernie, A.R.; Fisahn, J.; San Segundo, B.; Guiderdoni, E.; Schippers, J.H.; et al. Salt-responsive ERF1 regulates reactive oxygen species–dependent signaling during the initial response to salt stress in rice. Plant Cell 2013, 25, 2115–2131. [Google Scholar] [CrossRef]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 2018, 9, 204. [Google Scholar] [CrossRef]
- Kumar, M.; Lee, S.C.; Kim, J.Y.; Kim, S.J.; Aye, S.S.; Kim, S.R. Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J. Plant Biol. 2014, 57, 383–393. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, T.; Zhang, W.; Li, X. SOS3 mediates lateral root development under low salt stress through regulation of auxin redistribution and maxima in Arabidopsis. New Phytologist. 2011, 189, 1122–1134. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Hoefsloot, H.C.; Mol, S.; Feron, R.; de Boer, G.J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef]
- Koevoets, I.T.; Venema, J.H.; Elzenga, J.T.; Testerink, C. Roots withstanding their environment: Exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 2016, 7, 1335. [Google Scholar] [CrossRef]
- Martinuz, A.; Zewdu, G.; Ludwig, N.; Grundler, F.; Sikora, R.A.; Schouten, A. The application of Arabidopsis thaliana in studying tripartite interactions among plants, beneficial fungal endophytes and biotrophic plant-parasitic nematodes. Planta 2015, 241, 1015–1025. [Google Scholar] [CrossRef]
- Irizarry, I.; White, J.F. Application of bacteria from non-cultivated plants to promote growth, alter root architecture and alleviate salt stress of cotton. J. Appl. Microbiol. 2017, 122, 1110–1120. [Google Scholar] [CrossRef]
- Muthukumar, T.; Sulaiman, M.R. Root Endophytic Nectria haematococca Influences Growth, Root Architecture and Phosphorus Content of Green Gram in Different Substrates. Proc. Natl. Acad. Sci. India Sect. B 2021, 91, 131–138. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.L.; Touraine, B.; Moënne Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Zamioudis, C.; Korteland, J.; Van Pelt, J.A.; van Hamersveld, M.; Dombrowski, N.; Bai, Y.; Pieterse, C.M. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB 72 expression in Arabidopsis roots during onset of induced systemic resistance and iron deficiency responses. Plant J. 2015, 84, 309–322. [Google Scholar] [CrossRef]
- Hossain, M.T.; Khan, A.; Harun-Or-Rashid, M.; Chung, Y.R. A volatile producing endophytic Bacillus siamensis YC7012 promotes root development independent on auxin or ethylene/jasmonic acid pathway. Plant Soil 2019, 439, 309–324. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Qurashi, A.W.; Sabri, A.N. Osmolyte accumulation in moderately halophilic bacteria improves salt tolerance of chickpea. Pak. J. Bot. 2013, 45, 1011–1016. [Google Scholar]
- de Carvalho, K.; de Campos, M.K.F.; Domingues, D.S.; Pereira, L.F.P.; Vieira, L.G.E. The accumulation of endogenous proline induces changes in gene expression of several antioxidant enzymes in leaves of transgenic Swingle citrumelo. Mol. Biol. Rep. 2013, 40, 3269–3279. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Das Bhowmik, S.; Long, H.; Cheng, Y.; Mundree, S.; Hoang, L.T.M. Rapid accumulation of proline enhances salinity tolerance in Australian wild rice Oryza australiensis domin. Plants 2021, 10, 2044. [Google Scholar] [CrossRef]
- Ghosh, D.; Sen, S.; Mohapatra, S. Modulation of proline metabolic gene expression in Arabidopsis thaliana under water-stressed conditions by a drought-mitigating Pseudomonas putida strain. Ann. Microbiol. 2017, 67, 655–668. [Google Scholar] [CrossRef]
- Nawaz, A.; Rehman, A.U.; Haydar, Z.; Rehman, H.U.; Ahmad, S.; Hussain, M. techniques of rice nursery establishment and transplanting. In Modern Techniques of Rice Crop Production; Springer: Singapore, 2022; pp. 29–42. [Google Scholar]
- Witham, F.H.; Blaydes, D.F.; Dewlin, R.M. Experiments in Plant Physiology; Von Nostrand Reinhold Company: New York, NY, USA, 1971; pp. 55–56. [Google Scholar]
- Sadasivam, S.; Manickam, A. Carotenes. Biochemical Methods; New Age International Publishers: New Delhi, India, 1997; pp. 187–188. [Google Scholar]
- Khare, N.; Goyary, D.; Singh, N.K.; Shah, P.; Rathore, M.; Anandhan, S.; Sharma, D.; Arif, M.; Ahmed, Z. Transgenic tomato cv. Pusa Uphar expressing a bacterial mannitol-1-phosphate dehydrogenase gene confers abiotic stress tolerance. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 103, 267–277. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; Nuckles, E.M.; Kuć, J. Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant Pathol. 1982, 20, 73–82. [Google Scholar] [CrossRef]
- Luck, H. Estimation of Catalase, Methods in Enzymatic Analysis; Academic Press: New York, NY, USA, 1974. [Google Scholar]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Havir, E.A. l-Phenylalanine ammonia-lyase from soybean cell suspension cultures. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1987; Volume 142, pp. 248–253. [Google Scholar]
- Gauillard, F.; Richardforget, F.; Nicolas, J. New spectrophotometric assay for polyphenol oxidase activity. Anal. Biochem. 1993, 215, 59–65. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Maksimović, J.J.D.; Živanović, B.D. Quantification of the antioxidant activity in salt-stressed tissues. In Plant Salt Tolerance; Humana Press: Totowa, NJ, USA, 2012; pp. 237–250. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, D.; Li, X.F.; Zhou, Z.J.; Feng, X.P.; Yang, W.J.; Jiang, D.A. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiol. Plant. 2010, 139, 55–67. [Google Scholar] [CrossRef]
- Liu, H.Y.; Sun, W.N.; Su, W.A.; Tang, Z.C. Co-regulation of water channels and potassium channels in rice. Physiol. Plant. 2006, 128, 58–69. [Google Scholar] [CrossRef]
- Kim, J.H.; Chung, B.Y.; Kim, J.S.; Wi, S.G.; Yang, D.H.; Lee, C.H.; Lee, M.C. Construction of gene-specific primers for various antioxidant isoenzyme genes and their expressions in rice (Oryza sativa L.) seedlings obtained from gamma-irradiated seeds. J. Photoscience 2004, 11, 115–120. [Google Scholar]





| Treatments | Shoot Length (cm) | Root Length (cm) | Shoot Dry Weight (g) | Root Dry Weight (g) | ||||
|---|---|---|---|---|---|---|---|---|
| CO51 | PB1 | CO51 | PB1 | CO51 | PB1 | CO51 | PB1 | |
| T1 | 78.67 bc ± 1.15 | 68.33 b ± 1.53 | 34.33 a ± 1.15 | 37.33 b ± 0.58 | 6.63 d ± 0.26 | 7.39 e ± 0.14 | 6.03 bc ± 0.25 | 7.12 d ± 0.29 |
| T2 | 75.67 c ± 1.53 | 73.50 a ± 3.04 | 22.67 c ± 2.08 | 31.33 e ± 0.58 | 5.11 e ± 0.25 | 6.00 f ± 0.2 | 2.99 d ± 0.52 | 4.41 e ± 0.24 |
| T3 | 77.67 bc ± 1.53 | 71.67 ab ± 0.58 | 23.33 c ± 1.15 | 43.00 a ± 1.73 | 7.71 b ± 0.34 | 8.73 d ± 0.15 | 5.51 c ± 0.19 | 7.01 d ± 0.21 |
| T4 | 80.67 b ± 1.53 | 69.33 b ± 1.15 | 32.13 a ± 1.21 | 34.33 cd ± 1.53 | 8.10 ab ± 0.13 | 11.01 a ± 0.17 | 6.60 a ± 0.14 | 7.99 ab ± 0.15 |
| T5 | 80.67 b ± 1.15 | 73.67 a ± 1.53 | 28.33 b ± 1.53 | 34.83 cd ± 0.76 | 8.19 a ± 0.22 | 10.13 b ± 0.36 | 6.27 ab ± 0.3 | 7.61 bc ± 0.25 |
| T6 | 84.00 a ± 2 | 74.00 a ± 2.65 | 22.33 c ± 1.53 | 36.33 bc ± 1.15 | 7.28 c ± 0.36 | 10.27 b ± 0.1 | 5.71 c ± 0.19 | 7.31 cd ± 0.21 |
| T7 | 77.33 c ± 1.15 | 71.67 ab ± 2.31 | 33.67 a ± 0.58 | 33.67 d ± 1.53 | 7.27 c ± 0.22 | 9.59 c ± 0.38 | 6.26 ab ± 0.15 | 8.11 a ± 0.2 |
| Treatments | Number of Tillers | Number of Effective Tillers | Number of Non-Effective Tillers | |||
|---|---|---|---|---|---|---|
| CO51 | PB1 | CO51 | PB1 | CO51 | PB1 | |
| T1 | 6.58 c ± 0.38 | 7.33 ab ± 0.29 | 6.17 c ± 0.29 | 5.42 ab ± 0.38 | 0.42 a ± 0.14 | 1.92 a ± 0.14 |
| T2 | 3.92 e ± 0.25 | 5.62 e ± 0.27 | 3.67 e ± 0.25 | 3.95 d ± 0.21 | 0.25 a ± 0 | 1.67 a ± 0.29 |
| T3 | 4.97 d ± 0.38 | 6.42 d ± 0.29 | 4.55 d ± 0.38 | 5.25 bc ± 0.5 | 0.42 a ± 0.14 | 1.17 b ± 0.29 |
| T4 | 7.33 b ± 0.38 | 6.75 bcd ± 0.43 | 7.00 b ± 0.43 | 5.67 ab ± 0.38 | 0.33 a ± 0.29 | 1.08 b ± 0.29 |
| T5 | 7.67 b ± 0.52 | 6.67 cd ± 0.14 | 7.42 b ± 0.72 | 5.83 ab ± 0.14 | 0.25 a ± 0.25 | 0.83 b ± 0.14 |
| T6 | 7.58 b ± 0.52 | 7.25 abc ± 0.25 | 7.33 b ± 0.38 | 6.17 a ± 0.38 | 0.25 a ± 0.25 | 1.08 b ± 0.29 |
| T7 | 8.92 a ± 0.52 | 7.75 a ± 0.43 | 8.50 a ± 0.66 | 4.33 d ± 0.76 | 0.42 a ± 0.29 | 1.17 b ± 0.29 |
| Treatments | Length of Total Root (cm) * | Projection Area (cm2) | Surface Area (cm2) | Average Diameter (mm) | Root Volume (cm3) | Fractal Dimension | Tips | Forks |
|---|---|---|---|---|---|---|---|---|
| CO51 | ||||||||
| T1 | 2861.81 a ± 125.42 | 428.92 a ± 9.74 | 1327.15 a ± 17.17 | 4.35 c ± 0.04 | 48.92 a ± 0.26 | 5.20 b ± 0.02 | 2883.33 c ± 93.54 | 12122.64 a ± 226.35 |
| T2 | 1889.62 c ± 57.26 | 215.13 f ± 11.77 | 692.05 f ± 7.16 | 2.30 e ± 0.02 | 20.49 f ± 0.64 | 3.32 f ± 0.02 | 2583.00 d ± 23.3 | 6874.53 d ± 141.68 |
| T3 | 2241.92 b ± 71.66 | 314.81 d ± 17.5 | 967.69 d ± 5.13 | 4.17 d ± 0.05 | 34.16 d ± 0.5 | 4.76 e ± 0.05 | 3281.00 b ± 72.64 | 7065.30 d ± 228.98 |
| T4 | 2400.66 b ± 45.82 | 370.39 c ± 7.26 | 1185.51 c ± 11.08 | 4.75 b ± 0.03 | 46.75 b ± 1.45 | 5.24 b ± 0.04 | 1924.67 g ± 79.74 | 9689.01 c ± 47.54 |
| T5 | 2042.90 c ± 172.17 | 280.42 e ± 15.47 | 855.92 e ± 46.37 | 5.31 a ± 0.03 | 48.38 ab ± 1.73 | 6.53 a ± 0.06 | 2424.00 e ± 56.51 | 5977.80 e ± 184.96 |
| T6 | 1911.03 c ± 24.91 | 278.87 e ± 8.57 | 850.08 e ± 16.61 | 4.33 c ± 0.1 | 30.81 e ± 2.26 | 4.92 d ± 0.07 | 2256.33 f ± 49.05 | 4859.81 f ± 67.45 |
| T7 | 2931.01 a ± 108.1 | 405.36 b ± 11.89 | 1249.99 b ± 42.18 | 4.08 d ± 0.06 | 43.13 c ± 1.08 | 5.06 c ± 0.08 | 3629.00 a ± 66.55 | 9982.93 b ± 230.85 |
| PB1 | ||||||||
| T1 | 1303.12 e ± 80.02 | 241.45 c ± 14.48 | 643.98 f ± 14.91 | 3.26 bc ± 0.34 | 28.85 c ± 1.11 | 2.74 c ± 0.02 | 1468.67 e ± 34.95 | 3991.03 d ± 129.79 |
| T2 | 751.06 f ± 61.2 | 112.28 e ± 2.17 | 352.65 g ± 5.56 | 2.46 d ± 0.02 | 16.98 d ± 0.3 | 1.96 d ± 0.04 | 1178.00 f ± 15.52 | 2702.45 e ± 81.97 |
| T3 | 1471.05 d ± 22.76 | 231.16 c ± 12.07 | 715.19 e ± 7.8 | 3.16 bc ± 0.05 | 31.26 bc ± 1.92 | 3.30 b ± 0.11 | 1492.00 e ± 25.71 | 5280.76 b ± 65.02 |
| T4 | 1863.88 b ± 53.22 | 292.11 b ± 2.93 | 1047.92 b ± 11.26 | 3.38 b ± 0.02 | 35.80 a ± 1.71 | 3.59 b ± 0.12 | 2553.67 b ± 69.74 | 5202.17 b ± 238.94 |
| T5 | 1747.94 c ± 37.06 | 206.25 d ± 4.01 | 926.43 c ± 6.8 | 3.17 bc ± 0.06 | 30.91 bc ± 1.45 | 3.37 b ± 0.02 | 1763.67 d ± 49.24 | 4861.84 c ± 120.06 |
| T6 | 1427.39 d ± 70.47 | 236.92 c ± 8.84 | 759.30 d ± 22.91 | 3.03 c ± 0.08 | 32.24 b ± 0.87 | 3.42 b ± 0.3 | 2468.33 c ± 28.75 | 4811.04 c ± 99.7 |
| T7 | 2052.01 a ± 48.85 | 312.63 a ± 7.44 | 1075.60 a ± 21.55 | 4.88 a ± 0.04 | 36.41 a ± 0.82 | 3.93 a ± 0.21 | 2623.67 a ± 26.1 | 5795.27 a ± 79.6 |
| Sr. No. | Treatment | Details of the Treatments (Separately for Both Cultivars) |
|---|---|---|
| 1. | T1 | Negative control |
| 2. | T2 | Positive control (200 mM NaCl) |
| 3. | T3 | 200 mM NaCl + Trichoderma viride |
| 4. | T4 | 200 mM NaCl + Bacillus haynesii 2P2 |
| 5. | T5 | 200 mM NaCl + Bacillus safensis BTL5 |
| 6. | T6 | 200 mM NaCl + Brevibacterium frigoritolerans W19 |
| 7. | T7 | 200 mM NaCl + Pseudomonas fluorescens 1001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gupta, A.; Singh, A.N.; Tiwari, R.K.; Sahu, P.K.; Yadav, J.; Srivastava, A.K.; Kumar, S. Salinity Alleviation and Reduction in Oxidative Stress by Endophytic and Rhizospheric Microbes in Two Rice Cultivars. Plants 2023, 12, 976. https://doi.org/10.3390/plants12050976
Gupta A, Singh AN, Tiwari RK, Sahu PK, Yadav J, Srivastava AK, Kumar S. Salinity Alleviation and Reduction in Oxidative Stress by Endophytic and Rhizospheric Microbes in Two Rice Cultivars. Plants. 2023; 12(5):976. https://doi.org/10.3390/plants12050976
Chicago/Turabian StyleGupta, Amrita, Arvind Nath Singh, Rajesh Kumar Tiwari, Pramod Kumar Sahu, Jagriti Yadav, Alok Kumar Srivastava, and Sanjay Kumar. 2023. "Salinity Alleviation and Reduction in Oxidative Stress by Endophytic and Rhizospheric Microbes in Two Rice Cultivars" Plants 12, no. 5: 976. https://doi.org/10.3390/plants12050976
APA StyleGupta, A., Singh, A. N., Tiwari, R. K., Sahu, P. K., Yadav, J., Srivastava, A. K., & Kumar, S. (2023). Salinity Alleviation and Reduction in Oxidative Stress by Endophytic and Rhizospheric Microbes in Two Rice Cultivars. Plants, 12(5), 976. https://doi.org/10.3390/plants12050976







