Abstract
The bacteria harboring phoD encodes alkaline phosphatase (ALP), a secretory enzyme that hydrolyzes organic phosphorous (P) to a usable form in the soil. The impact of farming practices and crop types on phoD bacterial abundance and diversity in tropical agroecosystems is largely unknown. In this research, the aim was to study the effect of farming practices (organic vs. conventional) and crop types on the phoD-harboring bacterial community. A high-throughput amplicon (phoD gene) sequencing method was employed for the assessment of bacterial diversity and qPCR for phoD gene abundance. Outcomes revealed that soils treated for organic farming have high observed OTUs, ALP activity, and phoD population than soils managed under conventional farming with the trend of maize > chickpea > mustard > soybean vegetated soils. The relative abundance of Rhizobiales exhibited dominance. Ensifer, Bradyrhizobium, Streptomyces, and Pseudomonas were observed as dominant genera in both farming practices. Overall, the study demonstrated that organic farming practice favors the ALP activity, phoD abundance, and OTU richness which varied across crop types with maize crops showing the highest OTUs followed by chickpea, mustard, and least in soybean cropping.
1. Introduction
Phosphorous is one of the most important macronutrients essential for the growth and development of plants. Regardless of its abundance in soils, the availability of phosphorous is very limited in the terrestrial environment. Generally, two forms of P are present in the soil, organic and inorganic forms; however, only inorganic orthophosphate (PO43−) ions in the soil solution are easily accessible for plant use [1]. A huge amount of P fertilizers is utilized in agriculture as mineral P fertilizers and organic fertilizers (compost/manure) to maintain crop productivity. Soon after its application in the soil, a portion of inorganic P is readily used by the plants and microbes, whereas the residual P is immobilized as an insoluble form in the soil. Microbes play a vital role in recycling phosphorous from a recalcitrant unavailable form of P by solubilization and mineralization of inorganic and organic forms of P, respectively [2]. Inorganic P in soil mainly comprises minerals, such as apatite, oxyapatite, and hydroxyapatite, which are poorly soluble and assimilable. The inorganic P is solubilized by soil microbes by excreting organic acids [3]. The organic P mineralization is through the enzymatic process by the P-hydrolyzing extracellular enzyme phosphatases [1,2] which comprise phosphomonoesterases, phosphodiesterases, phosphotriesterases, etc. The phosphomonoesterases are composed of alkaline and acid phosphatases, nucleotidases, and phytases. Activities of both alkaline and acid phosphatases in soil have been analyzed to assess the organic phosphorous mineralization to inorganic phosphorous form [4]. Alkaline phosphatases (ALP) primarily originate from soil microorganisms, especially bacteria, which are involved in the hydrolysis of organic phosphorous [4]. To date, three homologous ALP encoding gene families have been identified, namely, phoA, phoD, and phoX, as a component of Pho regulon [5]. According to metagenomics datasets, based on the sequence similarity, phoD is the most frequently available gene than phoA and phoX. phoD-harboring bacteria have been widely identified and ubiquitously distributed among terrestrial and aquatic ecosystems [6]. Hence, phoD is considered a good biomarker to bestow an understanding of P transformation in an agroecosystem. Some studies showed a negative correlation between the activity of the ALP enzyme and available phosphorous [7,8]. There are reports that bacteria enhance the production of ALP during scarcity of P by upregulating the expression of functional gene encoding phosphatase enzyme [4]. Moreover, some studies reported a positive correlation between alkaline phosphatase activity and phoD copy number [9,10,11]. However, more evaluation of the origin/source of phoD is required to amplify our knowledge to understand the relationship between the potential activity of alkaline phosphatase and available P in the soil [12].
Researchers have shown the shift in phoD-gene-containing bacterial population under different environmental sites and experimental conditions, including fertilizer management practices in agriculture [6,8,11], soil pH [12], amendment of organic matter [11,13,14], and extreme environments [15]. Tan et al. studied that in pasture soil phosphorous fertilizer incorporation enhanced the phoD community [5]. Xie et al. carried out a field experiment in the temperate monsoon climate of China to study the effect of fertilization treatment, crop rotation, and wheat varieties on functional communities of the rhizosphere associated with P cycling [16]. They found that crop rotation changed the community composition of bacteria having phoD genes in the wheat rhizosphere, whereas fertilization management had no effect. A negative correlation was seen between the phoD abundance and available P, P uptake, and wheat biomass. In another study, Fraser et al. reported that phoD copy number correlated positively with ALP activity in manure- and mineral-phosphorous-treated soils [9]. Results from the Illumina high-throughput approach revealed that the response of phoD bacteria to the P status of the soil is asynchronous, and nitrogen, carbon, and phosphorus soil stoichiometric ratios were the most dominant regulatory parameters for the phoD bacterial population in soils of Inner Mongolia [17]. Most of the studies regarding ALP activity and phoD-containing bacterial diversity to date have been carried out in the temperate and subtropical zones. In a recent study, Hegyi et al. reported that Actinobacteria, Acidobacteria, Cholroflexi, Firmicutes, and Proteobacteria are the superior phyla analyzed through next-generation sequencing of the 16S rRNA gene in the agricultural soil of Vietnam [18]. The study showed a positive correlation between soil phosphatase activities and soil organic C and also between acid phosphatase and total P. Similarly, a significant positive relationship was found between the abundance of the phoD gene and the diversity of the bacterial community of soil. There are still very limited studies with reference to the influence of various factors on phoD soil bacterial abundance and community and ALP activities, especially in an agricultural system with different fertilizer management practices.
However, as per the reviewed literature, information regarding the phoD-containing bacterial distribution, diversity, and community composition is still lacking, particularly in the Indian tropical agroecosystem. To procure a better understanding of the process involved through which bacteria participate in P turnover, there is a need for more studies focusing on the phoD bacterial population in the soil. In this study, we employed a high-throughput targeted amplicon sequencing to investigate the phoD-containing bacterial population in response to different farming practices vegetated with varied crops. In the present study, we hypothesized that the impact of farming practices (organic vs. conventional) and crop types would alter the abundance and composition of phoD-gene-harboring bacterial communities and soil alkaline phosphatase activity. The pivotal goal of this study was to (1) evaluate the influence of farming practices (organic vs. conventional) on ALP enzyme activity, phoD bacterial abundance, and diversity in soil under the influence of different crops and (2) assess the relationship between the ALP activity and phoD abundance and community.
2. Results
2.1. Soil Variables and Crop Biomass
The value of pH of the soil was significantly increased in soils treated for organic farming than the conventional (Figure 1a). Mineral-N content was highest in conventional farming (Figure 1b). A significant increase in available P was detected in conventional farming compared to organic treatments, and the effect of crops on the available P was also significant (Figure 1c; p < 0.001). The microbial biomass P (MBP) showed a significantly higher value in organic farming, and the effect of the crop on microbial biomass P was also significant (Figure 1d; p < 0.001). The root and shoot biomass were observed highest in organic farming practice, and the effect of crops and farming practice on the root and shoot biomass was also significant (Figure 1e,f; p < 0.001).
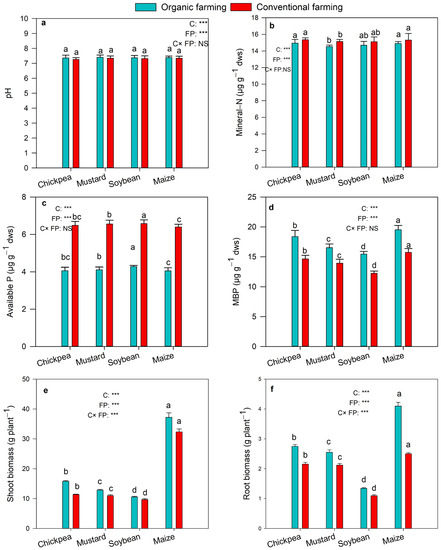
Figure 1.
pH value (a), content of mineral-N (b), available P (c), MBP (d), shoot biomass (e), and root biomass (f) in organic and conventional farming. Different lowercases indicate significant differences (Tukey’s post hoc test) between crops in the same farming practice (p < 0.05). The crops (C) and farming practice (FP) specify the results of MANOVA indicating p-value at different significance levels, *** p < 0.001, NS: not significant). The values are two-year average mean ± SD. MBP, microbial biomass P; ALP, alkaline phosphatase.
2.2. Soil Alkaline Phosphatase (ALP) and Abundance of phoD Gene
Variations in soil ALP activity and phoD gene copy number in rhizosphere soils of crops vegetated in both farming fields are depicted in Figure 2a,b. The ALP activity ranged from 2.45–3.59 µmol g−1soil h−1 in organic to 1.46–2.96 µmol g−1soil h−1 in conventional farming soils. Within the crops, maize showed the highest ALP activity and soybean the least (Figure 2a). MANOVA showed a significant (p < 0.001) effect of farming practices and crops on ALP enzyme activity. Tukey’s HSD test indicated significant variation in ALP activity between different crops in the same farming practice. qPCR analysis showed that, compared to conventional farming (4.5 × 106–1.1 × 107 copies g−1 dws), the phoD population was substantially higher among organic farming, which ranged from 6 × 106 to 1.3 × 107 copies g−1 dws (Figure 2b). The abundance of phoD gene copy was highest in soil samples of maize crops, followed by chickpea, mustard, and soybean soils. The MANOVA result showed a significant (p < 0.001) effect of farming practices and types of crops on the abundance of phoD gene copy. Tukey’s HSD test suggested phoD abundance to differ significantly between different crops.
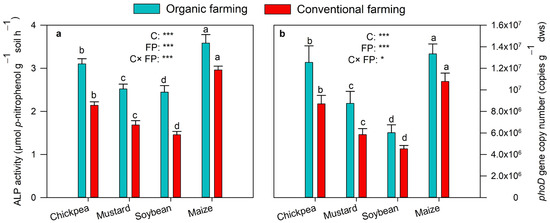
Figure 2.
ALP activity (a) and phoD gene copy number (b) in organic and conventional farming. Different lower cases indicate significant differences (Tukey’s post hoc test) between crops in the same farming practice (p < 0.05). The crops (C) and farming practice (FP) specify the results of MANOVA indicating p-value at different significance levels (* p < 0.05, *** p < 0.001). The values are two-year average mean ± SD. MBP, microbial biomass P; ALP, alkaline phosphatase.
2.3. phoD-Gene-Containing Bacterial Community and Relationship with Soil Parameters
Initially, a total of 674,076 raw reads were generated from phoD amplicon 2 × 300 pair-end sequencing, with an average of 84,260 reads per sample. The stringent poor-quality filtration resulted in a total of 254,895 high-quality reads subjected for downstream analyses. To compare phoD-gene-harboring bacterial community among different samples, rarefaction of sequences/reads was performed to obtain the OTUs assigned by an equal number of sequences. We rarified the sequences at minimum library size to 2629 reads for each sample. The phoD OTUs ranged between 346 and 672 with ~457 average OTUs per sample (Table 1). The observed OTUs composition of phoD gene bacteria was greater in soils treated for organic farming than the conventional one (Table 1). The highest OTUs were reported in soils of maize crop under organic farming, followed by chickpea-, mustard, and soybean-planted soils.

Table 1.
Diversity indices (rarefied at minimum library size, i.e., 2629 reads).
We estimated the Pearson correlation coefficient between the diversity indices, soil parameters, and phoD copy number (Table 2). The Pearson correlation showed a significant (p < 0.01) positive correlation between OTUs richness, Shannon and Simpson index with that of phoD abundance, and ALP activity of soil (Table 2).

Table 2.
Pearson correlation coefficient between diversity indices and soil parameters in different crops and farming practices (* p < 0.05, ** p < 0.01). ALP, alkaline phosphatase; MBP, microbial biomass P.
The results of CCA (canonical correspondence analysis) showed that the structure of phoD bacterial community significantly correlated with ALP activity, MBP, and available P and to a lesser extent with pH and mineral-N (Figure 3). The phoD community structure in different farming practices in all four crops changed along the first axis. The soil pH and mineral-N showed a strong correlation with the CCA2 axis that governs 24.69% of the overall variance in the phoD community.
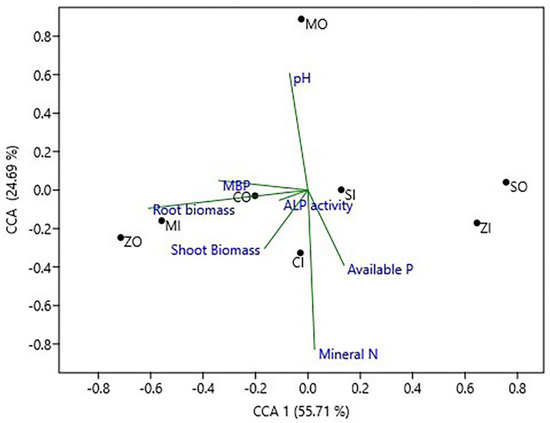
Figure 3.
Ordination plot of CCA (Canonical Correspondence Analysis) to exhibit the correlation between soil variables and phoD encoding community structure. CO, chickpea organic farming; CI, chickpea conventional farming; MO, mustard organic farming; MI, mustard conventional farming; SO, soybean organic farming; SI, soybean conventional farming; ZO, maize organic farming; ZI, maize conventional farming; MBP, microbial biomass P.
2.4. Relative Abundance of phoD Bacterial Community
All the phoD reads were affiliated with the phyla Proteobacteria and Actinobacteria. Proteobacteria was the dominant phylum accounting for 65.24–94.57% of all the sequences. These OTUs were classified into eight bacterial orders that include Rhizobiales, Streptomycetales, Pseudomonadales, Rhodobacteriales, Rhodospirallales, Burkholderiales, Pseudonocardiales, and Micromonosporales. The Rhizobiales were dominant among all samples and accounted for 37.28–78.27% (Figure 4a). The phoD-containing bacterial order varied among farming practice and crops, but no particular trend was observed. A total of 29 genera were identified, among which the dominant genera present were Bradyrhizobium (4.41–42.73%), Ensifer (1.04–49.65%), Streptomyces (3.29–37.08%), Mesorhizobium (3.36–29.12%), Sinorhizobium (2.28–23.55%), Pseudomonas (0.15–15.62%), and Skermanella (3.75–15.43%) (Figure 4b). The relative abundance of Bradyrhizobium was most predominant in organic farming practice under chickpea cropping. Moreover, Pseudomonas also showed higher relative abundance in organic treatment, whereas Streptomyces is enriched in soybean cropping with conventional farming soil.
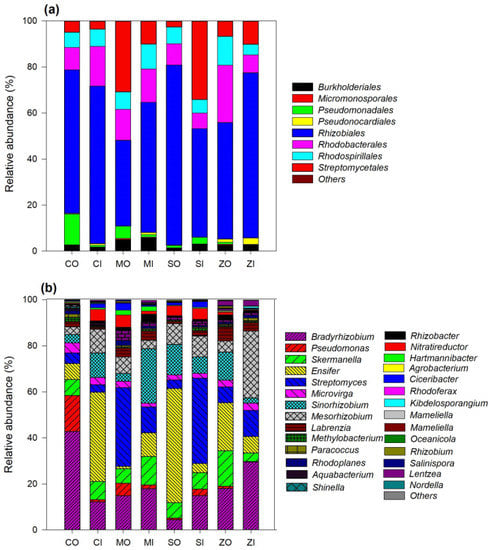
Figure 4.
(a) Relative abundance of phoD-harboring bacterial communities under organic and conventional farming practices and different crops at the order level. (b) Relative abundance of phoD-harboring bacterial communities under organic and conventional farming practices and different crops at the genera level. CO, chickpea organic farming; CI, chickpea conventional farming; MO, mustard organic farming; MI, mustard conventional farming; SO, soybean organic farming; SI, soybean conventional farming; ZO, maize organic farming; ZI, maize conventional farming.
3. Discussion
In the current study, organic farming practice stimulates TN, SOC, and TP [19]. Microbial biomass P (MBP) increases as a result of organic farming practice that stimulates the biological activity of soil in comparison to conventional farming. The organically amended soil is well established to increase organic matter in the fields [20]. Similar studies have shown an increase in organic matter content in organic farming and organic matter-rich soil to have long-term potential to sustain nutrient release [21,22]. The pH is increased in organic farming soil in comparison to the conventional counterpart. The lowering of pH in conventional farming soil maybe because of nitrification of NH4+ and thus H+ ion is produced which results in enhanced soil acidity. Chakraborty et al. showed a decrease in pH with increasing chemical fertilizer application [23]. In this study, the available P is significantly (p < 0.001) increased under the conventional farming practice. Similar results are also reported by Liu et al. [24]. Fraser et al. reported the same trend that indicated low available P in organic management [10]. This may be because fertilizer P administers orthophosphate usually amplifying labile inorganic P [25]. However, Sakurai et al. reported the opposite trend [13]. This discrepancy in available P maybe because of confounding features, including the quantity of phosphorous in fertilizers, crop P demand, and diverse agricultural practices.
Consistent with our hypothesis, the farming practices and crop types influenced the abundance and phoD gene bacterial communities. In the present investigation, organic farming stimulated the alkaline phosphatase activity of soil. Higher ALP activity was determined in organic farming than in the conventional one with significant differences (p < 0.001) (Figure 2a). It has been reported that organic matter application enhances the ALP activity in the soil [9,10,11,13]. The possible reason for the increased ALP activity in organic farming may be increased SOC that allows bacteria to proliferate due to an additional carbon source [6]. In addition, the amendment of cattle manure in soil results in a significant increase in alkaline phosphatase activity [26]. phoD gene copy number was significantly higher (p < 0.001) in soils of organic farming fields (Figure 2b). The earlier studies also reported similar results in the long-term manure-fertilized soil [8,9,10]. It is suggested that phoD abundance was highest in organic treatment with the highest alkaline phosphatase activity and SOC that increases organic matter suggesting higher nutrient content which accordingly increases bacterial abundance [27]. In organic farming practice, the organic fertilizer, low in available P content, and plenty of C-rich substrate possibly influenced the proliferation of various phoD-gene-containing bacterial population, hence increasing the phoD abundance and alkaline phosphatase activity [28]. In addition, we found a significant (p < 0.01) and strong positive correlation between OTUs, Shannon index, and Simpson index and alkaline phosphatase activity and phoD copy number (Table 2). The previous report indicated a significant positive correlation between phoD gene copy number and the bacterial community such as Shannon and Chao 1 diversity [18]. Zhu et al. showed a positive correlation between Chao 1 index and alkaline phosphatase activity, suggesting that phoD-gene-harboring bacteria may be highly activated while secreting ALP enzyme [17]. Similarly, a significant (p < 0.001) variation was observed among crop species for the activity of alkaline phosphatase and phoD abundance. The elevated ALP activity and the abundance of phoD bacteria exhibited a trend of maize > chickpea > mustard > soybean fields in soil samples (Figure 2a,b). This may be due to the influence of varied plant species on soil microbes that exhibit variability among plant physiological attributes including exudates of root [29] that possibly differ with crops and physiological attributes [30].
Recently, many studies have been reported demonstrating the shifts in phoD-containing bacterial population in soils with organic and inorganic fertilization [5,6,8,13,28]. As existing literature in the context of Indian agroecosystem suggests scarcity of knowledge about phoD encoding bacterial assemblages; however, agroecosystems situated in other countries have been analyzed for the same. Wei et al. studied that the P fertilization influences phosphorous mineralizing microbes in paddy soil, and all the sequences are classified into five classes: Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Cyanobacteria, and Actinobacteria [31]. The dominant genera of this study were Methylobacterium, Methylomonas, and Bradyrhizobium. Here, in this study, phoD taxonomic classification revealed that Proteobacteria and Actinobacteria were the two phyla among which Proteobacteria showed dominance in both farming practices. The investigations on phoD bacterial population identified similar genera [14,28]. However, Proteobacteria was the most dominant in organic farming samples. The previous study also detected Proteobacteria as the dominant phylum [6,8,28]. The dominance of phylum Proteobacteria may be because of the supplement of compost which contributed to an increased level of soil nutrients (e.g., SOC, TN, and TP) and stimulated the copiotrophic bacterial growth and resulted in a shift of phoD-harboring bacterial community. Tian et al. reported a higher abundance of Proteobacteria in swine manure added to soil with increased soil nutrients and promoted the proliferation of copiotrophic phyla, i.e., Proteobacteria [32]. We have also observed an increased level of Actinobacteria in conventional farming as compared to the organic amendment. Actinobacteria are more apparent to be an index for inorganic fertilization as compared to organic fertilization [33]. The genera Ensifer belonging to the phylum Proteobacteria was the most predominant genera and its relative abundance was highest in organic farming practice with soybean cropping. The previous studies on total and P-mineralizing bacterial communities in maize cropping detected similar genera Ensifer in soil fertilized with compost than chemical and nonfertilized soil [34].
The higher abundance of Ensifer sp. in soils managed with organic farming compared to the conventional suggests that organic fertilization perhaps enriched this genus in soil and accordingly strengthen their abundance in the rhizosphere [34]. The relative abundance of Bradyrhizobium, a free-living and nitrogen fixer that is present in all the samples, indicates that this genus perhaps plays a major role in linking nitrogen (N) and phosphorous (P) cycle [28]. Therefore, further study is imperative to acknowledge the role of Bradyrhizobium sp. in establishing the possible link between nitrogen cycling and phosphorous turnover under diverse conditions, which may contribute to accelerating ALP activity as well as phosphorous transport rates and also alter soil nitrogen pools [28]. In another study, Gitonga et al. [35] observed the effect of farm management (conventional and organic farming) system on Bradyrhizobia species abundance and diversity and reported proliferation of Bradyrhizobium in organic farming practice and hence enhanced Bradyrhizobial diversity. This may be because organic farming has been revealed to increase soil organic carbon which provides the required energy for microbes and thus increases their abundance and diversity as compared to conventional farming practice [36]. Here, in this study, we observed that organic farming stimulated the proliferation of Pseudomonas (relative abundance: 15.62%) in soils planted with chickpeas. Hu et al. also reported an increased relative abundance of Pseudomonas in the organic amendment [6]. It is reported that several species of Pseudomonas grow rapidly under straw as the only carbon source, on the basis of their plant lignin and hemicellulose degradation capacity [37]. Moreover, Pseudomonas sp. inhabits several P-mineralizing bacterial populations [38,39]. Another dominant genus identified in this study was Streptomyces. Hegyi et al. also reported Streptomyces as abundant, potential phosphate-solubilizing bacteria in soil and Streptomyces liacinus as a phoD gene encoding isolate [18]. It is established that apart from plant growth-promoting capabilities, Streptomyces species are generally associated with a phosphorous transformation, including phosphorous solubilization and mineralization [11,18]. Moreover, several Streptomyces species reported secreting alkaline phosphatase enzyme, namely, S. griseus, S. hiroshimensis, and S. hygroscopicus [40,41]. The organic farming practice showed the greater OTUs richness and Shannon diversity index compared to conventional farming (Table 1).
The observed OTUs were positively correlated with ALP activity and phoD abundance. These findings are in accordance with Sakurai et al. [13] showing that in organic matter amended soils phoD encoding bacterial assemblages shifted differently from those in conventional farming supplied with chemical fertilizers. Several earlier studies have also reported a change in phoD encoding bacterial assemblages in response to different fertilization management [11,12,42]. Our findings, as well as the previous reports, suggested that the change in phoD bacterial community by organic fertilizer might be caused by community composition turnover, i.e., existing OTUs replacement with the new OTUs. Moreover, organic farming showed higher alkaline phosphatase activity than conventional farming, and the variation in OTU composition of phoD may have enhanced enzymatic activity [42]. According to Watts et al. [43], the bacterial diversity in soil could be enhanced by the bacteria supplemented through the organic fertilizer.
Therefore, in the present study, we found that organic fertilizer incorporation strengthens the phoD-containing bacterial richness in comparison to conventional farming, suggesting a substantial effect of the introduced bacterial population from organic fertilizer on the alpha diversity of microbes. Our results revealed the effect of crops on the phoD bacterial population, but no particular trend was observed in test crops at the genera level. The OTU richness of phoD was highest in soils vegetated with maize compared to chickpea, mustard, and soybean field soils. Neal et al. have also reported that the effect of crop type on the phosphohydrolase genes was significant [44]. It is well-established fact that plants excrete a composite mixture of chemical compounds from the roots in the soil, which could possibly favor discrete microbial communities towards the rhizospheric zone [45,46,47]. Through enzymatic hydrolysis (phosphatase activity) of organic P and solubilization of mineral phosphate, these microbes can induce a supply of orthophosphate in soil [48].
4. Materials and Methods
4.1. Study Site and Experimental Design
The sampling sites were located at the agriculture field of Dagmagpur (Mirzapur district of Uttar Pradesh) (25°9′ N, 83°34′, 80 m above MSL), India. This zone has a seasonal tropical monsoonal climate with an average rainfall of 849.9 mm annually and the mean temperature (minimum to maximum) generally ranges between 8 °C and 10 °C in January and 38 °C and 42 °C in June. The soil of the study site was Alfisol with a silty sandy texture (32:64:4, sand:silt:clay). The agricultural farm was governed by the farmers, and the study site has a long (approximately 30 years) agricultural history. In this area, besides selected test crops for the present study, rice and wheat are the major crops. The soil total P was 152.17 µg g−1 in organic and 127.08 µg g−1 in conventional farming practice. The main soil physicochemical properties were given in [19].
Two agricultural fields having two different farming practices were selected: one field received compost (organic farming) and another field mineral fertilizer (conventional farming). The experimental plots were designed in randomized complete block design including three blocks (5 m × 4 m with 1 m gap) per site and a treatment combination of 4 crops × 2 farming practices. To avoid edge effects, the organic and conventional farming experimental plots were separated by a 100 m distance. In the organic farming plot, compost was added as an organic supplement. The compost applied is composed of crop residues and cow dung prepared by NADEP (Narayan Deorao Pandharipande) technique [49]. After maturation, the compost was dispersed manually at the rate of 15 tons ha−1 and plowed up to 15 cm depth to mix and homogenize before the cropping season of Rabi and Kharif. No other nutritional supplement was added apart from cow urine (1:50; urine:water dilution ratio), which is added twice, during the vegetative and flowering stages of the crops as a nitrogen source at the interval of 40 days. The cow urine used in the present study comprises 15 g N L−1. In conventional farming, as per the standard practice, NPK (chemical fertilizer) was applied at the rate of 120, 40, and 60 kg ha−1 in Rabi crops, and for the Kharif crops, the rate of NPK application was 20, 40, and 60 kg ha−1. This was executed twice annually, i.e., one-time application in both seasons as a basal dose. There were four crops selected in this study. Two each from Rabi (chickpea: Cicer arietinum L. var Pusa-256, and mustard: Brassica campestris var. T-151) and Kharif (maize: Zea mays var. Ganga II and soybean: Glycine max var. PS-1225). The experiments were conducted for two cropping seasons (2017–2019). No pesticides or fungicides were added, and weeds were removed manually.
4.2. Soil Sampling and Soil Physicochemical Analyses
Rhizosphere soil samples in triplicate (0–15 cm) from both (organic and conventional) farming fields were collected randomly at the mid-flowering stage of a crop growth cycle. Soil adhered to the rhizosphere zone was collected after tapping the root gently of each test crop in a plastic bag. Soil samples (in triplicate) were mixed and homogenized and sieved (2 mm mesh) to remove plant debris. The homogenized soil samples are divided into two parts: one part stored at −20 °C for downstream analyses of phoD-harboring bacterial community and qPCR experiments and the other kept (at 4 °C) for the analyses of ALP activity and soil properties. Soil physicochemical properties, alkaline phosphatase activity, and phoD gene abundance were measured for two consecutive years (2017–2019) in soil samples, and data were pooled as average data for two years and the phoD diversity analysis was performed only for second-year soil samples. All the basic physical and chemical properties of soil were analyzed following the standard protocols [50]. Total P was analyzed as per Allen et al. [51]. To measure soil available P, the method of Olsen et al. was followed [52]. Microbial biomass phosphorus (MBP) was measured following the standard protocol of Brookes et al. [53], and crop biomass was measured as per Neha et al. [19].
4.3. Assay of Soil Alkaline Phosphatase (ALP) Activity
Using p-nitrophenol phosphate (p-NPP) as substrate, ALP activity was measured [54]. The analysis was carried out by taking rhizosphere soil (1 g) with 1 mL modified universal buffer (pH 11), p-NPP, and incubating at 37 °C for 1 h. After 1 h, 0.5 M NaOH was mixed to terminate the reaction. The reaction mixture was filtered, and the p-nitrophenol (p-NP) was measured spectrophotometrically (at 420 nm). The activity was recorded as µmol of p-NP g−1 soil h−1.
4.4. Soil DNA Extraction and phoD Gene Quantification
Total genomic soil DNA was extracted from 0.5 g frozen soil by FastDNA Spin Kit. NanoDrop 2000 spectrophotometer was used to measure DNA concentration and quality. The phoD gene copy number (abundance) was quantified by qPCR (iCycler iQ5 thermocycler; Bio-Rad, Hercules, CA, USA). The extracted DNA was amplified for phoD using ALPS F-730 and ALPS R-1101 primers as described by Sakurai et al. [13]. The PCR reaction mixture (20 µL) contains 10 µL of PowerUpTM SYBR Green Master Mix, 0.5 µL of each primer concentration (10 µM), DNA template (2 µL), and water (nuclease-free) to make up the final volume to 20 µL. The PCR conditions for amplification of phoD gene were as follows: 3 min at 94 °C (initial denaturation), 40 cycles at 94 °C for 1 min (denaturation), at 61 °C for 45 s (annealing), and final extension at 72 °C for 45 s. Data were tested for PCR amplification efficiencies which were 114.5% and R2 = 0.963.
4.5. Illumina MiSeq High-Throughput Sequencing for phoD Gene Amplicons and Data Analysis
The Illumina Miseq 300 bp paired-end sequencing platform was used to assess the phoD-gene-containing bacterial population. The target gene (pho D) was amplified in rhizosphere soil DNA using ALPS-F730 and ALPS-R1101 primers [13]. The indexed paired-end library was prepared by adding Illumina Nextera XT compatible adapters to the forward and reverse primer sequences. The reaction mixture comprises template DNA (50 ng), KAPA Hifi HotStart Ready Mix (KAPA Biosystems, Wilmington, NC, USA), and modified primers (100 nM) ALPS-F730 and ALPS-R1101. PCR conditions were set as initial denaturation (at 94 °C for 3 min), followed by denaturation (30 cycles at 94 °C, 1 min), annealing (61 °C, 45 s), extension (72 °C, 30 s), and terminal extension (72 °C for 7 min). The amplicons were cleaned up and subjected to quantitate libraries using a quantitation assay of Qubit DNA high sensitivity (ThermoScientific, Grand Island, NE, USA). The quantity of the library was corroborated with the help of the D7500 DNA kit and Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Illumina MiSeq 2 × 300 bp platform (Illumina, San Diego, CA, USA) was used for sequencing as per the manufacturer’s protocol. The generated raw targeted phoD gene amplicon sequences have been deposited to the NCBI sequence read archive (SRA) database under the BioProject accession number PRJNA797670.
4.6. Sequence Analysis
The sequences were analyzed as per Bhardwaj et al. [55] with slight modifications for the phoD gene. The FastQC was used to overview raw forward (R1) and reverse (R2) reads for basic quality control [56]. Raw sequences were quality filtered and trimmed using Trimmomatic V0.35 with criteria such as (i) adaptor sequences removal and (ii) eliminating unclear reads (reads having undefined nucleotides “N” > 5%) and low-quality sequences. The quality-passed (forward and reverse) joining of paired reads was executed by PEAR (Paired-End reAd mergeR), and the remaining single reads were discarded [57]. The joined reads with a quality score of < 30 and sequence size with less than 250 bp and more than 380 bp sequences were filtered out to obtain high-quality sequences (HQS). An appropriate pipeline Quantitative Insights Into Microbial Ecology (QIIME), version 1.9.0, was used for the analysis of high-quality reads [58] for bacterial diversity estimation. Initially, the quality-passed reads were screened against the funGene database regarding phoD sequences using the HMMER model with default settings. This allowed us to obtain only the phoD-gene-specific amplicon reads and filtered the unwanted reads such as 16S rRNA contamination and chimeric reads. Then, the phoD-gene-featured sequences were subjected to operational taxonomic units (OTUs) determination at 75% similarity clustering employing pick_otus.py script [59]. The OTUs were rarefied to the least library size (2629 reads) to find out the taxonomic diversity alpha indices, such as observed OTUs, Good’s coverage, Chao1, Simpson, Shannon, after discarding OTUs with <10 reads count. Furthermore, the taxonomic classification of reads was obtained using a Kaiju metagenome classifier with the k-mer setting of 31, integrated into MGX software.
4.7. Statistical Analyses
To study the effect of farming practices and crops on the physicochemical and microbiological properties of soil, MANOVA (multivariate analysis of variance) was conducted with Tukey’s post hoc test (p < 0.05). Pearson correlation test was used to study the relationship between diversity indices and soil parameters (IBM SPSS Statistics 20). Canonical correlation analysis (CCA) was employed to evaluate the effect of farming practices and crops to explore the correlation between soil variables and phoD-gene-harboring bacterial community using PAST v 3.20.
5. Conclusions
The farming practices and crop types change the composition of phoD-containing bacterial communities. The present study suggests that alkaline phosphatase activity, phoD abundance, and OTUs richness were increased in response to organic farming practice. Rhizobiales showed dominance at the order level. Bradyrhizobium, Ensifer, Streptomyces, and Pseudomonas were detected as the dominant members. This study will provide a better understanding of the significance of the alkaline phosphatase enzyme and its role in P mineralization to improve P management strategy to maintain sustainable agriculture. Future research is needed to address the influence of various types and quantities of fertilizers under different cropping systems on phoD communities for stimulating P availability in agricultural soils.
Author Contributions
N. conducted wet labs, data analysis, and wrote the manuscript; Y.B. performed statistical and sequence analysis; B.R. carried out NGS data analysis; S.K.D. did conceptualization and reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The University Grant Commission, New Delhi-Centre for Advanced Study (UGC-CAS), Department of Botany, Banaras Hindu University, Varanasi, India, has provided fund (File No-R/Dev/IX-Sch.(SRF-JRF-CAS-Botany)/51186 in the form of JRF and SRF to Neha for this study.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The phoD gene amplicon sequences have been deposited to the NCBI sequence read archive (SRA) database under the BioProject accession number PRJNA797670.
Acknowledgments
We are thankful to the coordinator of the Centre of Advanced Study, Department of Botany, Banaras Hindu University, Varanasi, for desired facilities.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Condron, L.M.; Turner, B.L.; Cade-Menun, B.J. Chemistry and dynamics of soil organic phosphorus. In Phosphorus: Agriculture and the Environment; Sims, J., Sharpley, A., Eds.; John Wiley & Sons, Ltd.: Madison, WI, USA, 2005; Volume 46, pp. 87–121. [Google Scholar] [CrossRef]
- Richardson, A.E. Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Funct. Plant Biol. 2001, 28, 897–906. [Google Scholar] [CrossRef]
- Richardson, A.E.; Lynch, J.P.; Ryan, P.R.; Delhaize, E.; Smith, F.A.; Smith, S.E.; Harvey, P.R.; Ryan, M.H.; Veneklaas, E.J.; Lambers, H.; et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil 2011, 349, 121–156. [Google Scholar] [CrossRef]
- Nannipieri, P.; Giagnoni, L.; Landi, L.; Renella, G. Role of phosphatase enzymes in soil. In Phosphorus in Action; Springer: Berlin/Heidelberg, 2011; pp. 215–243. [Google Scholar] [CrossRef]
- Tan, H.; Barret, M.; Mooij, M.J.; Rice, O.; Morrissey, J.P.; Dobson, A.; Griffiths, B.; O’gara, F. Long-term phosphorus fertilisation increased the diversity of the total bacterial community and the phoD phosphorus mineralizer group in pasture soils. Biol. Fert. Soils 2013, 49, 661–672. [Google Scholar] [CrossRef]
- Hu, Y.; Xia, Y.; Sun, Q.; Liu, K.; Chen, X.; Ge, T.; Zhu, B.; Zhu, Z.; Zhang, Z.; Su, Y. Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Sci. Total Environ. 2018, 628, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Fraser, T.D.; Lynch, D.H.; Gaiero, J.; Khosla, K.; Dunfield, K.E. Quantification of bacterial non-specific acid (phoC) and alkaline (phoD) phosphatase genes in bulk and rhizosphere soil from organically managed soybean fields. Appl. Soil Ecol. 2017, 111, 48–56. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, N.; Condron, L.M.; Dunfield, K.E.; Chen, Z.; Wang, J.; Chen, L. Soil alkaline phosphatase activity and bacterial phoD gene abundance and diversity under long-term nitrogen and manure inputs. Geoderma 2019, 349, 36–44. [Google Scholar] [CrossRef]
- Fraser, T.; Lynch, D.H.; Entz, M.H.; Dunfield, K.E. Linking alkaline phosphatase activity with bacterial phoD gene abundance in soil from a long-term management trial. Geoderma 2015, 257, 115–122. [Google Scholar] [CrossRef]
- Fraser, T.D.; Lynch, D.H.; Bent, E.; Entz, M.H.; Dunfield, K. Soil bacterial phoD gene abundance and expression in response to applied phosphorus and long-term management. Soil Biol. Biochem. 2015, 88, 137–147. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, N.; Chen, Z.; Tian, J.; Sun, N.; Xu, M.; Chen, L. Response of soil phoD phosphatase gene to long-term combined applications of chemical fertilizers and organic materials. Appl. Soil Ecol. 2017, 119, 197–204. [Google Scholar] [CrossRef]
- Ragot, S.A.; Huguenin-Elie, O.; Kertesz, M.A.; Frossard, E.; Bünemann, E.K. Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil. Plant Soil 2016, 408, 15–30. [Google Scholar] [CrossRef]
- Sakurai, M.; Wasaki, J.; Tomizawa, Y.; Shinano, T.; Osaki, M. Analysis of bacterial communities on alkaline phosphatase genes in soil supplied with organic matter. Soil Sci. Plant Nutr. 2008, 54, 62–71. [Google Scholar] [CrossRef]
- Lagos, L.M.; Acuña, J.J.; Maruyama, F.; Ogram, A.; de la Luz Mora, M.; Jorquera, M.A. Effect of phosphorus addition on total and alkaline phosphomonoesterase-harboring bacterial populations in ryegrass rhizosphere microsites. Biol. Fertil. Soils 2016, 52, 1007–1019. [Google Scholar] [CrossRef]
- Acuña, J.J.; Durán, P.; Lagos, L.M.; Ogram, A.; de la Luz Mora, M.; Jorquera, M.A. Bacterial alkaline phosphomonoesterase in the rhizospheres of plants grown in Chilean extreme environments. Biol. Fert. Soils 2016, 52, 763–773. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, F.; Wang, K.; Yue, H.; Lan, X. Responses of bacterial phoD gene abundance and diversity to crop rotation and feedbacks to phosphorus uptake in wheat. Appl. Soil Ecol. 2020, 154, 103604. [Google Scholar] [CrossRef]
- Zhu, X.; Zhao, X.; Lin, Q.; Li, G. Distribution characteristics of phoD-harbouring bacterial community structure and its roles in phosphorus transformation in steppe soils in Northern China. J. Soil Sci. Plant Nutr. 2021, 21, 1531–1541. [Google Scholar] [CrossRef]
- Hegyi, A.; Nguyen, T.B.K.; Posta, K. Metagenomic Analysis of Bacterial Communities in Agricultural Soils from Vietnam with Special Attention to Phosphate Solubilizing Bacteria. Microorganisms 2021, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Neha; Bhardwaj, Y.; Sharma, M.P.; Pandey, J.; Dubey, S.K. Response of crop types and farming practices on soil microbial biomass and community structure in tropical agroecosystem by lipid biomarkers. J. Soil Sci. Plant Nutr. 2022, 22, 1618–1631. [Google Scholar] [CrossRef]
- Chaudhary, V.; Rehman, A.; Mishra, A.; Chauhan, P.S.; Nautiyal, C.S. Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments. Microb. Ecol. 2012, 64, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, M.M.; Steffen, J.M.; Yates, K.L. Sustainability of soil organic matter at organic mixed vegetable farms in Michigan, USA. Org. Agri. 2020, 10, 487–496. [Google Scholar] [CrossRef]
- Rochette, P.; Angers, D.A.; Flanagan, L.B. Maize residue decomposition measurement using soil surface carbon dioxide fluxes and natural abundance of carbon-13. Soil Sci. Soc. Am. J. 1999, 63, 1385–1396. [Google Scholar] [CrossRef]
- Chakraborty, A.; Chakrabarti, K.; Chakraborty, A.; Ghosh, S. Effect of long-term fertilizers and manure application on microbial biomass and microbial activity of a tropical agricultural soil. Biol. Fert. Soils 2011, 47, 227–233. [Google Scholar] [CrossRef]
- Liu, E.; Yan, C.; Mei, X.; He, W.; Bing, S.H.; Ding, L.; Liu, Q.; Liu, S.; Fan, T. Long-term effect of chemical fertilizer, straw, and manure on soil chemical and biological properties in northwest China. Geoderma 2010, 158, 73–180. [Google Scholar] [CrossRef]
- Fraser, T.D.; Lynch, D.H.; O’Halloran, I.P.; Voroney, R.P.; Entz, M.H.; Dunfield, K.E. Soil phosphorus bioavailability as influenced by long-term management and applied phosphorus source. Can. J. Soil Sci. 2019, 99, 292–304. [Google Scholar] [CrossRef]
- Saha, S.; Prakash, V.; Kundu, S.; Kumar, N.; Mina, B.L. Soil enzymatic activity as affected by long term application of farm yard manure and mineral fertilizer under a rainfed soybean–wheat system in NW Himalaya. Eur. J. Soil Biol. 2008, 44, 309–315. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, X.X.; Guo, X.; Wang, D.; Chu, H. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 2015, 88, 9–18. [Google Scholar] [CrossRef]
- Luo, G.; Ling, N.; Nannipieri, P.; Chen, H.; Raza, W.; Wang, M.; Guo, S.; Shen, Q. Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil fractions. Biol. Fert. Soils. 2017, 53, 375–388. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Lanoue, A.; Strecker, T.; Scheu, S.; Steinauer, K.; Thakur, M.P.; Mommer, L. Root biomass and exudates link plant diversity with soil bacterial and fungal biomass. Sci. Rep. 2017, 7, 44641. [Google Scholar] [CrossRef]
- Bardgett, R.; Mawdsley, J.; Edwards, S.; Hobbs, P.; Rodwell, J.; Davies, W.J. Plant species and nitrogen effects on soil biological properties of temperate upland grasslands. Funct. Ecol. 1999, 13, 650–660. [Google Scholar] [CrossRef]
- Wei, X.; Hu, Y.; Razavi, B.S.; Zhou, J.; Shen, J.; Nannipieri, P.; Wu, J.; Ge, T. Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol Biochem. 2019, 131, 62–70. [Google Scholar] [CrossRef]
- Tian, W.; Wang, L.; Li, Y.; Zhuang, K.; Li, G.; Zhang, J.; Xiao, X.; Xi, Y. Responses of microbial activity, abundance, and community in wheat soil after three years of heavy fertilization with manure-based compost and inorganic nitrogen. Agric. Ecosyst. Environ. 2015, 213, 219–227. [Google Scholar] [CrossRef]
- Fernandez, A.L.; Sheaffer, C.C.; Wyse, D.L.; Sadowsky, M.J. Bacterial community composition in agricultural soils under long-term organic and conventional management. Agrosyst. Geosci. Environ. 2020, 3, e20063. [Google Scholar] [CrossRef]
- Xin, Y.Y.; Rahman, A.; Li, H.X.; Ting, X.U.; Ding, G.C.; Ji, L. Modification of total and phosphorus mineralizing bacterial communities associated with Zea mays L. through plant development and fertilization regimes. J. Integ. Agric. 2021, 20, 3026–3038. [Google Scholar] [CrossRef]
- Gitonga, N.M.; Njeru, E.M.; Cheruiyot, R.; Maingi, J.M. Genetic and morphological diversity of indigenous Bradyrhizobium nodulating soybean in organic and conventional family farming systems. Front. Sustain. Food Syst. 2021, 4, 606618. [Google Scholar] [CrossRef]
- Liao, J.; Liang, Y.; Huang, D. Organic farming improves soil microbial abundance and diversity under greenhouse condition: A case study in Shanghai (Eastern China). Sustainability 2018, 10, 3825. [Google Scholar] [CrossRef]
- Jiménez, D.J.; Dini-Andreote, F.; Van Elsas, J.D. Metataxonomic profiling and prediction of functional behaviour of wheat straw degrading microbial consortia. Biotechnol. Biofuels 2014, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Rosas, S.B.; Andrés, J.A.; Rovera, M.; Correa, N.S. Phosphate-solubilizing Pseudomonas putida can influence the rhizobia–legume symbiosis. Soil Biol. Biochem. 2006, 38, 3502–3505. [Google Scholar] [CrossRef]
- Hamdali, H.; Hafidi, M.; Virolle, M.J.; Ouhdouch, Y. Rock phosphate-solubilizing Actinomycetes: Screening for plant growth-promoting activities. World J. Microbiol. Biotechnol. 2008, 24, 2565–2575. [Google Scholar] [CrossRef]
- Moura, R.S.; Martín, J.F.; Martín, A.; Liras, P. Substrate analysis and molecular cloning of the extracellular alkaline phosphatase of Streptomyces griseus the GenBank accession number for the sequence reported in this paper is AJ278740. Microbiology 2001, 147, 1525–1533. [Google Scholar] [CrossRef]
- Nitta, M.; Goto, M.; Shibuya, N.; Okawa, Y. A novel protein with alkaline phosphatase and protease inhibitor activities in Streptomyces hiroshimensis. Biol. Pharma Bull. 2002, 25, 833–836. [Google Scholar] [CrossRef]
- Matsuoka, S.; Fujinaga, S.; Kobayashi, Y.; Hobara, S.; Osono, T. Bacterial 16S rDNA and alkaline phosphatase gene diversity in soil applied with composted aquatic plants. Limnology 2020, 21, 357–364. [Google Scholar] [CrossRef]
- Watts, D.B.; Torbert, H.A.; Feng, Y.; Prior, S.A. Soil microbial community dynamics as influenced by composted dairy manure, soil properties, and landscape position. Soil Sci. 2010, 175, 474–486. [Google Scholar] [CrossRef]
- Neal, A.; McLaren, T.; Lourenço Campolino, M.; Hughes, D.; Coelho, A.M.; Gomes de Paula Lana, U.; Aparecida Gomes, E.; Morais de Sousa, S. Crop type exerts greater influence upon rhizosphere phosphohydrolase gene abundance and phylogenetic diversity than phosphorus fertilization. FEMS Microbiol. Ecol. 2021, 97, fiab033. [Google Scholar] [CrossRef]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract Pseudomonas putida to the rhizosphere. PloS ONE 2012, 7, e35498. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Robert, C.A.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; Van Der Heijden, M.G. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Na. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.B.; Heuberger, A.L.; Broeckling, C.D.; Jahn, C.E. Non-targeted metabolomics reveals sorghum rhizosphere-associated exudates are influenced by the belowground interaction of substrate and sorghum genotype. Int. J. Mol. Sci. 2019, 20, 431. [Google Scholar] [CrossRef]
- Richardson, A.E.; Simpson, R.J. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 2011, 156, 989–996. [Google Scholar] [CrossRef]
- Chandra, R.; Kumar, N.; Tyagi, A. Nutrient dynamics and decomposition rates during composting of sulphitation pressmud by different methods. J. Environ. Sci. Eng. 2007, 49, 183–188. [Google Scholar]
- Bhardwaj, Y.; Sharma, M.P.; Pandey, J.; Dubey, S.K. Variations in microbial community in a tropical dry deciduous forest across the season and topographical gradient assessed through signature fatty acid biomarkers. Ecol. Res. 2020, 35, 139–153. [Google Scholar] [CrossRef]
- Allen, S. Chemical analysis. In Methods in Plant Ecology; Blackwell: Hoboken, NJ, USA, 1986; pp. 285–344. [Google Scholar]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; U.S. Department of Agriculture: Washington, DC, USA, 1954.
- Brookes, P.C.; Powlson, D.S.; Jenkinson, D.S. Measurement of microbial biomass phosphorus in soil. Soil Biol. Biochem. 1982, 14, 319–329. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Bhardwaj, Y.; Reddy, B.; Dubey, S.K. Temporal shift in methanotrophic community and methane oxidation potential in forest soils of dry tropics: High-throughput metagenomic approach. Biol. Fert. Soils 2020, 56, 859–867. [Google Scholar] [CrossRef]
- Andrew, S. A Quality Control Tool for High Throughput Sequence Data. 2018. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 January 2019).
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).