Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter
Abstract
1. Introduction
2. Results
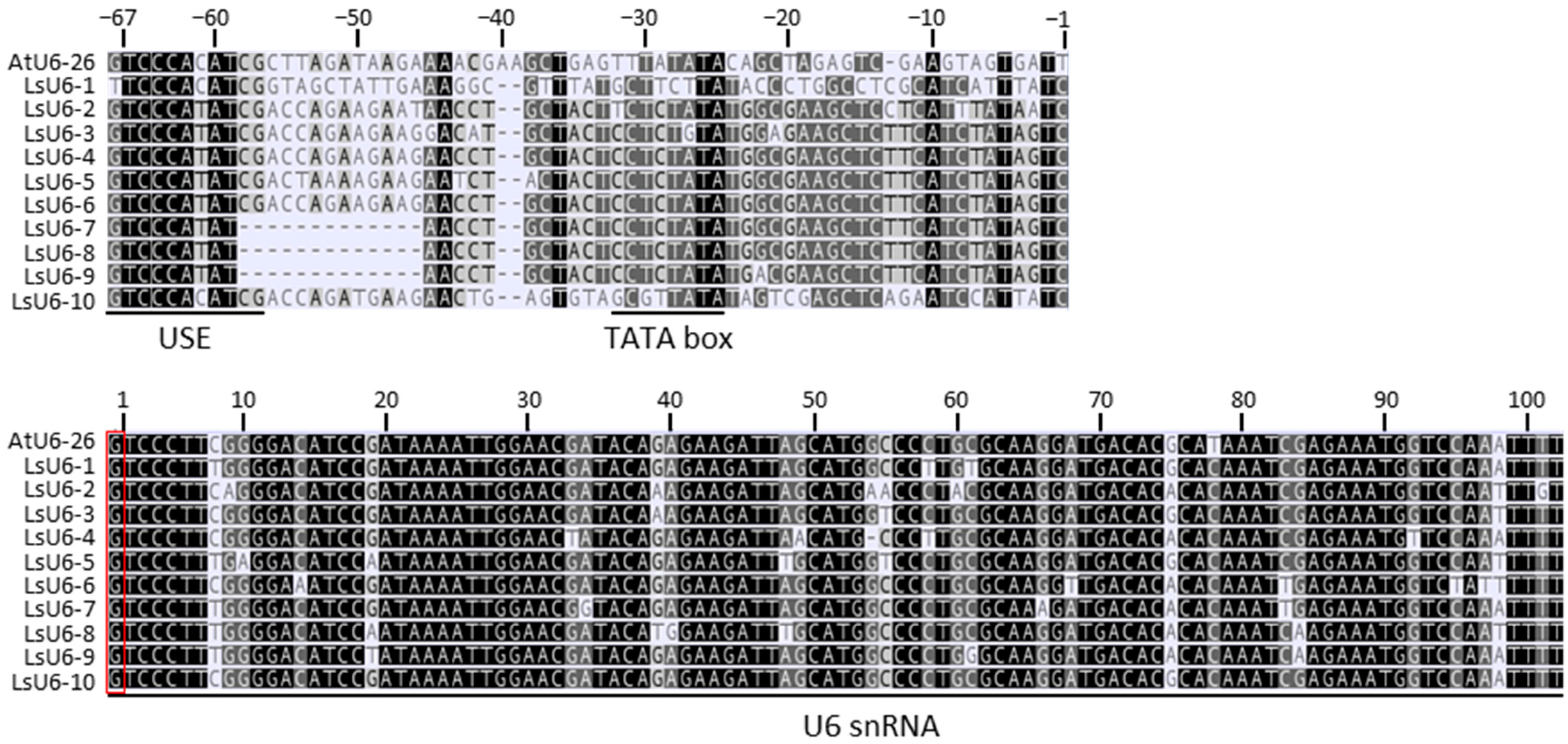
2.1. Identification of U6 Promoters in Leaf Lettuce
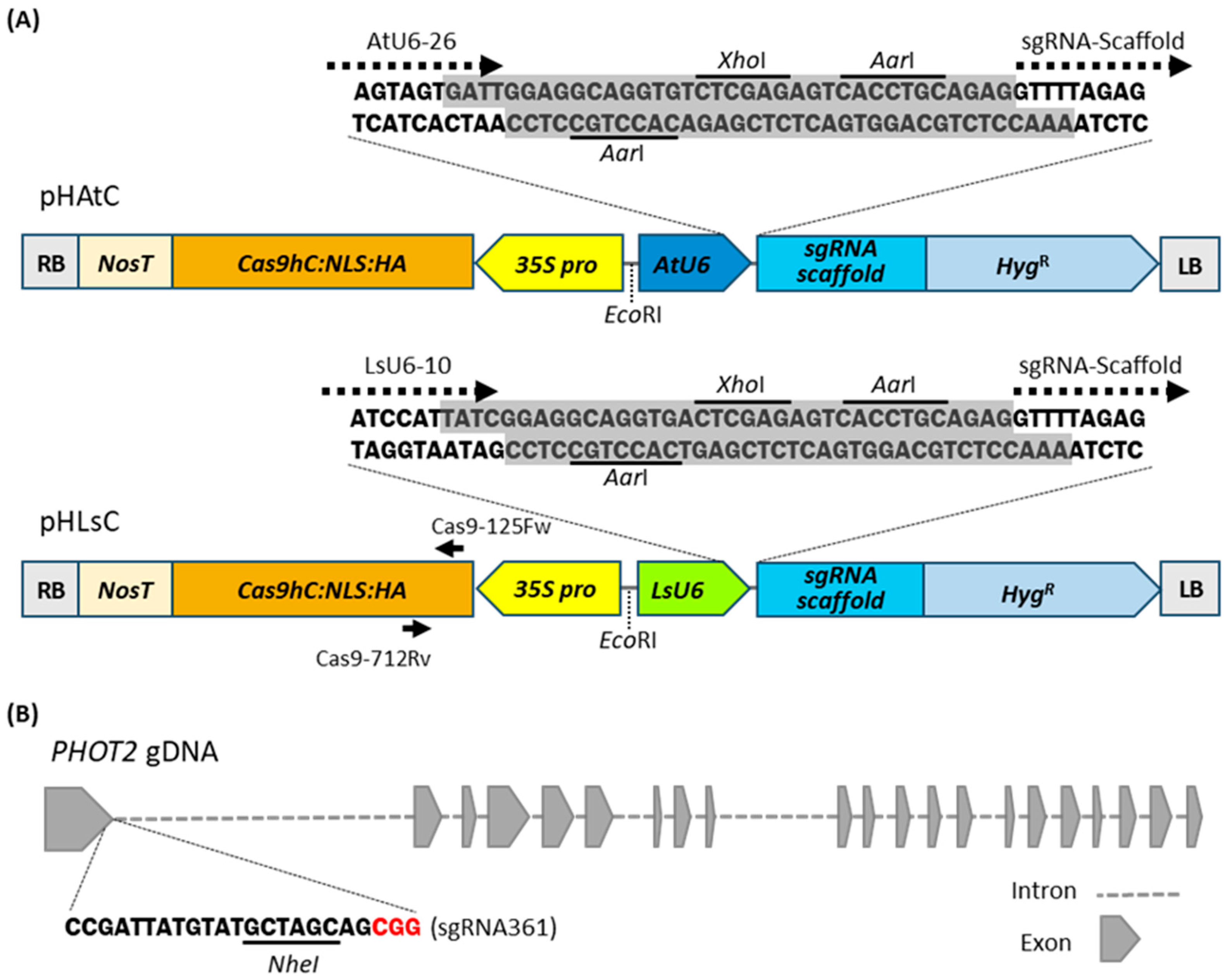
2.2. Plasmid Construction for the CRISPR/Cas9-Mediated Gene Editing System in Lettuce
2.3. The LsU6 Promoter Has Higher Efficiency Than the AtU6 Promoter in CRISPR/Cas9-Mediated Gene Editing in Lettuce
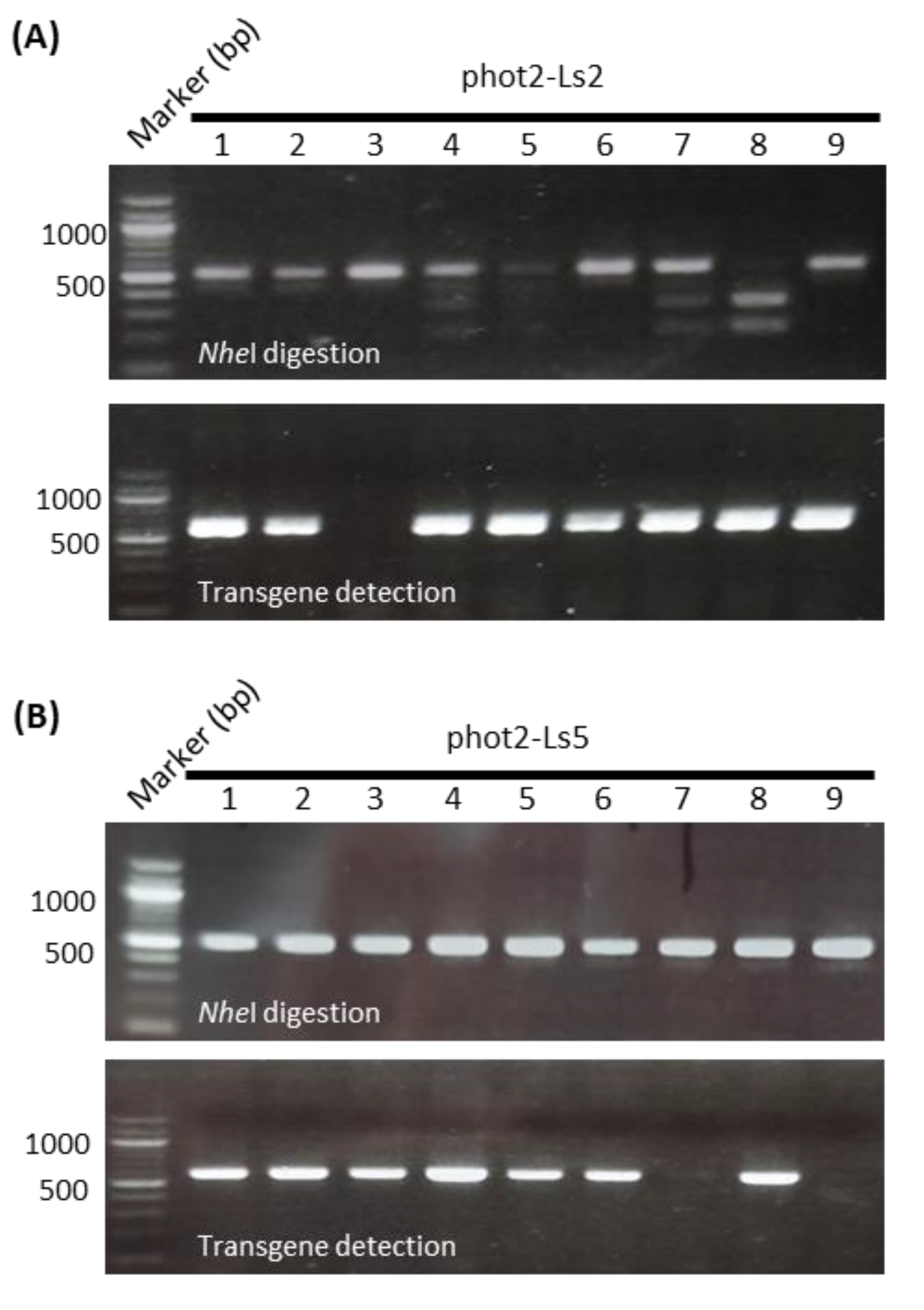
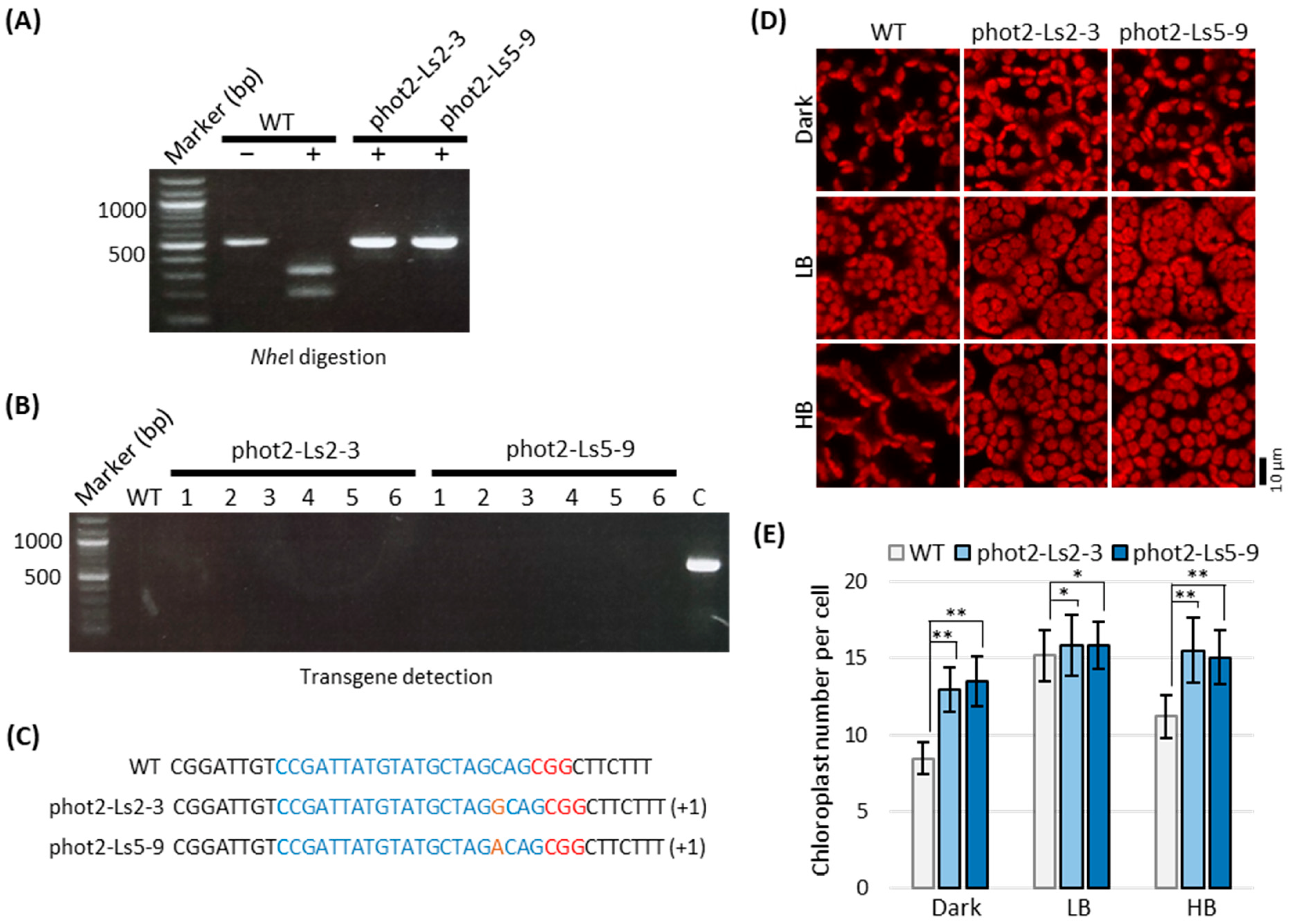
2.4. Lettuce Phot2 Mutants Are Defective in the Chloroplast Avoidance Response
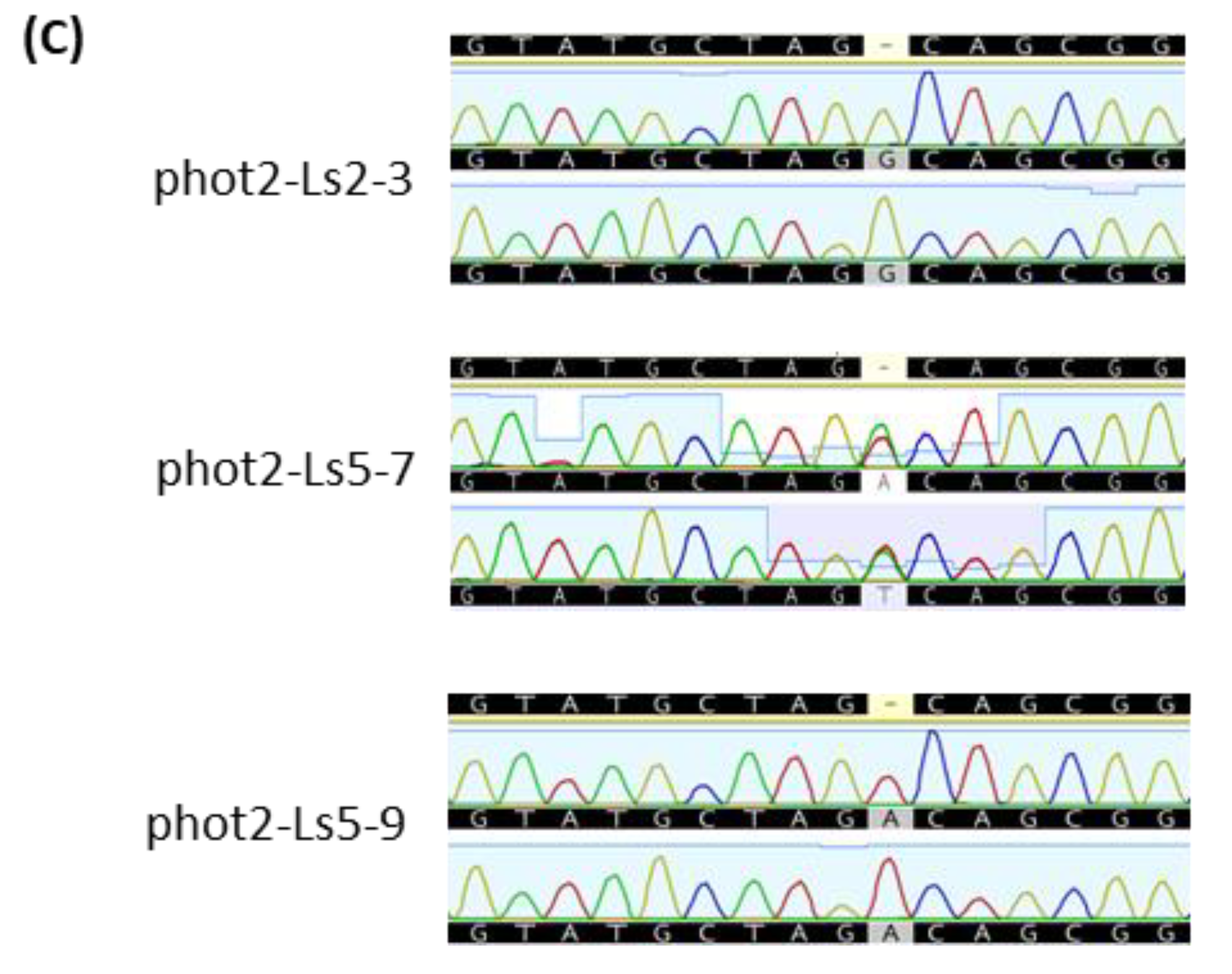
2.5. Selection of Transgene-Free and Genome-Edited Phot2 Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning of the LsU6-10 Promoter and Construction of the Binary Vectors
4.3. Agrobacterium-Mediated Lettuce Transformation
4.4. PCR-Based Restriction Enzyme (PCR/RE) Digestion Assay
4.5. Analysis of Chloroplast Photorelocation Movement
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef]
- Deveau, H.; Garneau, J.E.; Moineau, S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010, 64, 475–493. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., III. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 2014, 41, 63–68. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef]
- Satheesh, V.; Zhang, H.; Wang, X.; Lei, M. Precise editing of plant genomes–prospects and challenges. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 96, pp. 115–123. [Google Scholar]
- Fauser, F.; Schiml, S.; Puchta, H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Aarabidopsis thaliana. Plant J. 2014, 79, 348–359. [Google Scholar] [CrossRef]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana Benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Wang, M.-B.; Helliwell, C.A.; Wu, L.-M.; Waterhouse, P.M.; Peacock, W.J.; Dennis, E.S. Hairpin RNAs derived from RNA Polymerase II and Polymerase III promoter-directed transgenes are processed differently in plants. RNA 2008, 14, 903–913. [Google Scholar] [CrossRef]
- Long, L.; Guo, D.-D.; Gao, W.; Yang, W.-W.; Hou, L.-P.; Ma, X.-N.; Miao, Y.-C.; Botella, J.R.; Song, C.-P. Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 2018, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Di, Y.-H.; Sun, X.-J.; Hu, Z.; Jiang, Q.-Y.; Song, G.-H.; Zhang, B.; Zhao, S.-S.; Zhang, H. Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem. Biophys. Res. Commun. 2019, 519, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Bernard, G.; Gagneul, D.; Santos, H.A.D.; Etienne, A.; Hilbert, J.-L.; Rambaud, C. Efficient genome editing using CRISPR/Cas9 technology in chicory. Int. J. Mol. Sci. 2019, 20, 1155. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef]
- Ren, C.; Liu, Y.; Guo, Y.; Duan, W.; Fan, P.; Li, S.; Liang, Z. Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Hortic. Res. 2021, 8, 52. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.-J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471–482. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Park, J.-J.; Chen, M.; Cong, L.; Ge, Y.; Jiang, Q.; Debnath, S.; Li, G.; Wen, J.; Wang, Z. Improving the genome editing efficiency of CRISPR/Cas9 in Arabidopsis and Medicago truncatula. Planta 2020, 252, 15. [Google Scholar] [CrossRef]
- Christie, J.M. Phototropin blue-light receptors. Annu Rev Plant Biol 2007, 58, 21–45. [Google Scholar] [CrossRef]
- Kagawa, T.; Sakai, T.; Suetsugu, N.; Oikawa, K.; Ishiguro, S.; Kato, T.; Tabata, S.; Okada, K.; Wada, M. Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 2001, 291, 2138–2141. [Google Scholar] [CrossRef]
- Sakai, T.; Kagawa, T.; Kasahara, M.; Swartz, T.E.; Christie, J.M.; Briggs, W.R.; Wada, M.; Okada, K. Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6969–6974. [Google Scholar] [CrossRef]
- Jarillo, J.A.; Gabrys, H.; Capel, J.; Alonso, J.M.; Ecker, J.R.; Cashmore, A.R. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 2001, 410, 952–954. [Google Scholar] [CrossRef]
- Kim, M.J.; Moon, Y.; Tou, J.C.; Mou, B.; Waterland, N.L. Nutritional value, bioactive compounds and health benefits of lettuce (Lactuca sativa L.). J. Food Compos. Anal. 2016, 49, 19–34. [Google Scholar] [CrossRef]
- López, A.; Javier, G.-A.; Fenoll, J.; Hellín, P.; Flores, P. Chemical composition and antioxidant capacity of lettuce: Comparative study of regular-sized (Romaine) and baby-sized (Little Gem and Mini Romaine) types. J. Food Compos. Anal. 2014, 33, 39–48. [Google Scholar] [CrossRef]
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Bertier, L.D.; Ron, M.; Huo, H.; Bradford, K.J.; Britt, A.B.; Michelmore, R.W. High-resolution analysis of the efficiency, heritability, and editing outcomes of CRISPR/Cas9-induced modifications of NCED4 in lettuce (Lactuca sativa). G3 Genes Genomes Genet. 2018, 8, 1513–1521. [Google Scholar] [CrossRef]
- Luo, C.; Wang, S.; Ning, K.; Chen, Z.; Wang, Y.; Yang, J.; Wang, Q. LsAP2 regulates leaf morphology by inhibiting CIN-like TCP transcription factors and repressing LsKAN2 in lettuce. Hortic. Res. 2021, 8, 184. [Google Scholar] [CrossRef]
- Nguyen, C.D.; Li, J.; Mou, B.; Gong, H.; Huo, H. A case study of using an efficient CRISPR/Cas9 system to develop variegated lettuce. Veg. Res. 2021, 1, 4. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.-T.; Ryu, J.; Choi, M.K.; Kweon, J.; Kang, B.-C.; Ahn, H.-M.; Bae, S.; Kim, J.; Kim, J.-S. A simple, flexible and high-throughput cloning system for plant genome editing via CRISPR-Cas system. J. Integr. Plant Biol. 2016, 58, 705–712. [Google Scholar] [CrossRef]
- Suetsugu, N.; Kagawa, T.; Wada, M. An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 2005, 139, 151–162. [Google Scholar] [CrossRef]
- Hamdan, M.F.; Karlson, C.K.S.; Teoh, E.Y.; Lau, S.-E.; Tan, B.C. Genome editing for sustainable crop improvement and mitigation of biotic and abiotic stresses. Plants 2022, 11, 2625. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Briggs, W.R.; Beck, C.F.; Cashmore, A.R.; Christie, J.M.; Hughes, J.; Jarillo, J.A.; Kagawa, T.; Kanegae, H.; Liscum, E.; Nagatani, A.; et al. The phototropin family of photoreceptors. Plant Cell 2001, 13, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-W.; Rothfels, C.J.; Melkonian, M.; Villarreal, J.C.; Stevenson, D.W.; Graham, S.W.; Wong, G.K.-S.; Mathews, S.; Pryer, K.M. The origin and evolution of phototropins. Front. Plant Sci. 2015, 6, 637. [Google Scholar] [CrossRef]
- Wada, M.; Kong, S.-G. Analysis of chloroplast movement and relocation in Arabidopsis. In Chloroplast Research in Arabidopsis; Springer: Berlin/Heidelberg, Germany, 2011; pp. 87–102. [Google Scholar]
- Kasahara, M.; Kagawa, T.; Oikawa, K.; Suetsugu, N.; Miyao, M.; Wada, M. Chloroplast avoidance movement reduces photodamage in plants. Nature 2002, 420, 829–832. [Google Scholar] [CrossRef]
- Gotoh, E.; Suetsugu, N.; Yamori, W.; Ishishita, K.; Kiyabu, R.; Fukuda, M.; Higa, T.; Shirouchi, B.; Wada, M. Chloroplast accumulation response enhances leaf photosynthesis and plant biomass production. Plant Physiol. 2018, 178, 1358–1369. [Google Scholar] [CrossRef]
- Chen, H.; Nelson, R.S.; Sherwood, J.L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 1994, 16, 664–668. [Google Scholar]
- Ismail, H.; Dilshad, E.; Waheed, M.T.; Sajid, M.; Kayani, W.K.; Mirza, B. Transformation of Lactuca sativa L. with rol C gene results in increased antioxidant potential and enhanced analgesic, anti-inflammatory and antidepressant activities in vivo. 3 Biotech 2016, 6, 215. [Google Scholar] [CrossRef]
- Edwards, K.; Johnstone, C.; Thompson, C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991, 19, 1349. [Google Scholar] [CrossRef]
- Riu, Y.-S.; Song, H.-G.; Kim, H.-S.; Kong, S.-G. Guard-cell-specific expression of phototropin2 C-terminal fragment enhances leaf transpiration. Plants 2021, 11, 65. [Google Scholar] [CrossRef]

| Transgenic Line (R0) | NheI Digestion | Mutation Type | Chloroplast Positioning | Indel Pattern |
|---|---|---|---|---|
| phot2-At1 | +/− | Mo | Av | phot2-At1 |
| phot2-At2 | +/− | Mo | Av | phot2-At2 |
| phot2-At3 | − | Bi | Ac | phot2-At3 |
| phot2-At4 | − | Bi | Ac | phot2-At4 |
| phot2-At5 | + | WT | Av | WT |
| phot2-At6 | + | WT | Av | WT |
| phot2-At7 | + | WT | Av | WT |
| phot2-At8 | + | WT | Av | WT |
| phot2-At9 | +/− | Mo | Av | phot2-At1 |
| phot2-At10 | + | WT | Av | WT |
| phot2-At11 | + | WT | Av | WT |
| phot2-At12 | +/− | Mo | Av | phot2-At2 |
| phot2-At13 | + | WT | Av | WT |
| phot2-At14 | +/− | Mo | Av | phot2-At1 |
| phot2-At15 | +/− | Mo | Av | phot2-At1 |
| phot2-At16 | + | WT | Av | WT |
| phot2-At17 | − | Bi | Ac | phot2-At3 |
| phot2-At18 | + | WT | Av | WT |
| phot2-At19 | − | Bi | Ac | phot2-At4 |
| phot2-At20 | + | WT | Av | WT |
| phot2-At21 | + | WT | Av | WT |
| phot2-Ls1 | +/− | Mo | Av | phot2-Ls1 |
| phot2-Ls2 | +/− | Mo | Av | phot2-Ls2 |
| phot2-Ls3 | +/− | Mo | Av | phot2-Ls3 |
| phot2-Ls4 | − | Bi | Ac | phot2-Ls4 |
| phot2-Ls5 | − | Bi | Ac | phot2-Ls5 |
| phot2-Ls6 | − | Bi | Ac | phot2-Ls6 |
| phot2-Ls7 | − | Bi | Ac | phot2-Ls7 |
| phot2-Ls8 | + | WT | Av | WT |
| phot2-Ls9 | +/− | Mo | Av | phot2-Ls2 |
| phot2-Ls10 | − | Bi | Ac | phot2-Ls4 |
| phot2-Ls11 | + | WT | Av | WT |
| phot2-Ls12 | + | WT | Av | WT |
| phot2-Ls13 | − | Bi | Ac | phot2-Ls4 |
| phot2-Ls14 | +/− | Mo | Av | phot2-Ls3 |
| phot2-Ls15 | − | Bi | Ac | phot2-Ls6 |
| phot2-Ls16 | + | WT | Av | WT |
| phot2-Ls17 | +/− | Mo | Av | phot2-Ls3 |
| phot2-Ls18 | + | WT | Av | WT |
| phot2-Ls19 | − | Bi | Ac | phot2-Ls5 |
| phot2-Ls20 | + | WT | Av | WT |
| phot2-Ls21 | +/− | Mo | Av | phot2-Ls2 |
| phot2-Ls22 | − | Bi | Ac | phot2-Ls6 |
| Construct | Number of Transgenic Plants (R0 Generation) | Number of Transgenic Plants with Mutation | Mutation Frequency (%) | Mutation Type | |
|---|---|---|---|---|---|
| Number of Monoallelic (%) | Number of Biallelic (%) | ||||
| pHAtC | 21 | 10 | 48 | 6 (29) | 4 (19) |
| pHLsC | 22 | 16 | 73 | 7 (32) | 9 (41) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riu, Y.-S.; Kim, G.H.; Chung, K.W.; Kong, S.-G. Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants 2023, 12, 878. https://doi.org/10.3390/plants12040878
Riu Y-S, Kim GH, Chung KW, Kong S-G. Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants. 2023; 12(4):878. https://doi.org/10.3390/plants12040878
Chicago/Turabian StyleRiu, Young-Sun, Gwang Hoon Kim, Ki Wha Chung, and Sam-Geun Kong. 2023. "Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter" Plants 12, no. 4: 878. https://doi.org/10.3390/plants12040878
APA StyleRiu, Y.-S., Kim, G. H., Chung, K. W., & Kong, S.-G. (2023). Enhancement of the CRISPR/Cas9-Based Genome Editing System in Lettuce (Lactuca sativa L.) Using the Endogenous U6 Promoter. Plants, 12(4), 878. https://doi.org/10.3390/plants12040878






