A Novel and Highly Inclusive Quantitative Real-Time RT-PCR Method for the Broad and Efficient Detection of Grapevine Leafroll-Associated Virus 1
Abstract
:1. Introduction
2. Results
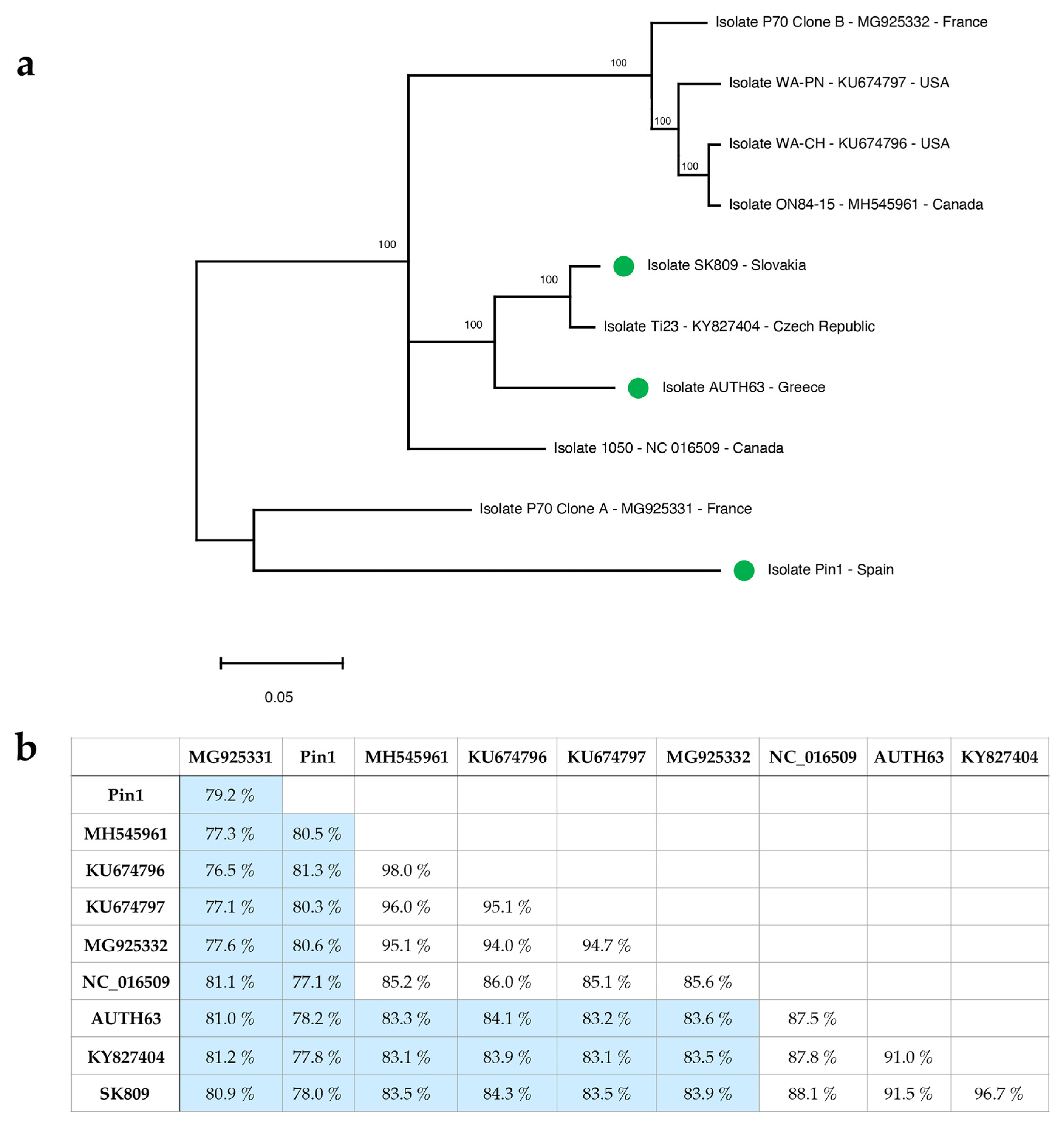
2.1. Molecular Characterization of Three GLRaV-1 near Full-Length Genomic Sequences by High-Throughput Sequencing (HTS)
2.2. GLRaV-1 Genetic Variability Analysis: Diagnostic Implications
2.3. Design of New and Highly Specific GLRaV-1 Primers and Probe
2.4. Detection of GLRaV-1 by Duplex Quantitative Real-Time RT-PCR
2.5. Validation of the Designed GLRaV-1 Real-Time RT-PCR
2.5.1. Validation of the Plant Internal Control
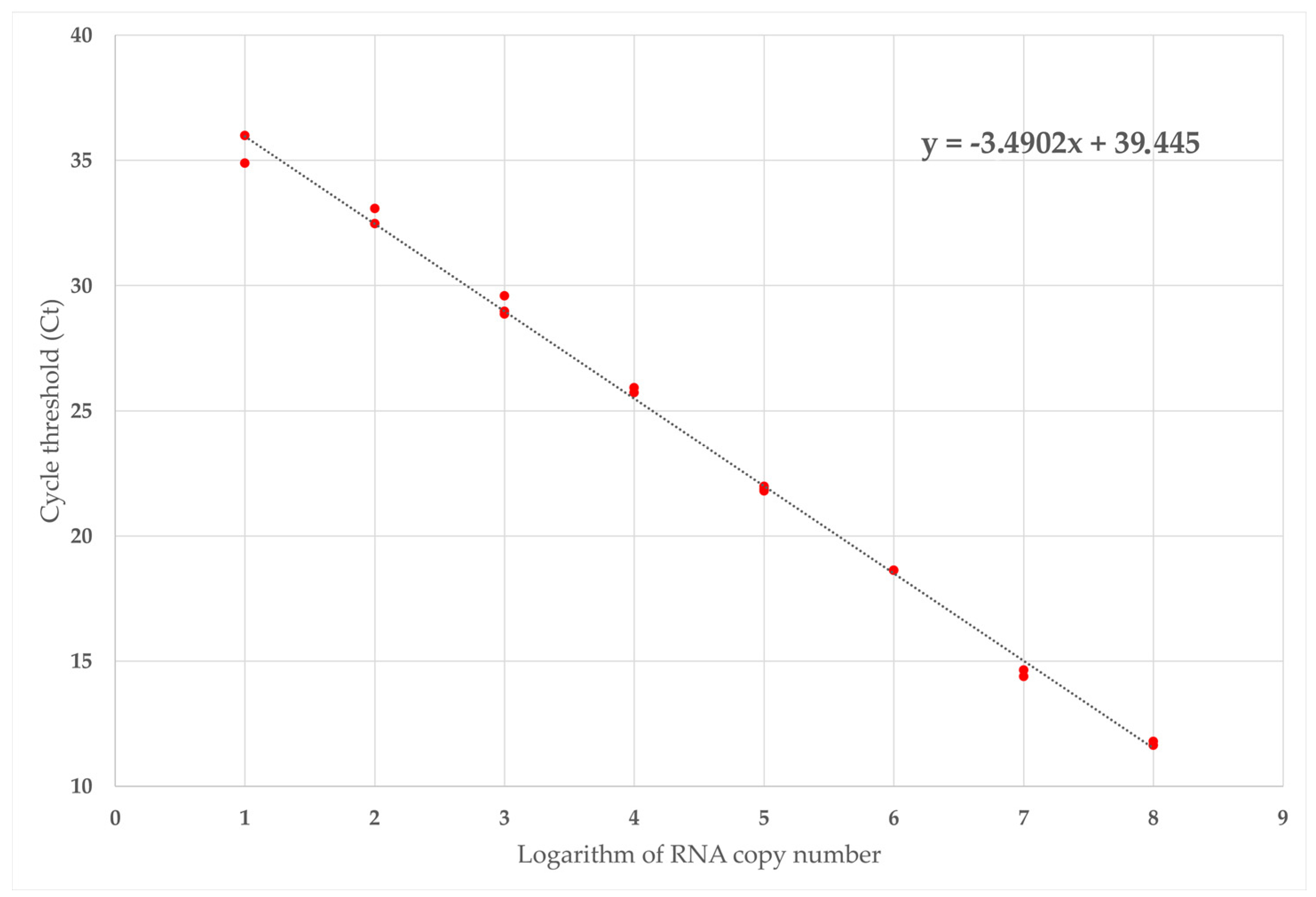
2.5.2. Technical Sensitivity and Absolute Quantification
2.5.3. Analytical Specificity and Selectivity
2.5.4. Repeatability and Reproducibility
2.6. Performance of the New Real-Time RT-PCR Method for GLRaV-1 Diagnosis in Field Samples
2.7. Quantitative Detection of GLRaV-1 in Planococcus Citri
3. Discussion
4. Material and Methods
4.1. Plant and Insect Material
4.2. Sample Preparation and RNA Purification
4.3. HTS Analysis and Genome Recovery
4.4. GLRaV-1 Detection Using Previously Reported RT-PCR Protocols
4.5. Phylogenetic Analysis of GLRaV-1 Full-Length Genomes
4.6. In Silico Evaluation of GLRaV-1 Detection Methods Specificity
4.7. GLRaV-1-Specific Primers and Probe Design
4.8. TaqMan Quantitative Real-Time GLRaV-1 RT-PCR Detection Method
4.9. Absolute Quantitation and Evaluation of Sensititvity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuchs, M. Grapevine Viruses: A Multitude of Diverse Species with Simple but Overall Poorly Adopted Management Solutions in the Vineyard. J. Plant Pathol. 2020, 102, 643–653. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Alabi, O.J.; Hwang, M.S.; Tian, T.; Mollov, D.; Golino, D. Characterization of a New Nepovirus Infecting Grapevine. Plant Dis. 2021, 105, 1432–1439. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Li, C.; Zhang, Z.; Ren, F.; Hu, G.; Shen, H.; Zhang, B.; Dong, Y. Identification and Characterization of a Novel Emaravirus from Grapevine Showing Chlorotic Mottling Symptoms. Front. Microbiol. 2021, 12, 694601. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, Z.; Li, C.; Ren, F.; Hu, G.; Zhang, B.; Dong, Y. High-Throughput Sequencing Indicates a Novel Marafivirus in Grapevine Showing Vein-Clearing Symptoms. Plants 2021, 10, 1487. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.F.; Fajardo, T.V.M.; Saldarelli, P. Grapevine Virus Diseases: Economic Impact and Current Advances in Viral Prospection and Management. Rev. Bras. Frutic. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Mannini, F.; Digiaro, M. The Effects of Viruses and Viral Diseases on Grapes and Wine. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 453–482. ISBN 9783319577067. [Google Scholar]
- Martelli, G.P. An Overview on Grapevine Viruses, Viroids, and the Diseases They Cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 31–46. ISBN 9783319577067. [Google Scholar]
- Maliogka, V.I.; Martelli, G.P.; Fuchs, M.; Katis, N.I. Control of Viruses Infecting Grapevine. Adv. Virus Res. 2015, 91, 175–227. [Google Scholar] [CrossRef]
- Poojari, S.; Boulé, J.; DeLury, N.; Lowery, D.T.; Rott, M.; Schmidt, A.M.; Úrbez-Torres, J.R. Epidemiology and Genetic Diversity of Grapevine Leafroll-Associated Viruses in British Columbia. Plant Dis. 2017, 101, 2088–2097. [Google Scholar] [CrossRef] [Green Version]
- Elçi, E. Genomic Variability and Recombination Analysis of Grapevine Leafroll-Associated Virus-1 Isolates from Turkey. J. Agric. Sci. 2019, 25, 319–327. [Google Scholar] [CrossRef]
- Habili, N.; Komínek, P.; Little, A. Grapevine Leafroll-Associated Virus 1 as a Common Grapevine Pathogen. Plant Viruses 2007, 1, 63–68. [Google Scholar]
- Predajňa, L.; Gažiová, A.; Holovičová, E.; Glasa, M. Analysis of a Short Genomic Region of Grapevine Leafroll-Associated Virus 1 (GLRaV-1) Reveals the Presence of Two Different Molecular Groups of Isolates in Slovakia. Acta Virol. 2013, 57, 353–356. [Google Scholar] [CrossRef]
- Herrbach, E.; Alliaume, A.; Prator, C.A.; Daane, K.M.; Cooper, M.L.; Almeida, R.P.P. Vector Transmission of Grapevine Leafroll-Associated Viruses. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International: Berlin/Heidelberg, Germany, 2017; pp. 483–503. ISBN 19577067_24. [Google Scholar]
- Hommay, G.; Alliaume, A.; Reinbold, C.; Herrbach, E. Transmission of Grapevine leafroll-associated virus-1 (Ampelovirus) and Grapevine virus A (Vitivirus) by the Cottony Grape Scale, Pulvinaria vitis (Hemiptera: Coccidae). Viruses 2021, 15, 2081. [Google Scholar] [CrossRef]
- Bertin, S.; Pacifico, D.; Cavalieri, V.; Marzachi, C.; Bosco, D. Transmission of Grapevine virus A and Grapevine leafroll-associated viruses 1 and 3 by Planococcus ficus and Planococcus citri fed on mixed-infected plants. Ann. Appl. Biol. 2016, 169, 53–63. [Google Scholar] [CrossRef]
- Donda, B.P.; Jarugula, S.; Naidu, R.A. An Analysis of the Complete Genome Sequence and Subgenomic RNAs Reveals Unique Features of the Ampelovirus, Grapevine leafroll-associated virus 1. Phytopathology 2017, 107, 1069–1079. [Google Scholar] [CrossRef]
- Esteves, F.; Teixeira Santos, M.; Eiras-Dias, J.E.; Fonseca, F. Molecular Data Mining to Improve Antibody-Based Detection of Grapevine leafroll-associated virus 1 (GLRaV-1). J. Virol. Methods 2013, 194, 258–270. [Google Scholar] [CrossRef]
- Kumar, S.; Sawant, S.D.; Sawant, I.S.; Prabha, K.; Jain, R.K.; Baranwal, V.K. First Report of Grapevine leafroll-associated virus 1 Infecting Grapevines in India. Plant Dis. 2012, 96, 1828. [Google Scholar] [CrossRef]
- Immanuel, T.M.; Delmiglio, C.; Ward, L.I.; Denton, J.O.; Clover, G.R.G. First Reports of Grapevine virus A, Grapevine fleck virus, and Grapevine leafroll-associated virus 1 in the United Kingdom. Plant Dis. 2015, 99, 898. [Google Scholar] [CrossRef]
- Zongoma, A.M.; Dangora, D.B.; Al Rwahnih, M.; Bako, S.P.; Alegbejo, M.D.; Alabi, O.J. First Report of Grapevine leafroll-associated virus 1 Infecting Grapevines (Vitis Spp.) in Nigeria. Plant Dis. 2017, 102, 258. [Google Scholar] [CrossRef]
- Karthikeyan, G.; Alabi, O.J.; Naidu, R.A. Occurrence of Grapevine leafroll-associated virus 1 in Two Ornamental Grapevine Cultivars in Washington State. Plant Dis. 2011, 95, 613. [Google Scholar] [CrossRef]
- Escobar, P.F.; Fiore, N.; Valenzuela, P.D.T.; Engel, E.A. First Detection of Grapevine leafroll-associated virus 4 in Chilean Grapevines. Plant Dis. 2008, 92, 1474. [Google Scholar] [CrossRef] [Green Version]
- Hommay, G.; Wiss, L.; Reinbold, C.; Chadoeuf, J.; Herrbach, E. Spatial Distribution Patterns of Parthenolecanium corni (Hemiptera, Coccidae) and of the Ampelovirus GLRaV-1 and the Vitivirus GVA in a Commercial Vineyard. Viruses 2020, 12, 1447. [Google Scholar] [CrossRef]
- Messmer, N.; Bohnert, P.; Schumacher, S.; Fuchs, R. Studies on the Occurrence of Viruses in Planting Material of Grapevines in Southwestern Germany. Viruses 2021, 13, 248. [Google Scholar] [CrossRef]
- Naidu, R.A.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine leafroll: A complex viral disease affecting a high-value fruit crop. Plant Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef] [Green Version]
- Martelli, G.P. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Gambino, G.; Gribaudo, I. Simultaneous Detection of Nine Grapevine Viruses by Multiplex Reverse Transcription-Polymerase Chain Reaction with Coamplification of a Plant RNA as Internal Control. Phytopathology 2006, 96, 1223–1229. [Google Scholar] [CrossRef] [Green Version]
- Little, A.; Rezaian, M.A. Improved Detection of Grapevine Leafroll-Associated Virus 1 by Magnetic Capture Hybridisation RT-PCR on a Conserved Region of Viral RNA. Arch. Virol. 2006, 151, 753–761. [Google Scholar] [CrossRef]
- Alabi, O.J.; Al Rwahnih, M.; Karthikeyan, G.; Poojari, S.; Fuchs, M.; Rowhani, A.; Naidu, R.A. Grapevine Leafroll-Associated Virus 1 Occurs as Genetically Diverse Populations. Phytopathology 2011, 101, 1446–1456. [Google Scholar] [CrossRef] [Green Version]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Real-Time RT-PCR (TaqMan®) Assays for the Detection of Grapevine leafroll associated viruses 1–5 and 9. J. Virol. Methods 2007, 141, 22–29. [Google Scholar] [CrossRef]
- Bertolini, E.; García, J.; Yuste, A.; Olmos, A. High Prevalence of Viruses in Table Grape from Spain Detected by Real-Time RT-PCR. Eur J. Plant Pathol. 2010, 128, 283–287. [Google Scholar] [CrossRef]
- Pacifico, D.; Caciagli, P.; Palmano, S.; Mannini, F.; Marzachì, C. Quantitation of Grapevine leafroll associated virus-1 and -3, Grapevine virus A, Grapevine fanleaf virus and Grapevine fleck virus in Field-Collected Vitis vinifera L. ‘Nebbiolo’ by Real-Time Reverse Transcription-PCR. J. Virol. Methods 2011, 172, 1–7. [Google Scholar] [CrossRef]
- López-Fabuel, I.; Wetzel, T.; Bertolini, E.; Bassler, A.; Vidal, E.; Torres, L.B.; Yuste, A.; Olmos, A. Real-Time Multiplex RT-PCR for the Simultaneous Detection of the Five Main Grapevine Viruses. J. Virol. Methods 2013, 188, 21–24. [Google Scholar] [CrossRef]
- Dubiela, C.R.; Fajardo, T.V.M.; Souto, E.R.; Nickel, O.; Eiras, M.; Fernando Revers, L. Simultaneous Detection of Brazilian Isolates of Grapevine Viruses by TaqMan Real-Time RT-PCR. Trop. Plant Pathol. 2013, 38, 158–165. [Google Scholar] [CrossRef] [Green Version]
- Bruisson, S.; Lebel, S.; Walter, B.; Prevotat, L.; Seddas, S.; Schellenbaum, P. Comparative Detection of a Large Population of Grapevine Viruses by TaqMan® RT-QPCR and ELISA. J. Virol. Methods 2017, 240, 73–77. [Google Scholar] [CrossRef]
- Aloisio, M.; Morelli, M.; Elicio, V.; Saldarelli, P.; Ura, B.; Bortot, B.; Severini, G.M.; Minafra, A. Detection of Four Regulated Grapevine Viruses in a Qualitative, Single Tube Real-Time PCR with Melting Curve Analysis. J. Virol. Methods 2018, 257, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Sabella, E.; Pierro, R.; Luvisi, A.; Panattoni, A.; D’Onofrio, C.; Scalabrelli, G.; Nutricati, E.; Aprile, A.; de Bellis, L.; Materazzi, A. Phylogenetic Analysis of Viruses in Tuscan Vitis vinifera sylvestris (Gmeli) Hegi. PLoS ONE 2018, 13, e0200875. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Hong, N.; Dong, Y.; Ma, Y.; Zhang, Z.P.; Ren, F.; Hu, G.; Zhou, J.; Wang, G. Genetic Diversity and Recombination Analysis of Grapevine leafroll-associated virus 1 from China. Arch. Virol. 2015, 160, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Katsiani, A.; Maliogka, V.I.; Katis, N.; Svanella-Dumas, L.; Olmos, A.; Ruiz-García, A.B.; Marais, A.; Faure, C.; Theil, S.; Lotos, L.; et al. High-Throughput Sequencing Reveals Further Diversity of Little Cherry Virus 1 with Implications for Diagnostics. Viruses 2018, 10, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Lara, A.; Erickson, T.M.; Golino, D.; Al Rwahnih, M. Development of a Universal RT-PCR Assay for Grapevine Vitiviruses. PLoS ONE 2020, 15, e0239522. [Google Scholar] [CrossRef]
- EPPO. PM 7/98 (5) Specific Requirements for Laboratories Preparing Accreditation for a Plant Pest Diagnostic Activity. EPPO Bulletin 2021, 51, 468–498. [Google Scholar] [CrossRef]
- Morán, F.; Olmos, A.; Lotos, L.; Predajňa, L.; Katis, N.; Glasa, M.; Maliogka, V.; Ruiz-García, A.B. A Novel Specific Duplex Real-Time RT-PCR Method for Absolute Quantitation of Grapevine Pinot gris virus in Plant Material and Single Mites. PLoS ONE 2018, 13, e0197237. [Google Scholar] [CrossRef]
- Caruso, A.G.; Bertacca, S.; Ragona, A.; Matić, S.; Davino, S.; Panno, S. Epidemiological Survey of Grapevine Leafroll-Associated Virus 1 and 3 in Sicily (Italy): Genetic Structure and Molecular Variability. Agriculture 2022, 12, 647. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Comparison of low density arrays, RT-PCR and real-time RT-PCR TaqMan-assays for the detection of Grapevine leafroll-associated viruses 1-5 and 9. J. Virol. Methods 2008, 149, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Sefc, K.M.; Leonhadrt, W.; Steinkellner, H. Partial sequence identification of Grapevine leafroll-associated virus-1 and development of a highly sensitive IC-RT-PCR detection method. J. Virol. Methods 2000, 86, 101–106. [Google Scholar] [CrossRef]
- Margaria, P.; Turina, M.; Palmano, S. Detection of Flavescence dorée and Bois noir phytoplasmas, Grapevine leafroll-associated virus-1 and -3 and Grapevine virus A from the same crude extract by reverse transcription-Real Time TaqMan assays. Plant Pathol. 2009, 58, 838–845. [Google Scholar] [CrossRef]
- Naidu, R.A. Grapevine leafroll-associated virus 1. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 128–139. ISBN 9783319577067. [Google Scholar]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of HTS for routine plant virus diagnostics: State of the art and challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maliogka, V.I.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.B.; Glasa, M.; Katis, N.; Olmos, A. Recent advances on detection and characterization of fruit tree viruses using high-throughput sequencing technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villamor, D.E.V.; Ho, T.; Al Rwahnih, M.; Martin, R.R.; Tzanetkis, I.E. High throughput sequencing for plant virus detection and discovery. Phytopathology 2019, 109, 716–725. [Google Scholar] [CrossRef]
- Mehetre, G.T.; Leo, V.V.; Singh, G.; Sorokan, A.; Maksimov, I.; Yadav, M.K.; Upadhyaya, K.; Hashem, A.; Alsaleh, A.N.; Dawoud, T.M.; et al. Current Developments and Challenges in Plant Viral Diagnostics: A Systematic Review. Viruses 2021, 13, 412. [Google Scholar] [CrossRef]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Avogadro, A. Essai d’une manière de determiner les masses relatives des molecules elementaires des corps, et les proportions se lon lesquelles elles entrent dans ces combinaisons (Essay on a manner of determinig the relative masses of the elementary molecules of bodies, and the proportions in which they enter into these compounds). J. Phys. 1811, 73, 58. (In French) [Google Scholar]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [Green Version]
- Komínek, P.; Glasa, M.; Bryxiová, M. Analysis of the molecular variability of Grapevine leafroll-associated virus 1 reveals the presence of two distinct virus groups and their mixed occurrence in grapevines. Virus Genes 2005, 31, 247–255. [Google Scholar] [CrossRef] [PubMed]


| Sample | |||
|---|---|---|---|
| Number | PIN1 | AUTH63 | SK809 |
| Total raw reads | 60,630,138 | 62,528,966 | 1,471,284 |
| Reads after QC | 60,597,198 | 40,061,946 | 1,269,123 |
| Reads after grapevine genome subtraction | 8,642,008 | 827,368 | 8360 |
| GLRaV-1 related contigs | 1 | 4 | 1 |
| Number of Target Copies | Ct Value Replicate 1 | Ct Value Replicate 2 | Ct Value Replicate 3 | Average ± SD |
|---|---|---|---|---|
| 4.4 × 108 | 12.28 | 11.55 | 11.69 | 11.84 ± 0.39 |
| 4.4 × 107 | 14.55 | 14.01 | 14.29 | 14.28 ± 0.27 |
| 4.4 × 106 | 19.30 | 18.53 | 18.53 | 18.79 ± 0.44 |
| 4.4 × 105 | 21.89 | 21.71 | 21.78 | 21.79 ± 0.09 |
| 4.4 × 104 | 25.91 | 25.82 | 25.63 | 25.79 ± 0.14 |
| 4.4 × 103 | 28.77 | 28.88 | 29.50 | 29.05 ± 0.39 |
| 4.4 × 102 | 32.98 | 32.37 | 33.46 | 32.94 ± 0.55 |
| 4.4 × 10 | 34.80 | 35.90 | 35.59 | 35.43 ± 0.57 |
| Sample | Origin | Virome (1) |
|---|---|---|
| 33.17 | Spain | GLRaV-3; GRSPaV; GRVFV; GAMaV; GFkV; GVA; GYSVd-1 |
| 33.24 | Spain | GLRaV-4; GLRaV-3; GYSVd-1 |
| 33.28 | Spain | GLRaV-3; GRSPaV; GRVFV; GAMaV; GVA; GYSVd-1 |
| 33.35 | Spain | GLRaV-4; GLRaV-3; GYSVd-1; GRVFV |
| 33.47 | Spain | GLRaV-3; GRSPaV; GRVFV; GYSVd-1 |
| 30T | Spain | GLRaV-3; GLRaV-2; GLRaV-4; GRSPaV; GVA; GFkV; GPGV; GYSVd-1 |
| U24 | Spain | GLRaV-3; GLRaV-4; GRSPaV; HSVd; GYSVd-1 |
| 29.9 | Greece | GLRaV-2; GLRaV-4; GRLDaV; GRVFV; HSVd; GYSVd-1 |
| Samples | StepOne Plus | QuantStudio | Roche 480 | All Equipments | ||||
|---|---|---|---|---|---|---|---|---|
| Mean Ct ± SD | CV (%) | Mean Ct ± SD | CV (%) | Mean Ct ± SD | CV (%) | Mean Ct ± SD | CV (%) | |
| 91.1 | 32.34 ± 2.74 | 8.5 | 27.15 ± 1.79 | 6.6 | 31.37 ± 1.55 | 5.0 | 30.29 ± 2.79 | 9.2 |
| 91.2 | 31.40 ± 1.40 | 4.5 | 29.33 ± 0.95 | 3.3 | 31.54 ± 0.50 | 1.6 | 30.76 ± 1.58 | 5.1 |
| 91.10 | 33.62 ± 1.01 | 3.0 | 32.43 ± 0.58 | 1.8 | 32.43 ± 1.71 | 5.3 | 32.83 ± 0.68 | 2.1 |
| 91.11 | 33.68 ± 1.69 | 5.0 | 30.61 ± 0.73 | 2.4 | 33.84 ± 0.68 | 2.0 | 32.71 ± 1.82 | 5.6 |
| 98.16 | 34.29 ± 0.98 | 2.9 | 33.64 ± 0.84 | 2.5 | 32.65 ± 0.75 | 2.3 | 33.53 ± 0.83 | 2.5 |
| 102.18 | 24.05 ± 1.91 | 7.9 | 21.49 ± 1.53 | 7.1 | 26.48 ± 1.71 | 6.5 | 24.00 ± 2.50 | 10.4 |
| 102.20 | 25.64 ± 1.08 | 4.2 | 20.48 ± 0.39 | 1.9 | 24.50 ± 0.49 | 2.0 | 23.54 ± 2.71 | 11.5 |
| Origin | Collection Date | Number of Samples | Positive | Negative |
|---|---|---|---|---|
| Utiel-Requena | 2015 | 48 | 5 (10.4%) | 43 (89.6%) |
| Priorato | 2016 | 29 | 0 (0.0%) | 29 (100.0%) |
| Manchuela | 2016 | 26 | 3 (11.5%) | 23 (88.5%) |
| Utiel-Requena | 2019 | 52 | 0 (0.0%) | 52 (100.0%) |
| Utiel-Requena | 2021 | 86 | 16 (18.6%) | 70 (81.4%) |
| Total | 241 | 24 (10.0%) | 217 (90.0%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morán, F.; Olmos, A.; Glasa, M.; Silva, M.B.D.; Maliogka, V.; Wetzel, T.; Ruiz-García, A.B. A Novel and Highly Inclusive Quantitative Real-Time RT-PCR Method for the Broad and Efficient Detection of Grapevine Leafroll-Associated Virus 1. Plants 2023, 12, 876. https://doi.org/10.3390/plants12040876
Morán F, Olmos A, Glasa M, Silva MBD, Maliogka V, Wetzel T, Ruiz-García AB. A Novel and Highly Inclusive Quantitative Real-Time RT-PCR Method for the Broad and Efficient Detection of Grapevine Leafroll-Associated Virus 1. Plants. 2023; 12(4):876. https://doi.org/10.3390/plants12040876
Chicago/Turabian StyleMorán, Félix, Antonio Olmos, Miroslav Glasa, Marilia Bueno Da Silva, Varvara Maliogka, Thierry Wetzel, and Ana Belén Ruiz-García. 2023. "A Novel and Highly Inclusive Quantitative Real-Time RT-PCR Method for the Broad and Efficient Detection of Grapevine Leafroll-Associated Virus 1" Plants 12, no. 4: 876. https://doi.org/10.3390/plants12040876
APA StyleMorán, F., Olmos, A., Glasa, M., Silva, M. B. D., Maliogka, V., Wetzel, T., & Ruiz-García, A. B. (2023). A Novel and Highly Inclusive Quantitative Real-Time RT-PCR Method for the Broad and Efficient Detection of Grapevine Leafroll-Associated Virus 1. Plants, 12(4), 876. https://doi.org/10.3390/plants12040876








