Transcriptomic and QTL Analysis of Seed Germination Vigor under Low Temperature in Weedy Rice WR04-6
Abstract
1. Introduction
2. Results
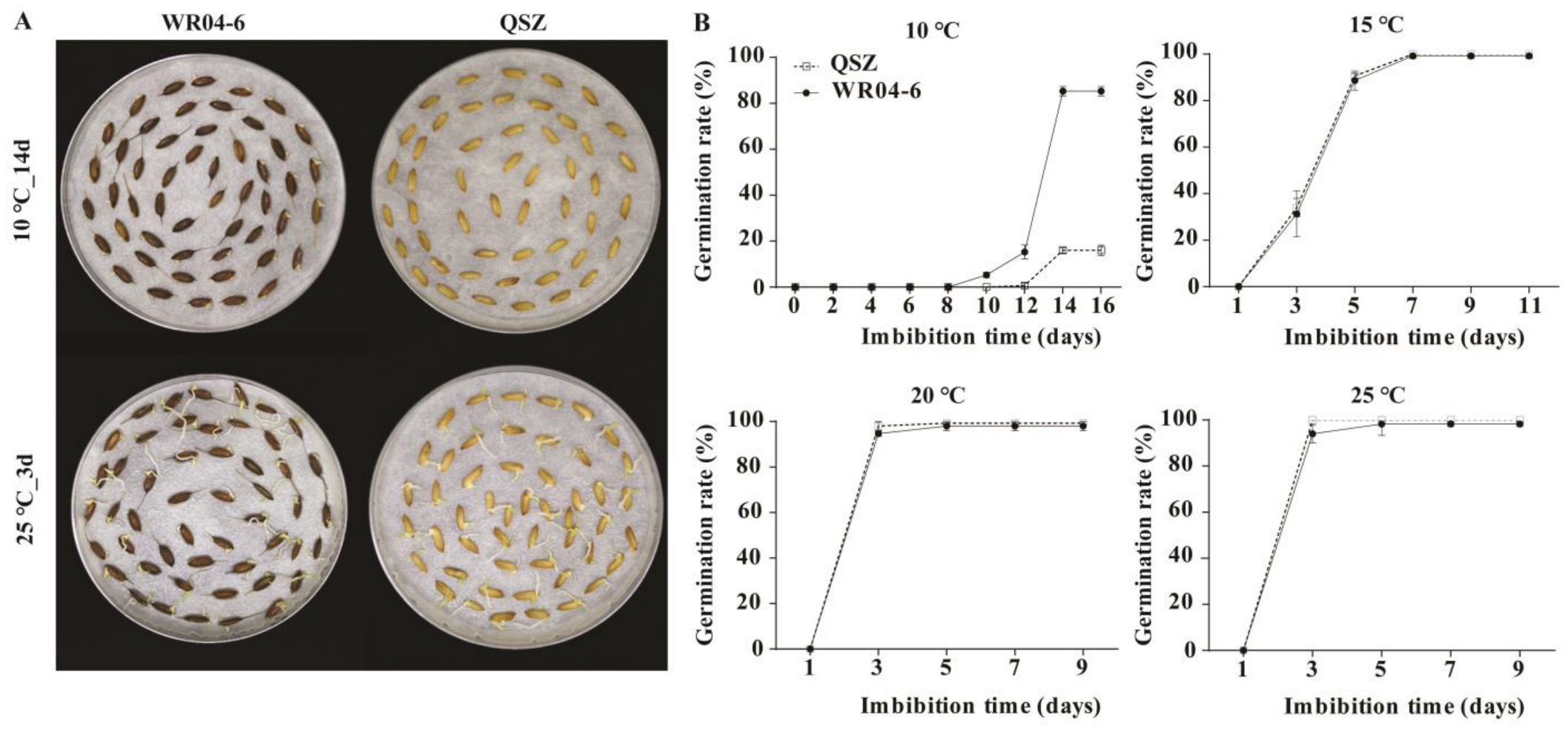
2.1. LTG Ability of WR04-6 and QSZ
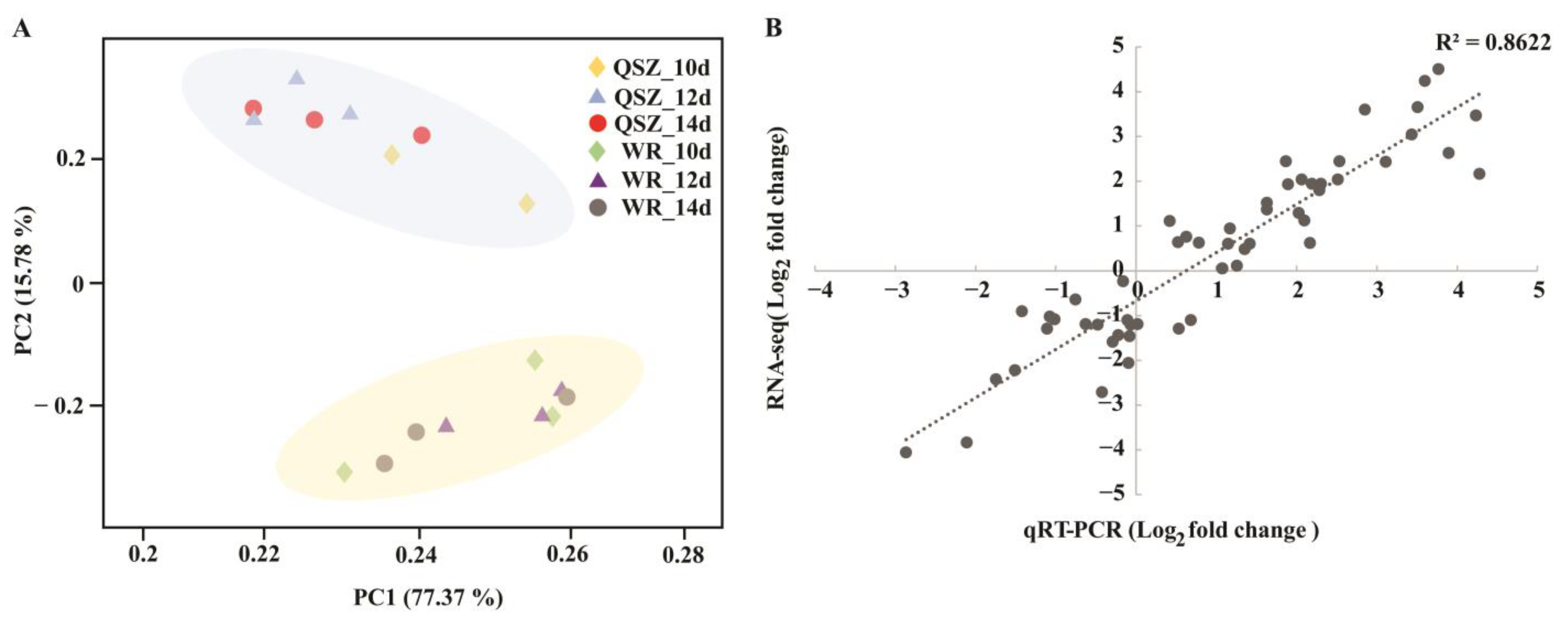
2.2. RNA-seq Data Overview
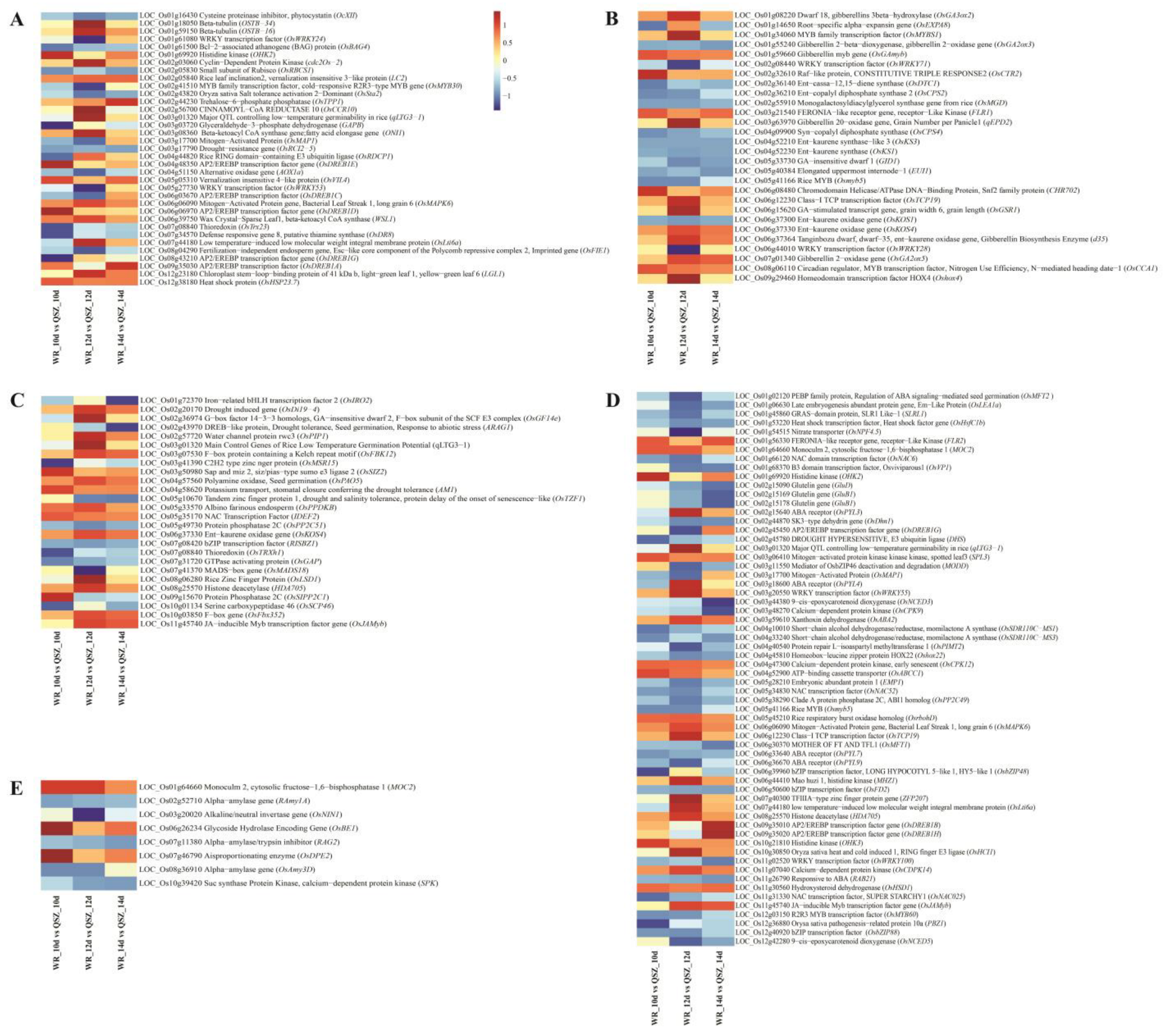
2.3. Functional Classification of DEGs of WR04-6 vs. QSZ during LTG
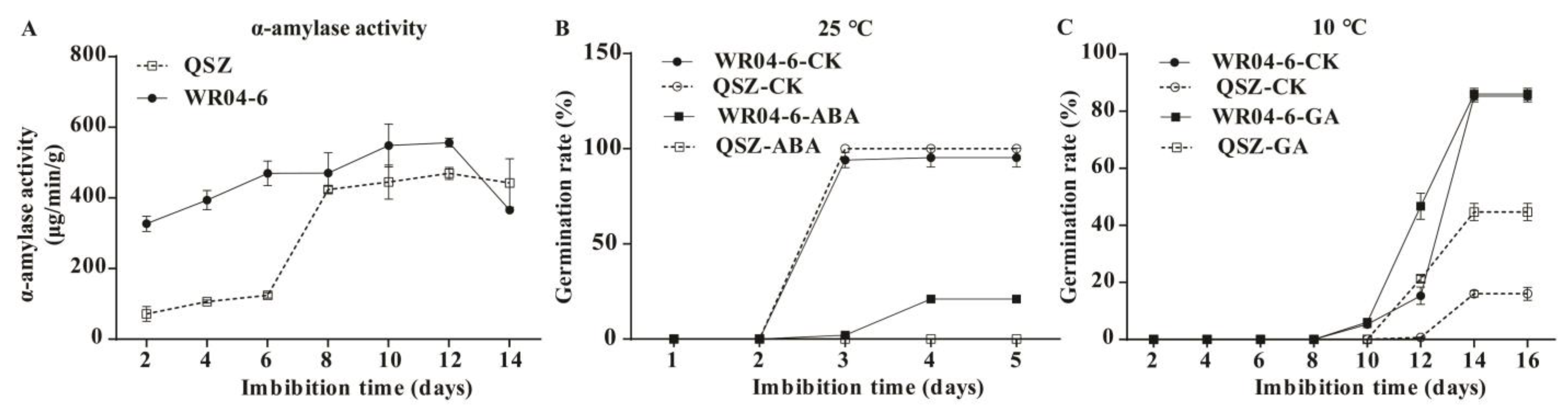
2.4. α-Amylase Activities of WR04-6 and QSZ during LTG
2.5. Effect of ABA and GA on Seed Germination in WR04-6 and QSZ
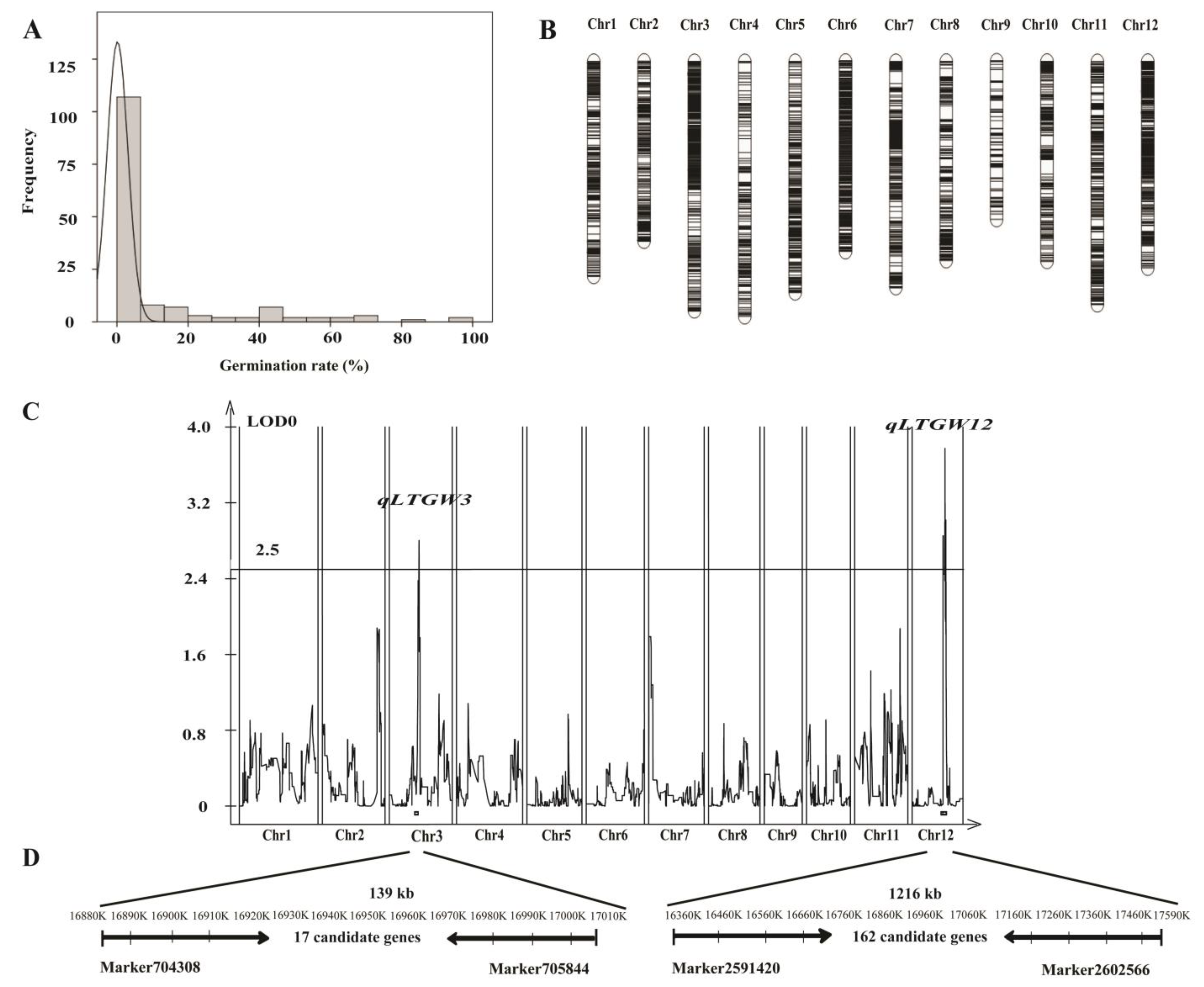
2.6. Identification of QTLs for LTG
2.7. Candidate Gene Prediction
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Assay of Seed Germination
4.3. RNA-seq
4.4. RNA Extraction and qRT-PCR
4.5. α-Amylase Activity Determination
4.6. Characteristics of Seed Germination under ABA or GA Treatment
4.7. QTL Mapping
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Kayess, M.O.; Hassan, M.M.; Nurhasan, M.; Ahmed, K. Effect of low temperature on chlorophyll and carotenoid content on the seedlings of some selected Boro rice varieties. Am. J. Plant Sci. 2020, 11, 125–136. [Google Scholar] [CrossRef]
- Byun, M.Y.; Cui, L.H.; Oh, T.K.; Jung, Y.-J.; Lee, A.; Park, K.Y.; Kang, B.G.; Kim, W.T. Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 16. [Google Scholar] [CrossRef]
- Xie, H.; Han, Y.; Li, X.; Dai, W.; Song, X.; Olsen, K.M.; Qiang, S. Climate-dependent variation in cold tolerance of weedy rice and rice mediated by OsICE1 promoter methylation. Mol. Ecol. 2020, 29, 121–137. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, V.; Saharawat, Y.; Gathala, M. Weedy rice: An emerging threat for direct-seeded rice production systems in India. Rice Res. 2013, 1, 1. [Google Scholar] [CrossRef]
- Rihan, H.Z.; Al-Issawi, M.; Fuller, M.P. Advances in physiological and molecular aspects of plant cold tolerance. J. Plant Interact. 2017, 12, 143–157. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H.; Sato, T.; Kiuchi, H.; Nonoue, Y.; Takeuchi, Y.; Ando, T.; Lin, S.Y.; Yano, M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 2004, 108, 794–799. [Google Scholar] [CrossRef]
- Cruz, R.P.d.; Sperotto, R.A.; Cargnelutti, D.; Adamski, J.M.; de FreitasTerra, T.; Fett, J.P. Avoiding damage and achieving cold tolerance in rice plants. Food Energy Secur. 2013, 2, 96–119. [Google Scholar] [CrossRef]
- Liu, C.; Schläppi, M.R.; Mao, B.; Wang, W.; Wang, A.; Chu, C. The bZIP 73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 2019, 17, 1834–1849. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H. Origins of functional nucleotide polymorphisms in a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Plant Mol. Biol. 2011, 75, 1–10. [Google Scholar] [CrossRef]
- Li, C.; Dong, S.; Beckles, D.M.; Miao, H.; Sun, J.; Liu, X.; Wang, W.; Zhang, S.; Gu, X. The qLTG1.1 candidate gene CsGAI regulates low temperature seed germination in cucumber. Theor. Appl. Genet. 2022, 135, 2593–2607. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Shi, S.; Shi, H.; Khanzada, H.; Wassan, G.M.; Zhu, C.; Peng, X.; Yu, Q.; Chen, X.; He, X.; et al. Mapping QTL for seed germinability under low temperature using a new high-density genetic map of rice. Front. Plant Sci. 2017, 8, 1223. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.; Tezuka, K.; Kawamoto, T.; Matsumoto, S.; Satoh-Nagasawa, N.; Ueda, K.; Sakurai, K.; Watanabe, A.; Takahashi, H.; Akagi, H. Identification of QTLs controlling low-temperature germination of the East European rice (Oryza sativa L.) variety Maratteli. Euphytica 2016, 207, 245–254. [Google Scholar] [CrossRef]
- Borjas, A.H.; De Leon, T.B.; Subudhi, P.K. Genetic analysis of germinating ability and seedling vigor under cold stress in US weedy rice. Euphytica 2016, 208, 251–264. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef] [PubMed]
- Shim, K.-C.; Kim, S.H.; Lee, H.-S.; Adeva, C.; Jeon, Y.-A.; Luong, N.H.; Kim, W.-J.; Akhtamov, M.; Park, Y.-J.; Ahn, S.-N. Characterization of a new qLTG3–1 allele for low-temperature germinability in rice from the wild species Oryza rufipogon. Rice 2020, 13, 10. [Google Scholar] [CrossRef]
- Lu, G.; Wu, F.Q.; Wu, W.; Wang, H.J.; Zheng, X.M.; Zhang, Y.; Chen, X.; Zhou, K.; Jin, M.; Cheng, Z.; et al. Rice LTG1 is involved in adaptive growth and fitness under low ambient temperature. Plant J. 2014, 78, 468–480. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Xie, K.; Wang, Y.; Liu, F.; Lin, Q.; Wang, W.; Yang, C.; Lu, B.; Liu, S.; et al. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.). Theor. Appl. Genet. 2013, 126, 2313–2322. [Google Scholar] [CrossRef]
- Yang, T.; Zhou, L.; Zhao, J.; Dong, J.; Liu, Q.; Fu, H.; Mao, X.; Yang, W.; Ma, Y.; Chen, L.; et al. The candidate genes underlying a stably expressed QTL for low temperature germinability in rice (Oryza sativa L.). Rice 2020, 13, 74. [Google Scholar] [CrossRef]
- Wang, X.; Zou, B.H.; Shao, Q.L.; Cui, Y.M.; Lu, S.; Zhang, Y.; Huang, Q.S.; Huang, J.; Hua, J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 2018, 3, 413–421. [Google Scholar] [CrossRef]
- Asano, K.; Yamasaki, M.; Takuno, S.; Miura, K.; Katagiri, S.; Ito, T.; Doi, K.; Wu, J.; Ebana, K.; Matsumoto, T. Artificial selection for a green revolution gene during japonica rice domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11034–11039. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L. Assessment of artificial selection in maize (Zea mays L.) and Asian rice (Oryza sativa L.) using QTL data. Genet. Resour. Crop Evol. 2017, 64, 1561–1568. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; Susan, M.C. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, M.; Zhang, G.; Liu, Z.; Hua, Y.; Jia, X.; Song, J.; Ma, D.; Sun, J. Weedy rice as a novel gene resource: A genome-wide association study of anthocyanin biosynthesis and an evaluation of nutritional quality. Front. Plant Sci. 2020, 11, 878. [Google Scholar] [CrossRef]
- Chauhan, B.S. Strategies to manage weedy rice in Asia. Crop Prot. 2013, 48, 51–56. [Google Scholar] [CrossRef]
- Sun, J.; Qian, Q.; Ma, D.R.; Xu, Z.J.; Liu, D.; Du, H.B.; Chen, W.F. Introgression and selection shaping the genome and adaptive loci of weedy rice in northern China. New Phytol. 2013, 197, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Li, L.F.; Li, Y.L.; Jia, Y.; Caicedo, A.L.; Olsen, K.M. Signatures of adaptation in the weedy rice genome. Nat. Genet. 2017, 49, 811. [Google Scholar] [CrossRef]
- Qiu, J.; Zhou, Y.; Mao, L.; Ye, C.; Wang, W.; Zhang, J.; Yu, Y.; Fu, F.; Wang, Y.; Qian, F.; et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Commun. 2017, 8, 15323. [Google Scholar] [CrossRef]
- Sun, J.; Ma, D.; Tang, L.; Zhao, M.; Zhang, G.; Wang, W.; Song, J.; Li, X.; Liu, Z.; Zhang, W.; et al. Population genomic analysis and de novo assembly reveal the origin of weedy rice as an evolutionary game. Mol. Plant. 2019, 12, 632–647. [Google Scholar] [CrossRef]
- Song, Z.J.; Wang, Z.; Feng, Y.; Yao, N.; Yang, J.; Lu, B.R. Genetic divergence of weedy rice populations associated with their geographic location and coexisting conspecific crop: Implications on adaptive evolution of agricultural weeds. J. Syst. Evol. 2015, 53, 330–338. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, G.; Cui, Z.; Kong, X.; Yu, X.; Gui, R.; Han, Y.; Li, Z.; Lang, H.; Hua, Y.; et al. Regain flood adaptation in rice through a 14-3-3 protein OsGF14h. Nat. Commun. 2022, 13, 5664. [Google Scholar] [CrossRef] [PubMed]
- Iwata, N.; Shinada, H.; Kiuchi, H.; Sato, T.; Fujino, K. Mapping of QTLs controlling seedling establishment using a direct seeding method in rice. Breed. Sci. 2010, 60, 353–360. [Google Scholar] [CrossRef]
- Okuno, K. Molecular mechanisms of cold tolerance in rice and wheat. Therm. Med. 2004, 20, 51–60. [Google Scholar] [CrossRef]
- Pipatpongpinyo, W.; Korkmaz, U.; Wu, H.; Kena, A.; Ye, H.; Feng, J.; Gu, X.-Y. Assembling seed dormancy genes into a system identified their effects on seedbank longevity in weedy rice. Heredity 2019, 124, 135–145. [Google Scholar] [CrossRef]
- Gross, B.L.; Michael, R.; Shih-Chung, H.; Caicedo, A.L.; Yulin, J.; Olsen, K.M. Seeing red: The origin of grain pigmentation in US weedy rice. Mol. Ecol. 2010, 19, 3380–3393. [Google Scholar] [CrossRef]
- Xiong, M.; Yu, J.; Wang, J.; Gao, Q.; Huang, L.; Chen, C.; Zhang, C.; Fan, X.; Zhao, D.; Liu, Q.-Q.; et al. Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 2022, 189, 402–418. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, X.; Zhou, D.; Ouyang, Y.; Yao, J. Overexpression of the 16-kDa α-amylase/trypsin inhibitor RAG2 improves grain yield and quality of rice. Plant Biotechnol. J. 2017, 15, 568–580. [Google Scholar] [CrossRef]
- Sugimoto, N.; Takeda, G.; Nagato, Y.; Yamaguchi, J. Temporal and Spatial Expression of the a-Amylase Gene during Seed Germination in Rice and Barley. Plant Cell Physiol. 1998, 39, 323–333. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, Y.; Hu, G.; Wang, X.; Chen, H.; Shi, Q.; Xiang, J.; Zhang, Y.; Zhu, D.; Zhang, Y. Reduced bioactive gibberellin content in rice seeds under low temperature leads to decreased sugar consumption and low seed germination rates. Plant Physiol. Biochem. 2018, 133, 1–10. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Ye, N.; Huang, M.; Feng, L.; Li, H.; Zhang, J. OsTPP1 regulates seed germination through the crosstalk with abscisic acid in rice. New Phytol. 2021, 230, 1925–1939. [Google Scholar] [CrossRef]
- Song, S.; Wang, G.; Wu, H.; Fan, X.; Liang, L.; Zhao, H.; Li, S.; Hu, Y.; Liu, H.; Ayaad, M.; et al. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 2020, 103, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Hirano, K.; Yano, K.; Wang, F.; Mori, M.; Kawamura, M.; Koketsu, E.; Hattori, M.; Ordonio, R.L.; Huang, P.; et al. Genome-wide association study identifies a gene responsible for temperature-dependent rice germination. Nat. Commun. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Oliver, S.N.; Dennis, E.S.; Dolferus, R. ABA regulates apoplastic sugar transport and is a potential signal for cold-induced pollen sterility in rice. Plant Cell Physiol. 2007, 48, 1319–1330. [Google Scholar] [CrossRef]
- Yu, J.; Lai, Y.; Wu, X.; Wu, G.; Guo, C. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem. Biophys. Res. Commun. 2016, 478, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.-h.D.; Shen, Q.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-Y.; Vo, K.T.X.; Nguyen, C.D.; Jeong, D.-H.; Lee, S.-K.; Kumar, M.; Kim, S.-R.; Park, S.-H.; Kim, J.-K.; Jeon, J.-S. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 2015, 8, 1–13. [Google Scholar] [CrossRef]
- Holdsworth, M.J.; Bentsink, L.; Soppe, W. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008, 179, 33–54. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Zhai, H.; Bai, X.; Zhu, Y.M.; Li, Y.; Cai, H.; Ji, W.; Ji, Z.J.; Liu, X.F.; Liu, X.; Li, J. A single-repeat R3-MYB transcription factor e negatively regulates freezing tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2010, 394, 1018–1023. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Jiang, N.; Hou, X.; Wu, S.; Zhang, Q.; Meng, J.; Luan, Y. Genome-wide identification of lncRNAs and analysis of ceRNA networks during tomato resistance to Phytophthora infestans. Phytopathology 2020, 110, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gong, B.; Bushel, P.R.; Thierry-Mieg, J.; Thierry-Mieg, D.; Xu, J.; Fang, H.; Hong, H.; Shen, J.; Su, Z. A comprehensive study design reveals treatment-and transcript abundance–dependent concordance between RNA-seq and microarray data. Nat. Biotechnol. 2014, 32, 926. [Google Scholar] [CrossRef]
- Li, M.; Chen, X.; He, D.; Yang, P. Proteomic analysis reveals that calcium channel blockers affect radicle protrusion during rice seed germination. Plant Growth Regul. 2020, 90, 393–407. [Google Scholar] [CrossRef]
- Xiaowen, S.; Dongyuan, L.; Xiaofeng, Z.; Wenbin, L.; Hui, L.; Weiguo, H.; Chuanbei, J.; Ning, G.; Chouxian, M.; Huaping, Z. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Wu, Y.; Bhat, P.R.; Close, T.J.; Lonardi, S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 2008, 4, e1000212. [Google Scholar] [CrossRef]
- Paterson, A.H.; Bowers, J.E.; Van de Peer, Y.; Vandepoele, K. Ancient duplication of cereal genomes. New Phytol. 2005, 165, 658–661. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Huang, R.; Wu, G.; Sun, J.; Zhu, Y.; Wang, H. Transcriptomic and QTL Analysis of Seed Germination Vigor under Low Temperature in Weedy Rice WR04-6. Plants 2023, 12, 871. https://doi.org/10.3390/plants12040871
Wang W, Huang R, Wu G, Sun J, Zhu Y, Wang H. Transcriptomic and QTL Analysis of Seed Germination Vigor under Low Temperature in Weedy Rice WR04-6. Plants. 2023; 12(4):871. https://doi.org/10.3390/plants12040871
Chicago/Turabian StyleWang, Wenjia, Ruizhi Huang, Gengwei Wu, Jian Sun, Ying Zhu, and Hua Wang. 2023. "Transcriptomic and QTL Analysis of Seed Germination Vigor under Low Temperature in Weedy Rice WR04-6" Plants 12, no. 4: 871. https://doi.org/10.3390/plants12040871
APA StyleWang, W., Huang, R., Wu, G., Sun, J., Zhu, Y., & Wang, H. (2023). Transcriptomic and QTL Analysis of Seed Germination Vigor under Low Temperature in Weedy Rice WR04-6. Plants, 12(4), 871. https://doi.org/10.3390/plants12040871






