Morphology and Phylogeny Reveal Three Montagnula Species from China and Thailand
Abstract
1. Introduction
2. Results
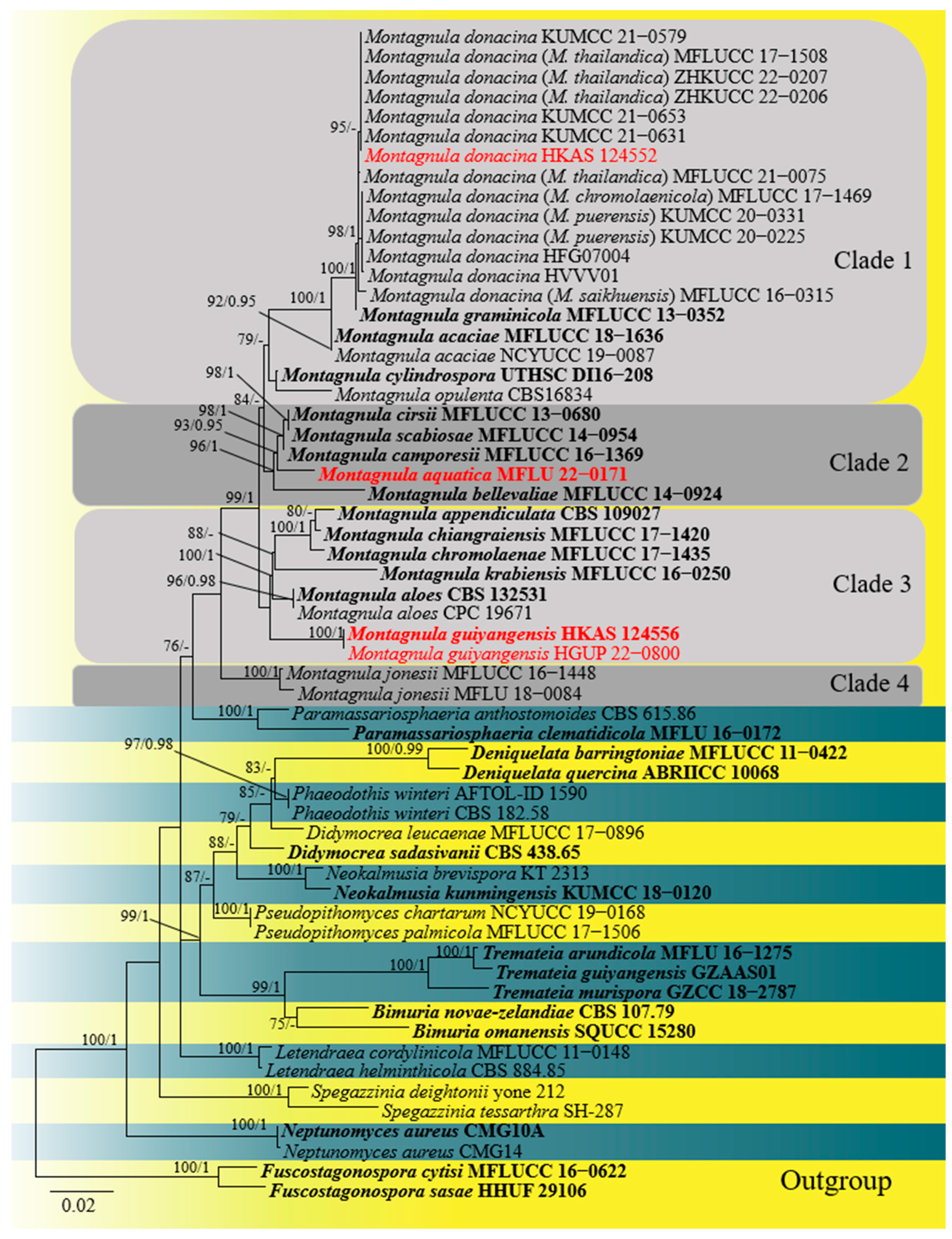
2.1. Phylogenetic Analyses
2.2. Taxonomy
- Montagnula aquatica Y.R. Sun, Yong Wang bis and K.D. Hyde, sp. nov. Figure 2.
- Index Fungorum number: IF900129; Facesoffungi number: FoF 12922.
- Holotype: MFLU 22−0171.
- Etymology: Referring to the aquatic habitat of the fungus.
- Montagnula guiyangensis Y.R. Sun, Yong Wang bis and K.D. Hyde, sp. nov. Figure 3.
- Index Fungorum number: IF900130; Facesoffungi number: FoF 12923.
- Holotype: HKAS 124556.
- Etymology: Referring to the location in which the fungus was collected.
- Montagnula donacina (Niessl) Wanas., E.B.G. Jones and K.D. Hyde, Fungal Biology 120 (11): 1365 (2016) Figure 4.
- =Montagnula chromolaenicola Mapook and K.D. Hyde.
- =Montagnula puerensis Tibpromma and Du.
- =Montagnula saikhuensis Wanas., E.B.G. Jones and K.D. Hyde.
- =Montagnula thailandica Mapook and K.D. Hyde.
- Index Fungorum number: IF557299; Facesoffungi number: FoF 07792.
3. Discussion
4. Materials and Methods
4.1. Collection, Examination, and Isolation
4.2. DNA Extraction, PCR Amplification, and Sequencing
4.3. Phylogenetic Analyses
| 1. Ascospores are didymosporous | 2 |
| 1. Ascospores are phragmosporous | 13 |
| 1. Ascospores are dictyosporous | 20 |
| 2. Didymospores with sheath | 3 |
| 2. Didymospores without sheath | 10 |
| 3. Didymospores surrounded by a mucilaginous sheath | 4 |
| 3. Sheath was drawn out to form polar appendages | 8 |
| 4. Ascospores are fusiform | M. krabiensis |
| 4. Ascospores are ellipsoidal | 5 |
| 5. Ascospores are asymmetrical | M. vakrabeejae |
| 5. Ascospores are symmetrical | 6 |
| 6. Asci are (4–)6–8-spored | M. chromolaenae |
| 6. Asci are 8-spored | 7 |
| 7. Ascospores brown, slightly constricted at the septum | M. graminicola |
| 7. Ascospores dark brown, not constricted at the septum | M. palmacea |
| 8. Ascospores 1-seriate, yellowish brown to brown | M. appendiculata |
| 8. Ascospores 2–3-seriate | 9 |
| 9. Ascomata 300–400 × 350–400 μm, asci 84–135 × 10–15 μm | M. guiyangensis |
| 9. Ascomata 150–220 × 200–230 μm, asci 60–75 × 8–11 μm | M. chiangraiensis |
| 10. Ascomata superficial | M. longipes |
| 10. Ascomata immersed or erumpent | 11 |
| 11. Ascomata 140–180 × 150–200 µm, not more than 200 µm | M. acacia |
| 11. Ascomata greater than 200 μm | 12 |
| 12 Ascospores brown, 12–17 × 4–6.5 μm | M. donacina |
| 12 Ascospores pale brown, 19–25 × 9–13 μm | M. opulenta |
| 13. Ascomata superficial | M. camporesii |
| 13. Ascomata immersed or erumpent | 14 |
| 14. Asci with short stalks | 15 |
| 14. Asci with long pedicellate | 16 |
| 15. Ascospores 5 transverse septa, 21–25 × 5–7 μm | M. subsuperficialis |
| 15. Ascospores 3 transverse septa, 24–35 × 7.5–14 μm | M. aquatica |
| 16. Ascospores with 2 transverse septa | M. bellevaliae |
| 16. Ascospores with 3 transverse septa | 17 |
| 17. Asci not more than 100 μm | M. jonesii |
| 17. Asci greater than 100 μm | 18 |
| 18. Ascospores greater than 30 μm, ovoid to ellipsoid | M. aloes |
| 18. Ascospores not more than 30 μm, ellipsoid to fusiform | 19 |
| 19. Ascomata 385–415 × 510–525 μm, asci 84.5–119.5 × 10.5–13.5 μm | M. cirsii |
| 19. Ascomata 300–320 × 300–360 μm, asci 110–130 × 14–20 μm | M. scabiosae |
| 20. Sporous transverse septa more than 10 | 21 |
| 20. Sporous transverse septa not more than 10 | 23 |
| 21. Sporous transverse septa more than 15, ascospores 40–45 × 15–17 μm | M. gigantea |
| 21. Sporous transverse septa not more than 15 | 22 |
| 22. Ascospores 32–40 × 8–9.8 μm, asci 80–110 × 13–15 μm | M. dura |
| 22. Ascospores 31–45 × 13.5–16.5 μm, asci 110–160 × 13–6 μm | M. triseti |
| 23. Sporous transverse septa not more than 5 | 24 |
| 23. Sporous transverse septa more than 5 | 27 |
| 24. Ascospores without sheath | 25 |
| 24. Ascospores with sheath | 26 |
| 25. Ascospores 2–3 transverse septa, 0–1 longitudinal septum, 12.5–16.5 × 4.8–6.5 μm | M. baatanensis |
| 25. Ascospores 5 transverse septa, 1 longitudinal septum, 24–29 × 9–11 μm | M. infernalis |
| 26. Ascospores 17.5–23 × 5.5–8.5 μm, fusiform to somewhat broadly fusiform | M. opuntiae |
| 26. Ascospores 16–18 × 6–7.5 μm, ellipsoid fusoid | M. thuemeniana |
| 27. Ascospores without sheath | 28 |
| 27. Ascospores with sheath | 29 |
| 28. Ascospores 17–25 × 7.5–10 μm, 5–7 transverse septa | M. obtusa |
| 28. Ascospores 39–47 × 15–19 μm, 7–9 transverse septa | M. opaca |
| 29. Ascospores broadly ellipsoid, 5–7 transverse septa | M. phragmospora |
| 29. Ascospores obovoid fusoid, 7(–10) transverse septa | 30 |
| 30. Ascospores 2–3 longitudinal septa, 40.8–52 × 17.6–22.4 μm | M. mohavensis |
| 30. Ascospores 1–2 longitudinal septa | 31 |
| 31. Ascospores 35–50 × 16–20 μm, asci 2–8-spored | M. dasylirionis |
| 31. Ascospores 27–42 × 12–15 μm, asci 4–8-spored | M. yuccigena |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munk, A. The system of the Pyrenomycetes. Dan. Bot. Ark. 1953, 15, 1–163. [Google Scholar]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Boonmee, S.; Lücking, R.; Bhat, D.J.; Liu, N.G.; et al. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 2020, 11, 1553–2107. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.; Saxena, R.; Erdogdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa-2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.A.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Camporesi, E.; Thambugala, K.M.; Mapook, A.; Kang, J.C.; Alias, S.A.; Chukeatirote, E.; Thines, M.; McKENZIE, E.H.C.; Hyde, K.D. Confusion surrounding Didymosphaeria—Phylogenetic and morphological evidence suggest Didymosphaeriaceae is not a distinct family. Phytotaxa 2014, 176, 102–119. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Tanaka, K.; Thambugala, K.M.; Phookamsak, R.; Tian, Q.; Camporesi, E.; Hongsanan, S.; Monkai, J.; Wanasinghe, D.N.; Mapook, A.; et al. A molecular phylogenetic reappraisal of the Didymosphaeriaceae (=Montagnulaceae). Fungal Divers. 2014, 68, 69–104. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Ren, G.; Wanasinghe, D.N.; de Farias, A.R.G.; Hyde, K.D.; Yasanthika, E.; Xu, J.C.; Balasuriya, A.; Chethana, K.W.T.; Gui, H. Taxonomic Novelties of Woody Litter Fungi (Didymosphaeriaceae, Pleosporales) from the Greater Mekong Subregion. Biology 2022, 11, 1660. [Google Scholar] [CrossRef]
- Berlese, A.N. Icones Fungorum. Pyrenomycetes; Kessinger Publishing, LLC.: Whitefish, MT, USA, 1896; pp. 1–216. [Google Scholar]
- Crivelli, P.G. Ueber die Heterogene Ascomycetengattung Pleospora rabh.: Vorschlag für eine Aufteilung. Doctoral Thesis, Dissertation ETH Zürich, Zürich, Switzerland, 1983. [Google Scholar] [CrossRef]
- Leuchtmann, A. Über Phaeosphaeria Miyake und andere bitunicate Ascomyceten mit mehrfach querseptierten Ascosporen. Sydowia 1984, 37, 75–194. [Google Scholar]
- Aptroot, A. Redisposition of some species excluded from Didymosphaeria (Ascomycotina). Nova Hedwig. 1995, 60, 325–380. [Google Scholar]
- Wanasinghe, D.N.; Jones, E.B.G.; Camporesi, E.; Dissanayake, A.J.; Kamolhan, S.; Mortimer, P.E.; Xu, J.C.; Abd-Elsalam, K.A.; Hyde, K.D. Taxonomy and phylogeny of Laburnicola gen. nov. and Paramassariosphaeria gen. nov. (Didymosphaeriaceae, Massarineae, Pleosporales). Fungal Biol. 2016, 120, 1354–1373. [Google Scholar] [CrossRef]
- Species Fungorum. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 27 January 2023).
- Tennakoon, D.S.; Hyde, K.D.; Wanasinghe, D.N.; Bahkali, A.H.; Camporesi, E.; Khan, S.; Phookamsak, R. Taxonomy and phylogenetic appraisal of Montagnula jonesii sp. nov. (Didymosphaeriaceae, Pleosporales). Mycosphere 2016, 7, 1346–1356. [Google Scholar] [CrossRef]
- Lu, L.; Karunarathna, S.C.; Dai, D.-Q.; Xiong, Y.-R.; Suwannarach, N.; Stephenson, S.L.; Elgorban, A.M.; Al-Rejaie, S.; Jayawardena, R.S.; Tibpromma, S. Description of Four Novel Species in Pleosporales Associated with Coffee in Yunnan, China. J. Fungi 2022, 8, 1113. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal Diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.P.; et al. Fungal Diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.U.; Jones, E.B.G.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal Diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar] [CrossRef]
- Du, T.Y.; Hyde, K.D.; Mapook, A.; Mortimer, P.E.; Xu, J.C.; Karunarathna, S.C.; Tibpromma, S. Morphology and phylogenetic analyses reveal Montagnula puerensis sp. nov. (Didymosphaeriaceae, Pleosporales) from southwest China. Phytotaxa 2021, 514, 1–25. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Shivas, R.G.; Burgess, T.I.; Decock, C.A.; Dreyer, L.L.; Granke, L.L.; Guest, D.I.; Hardy, G.S.; Hausbeck, M.K.; et al. Fungal Planet description sheets: 107–127. Persoonia 2012, 28, 138–182. [Google Scholar] [CrossRef]
- Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Ariyawansa, H.A.; Bhat, D.J.; Boonmee, S.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Phookamsak, R.; Phukhamsakda, C.; et al. Fungal Diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015, 72, 1–197. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Bahkali, A.H.; Camporesi, E.; Chomnunti, P.; Ekanayaka, H.; Gomes, A.A.; Hofstetter, V.; Jones, E.B.G.; Pinho, D.B.; et al. Fungal biodiversity profiles 11–20. Cryptogam. Mycol. 2015, 36, 355–380. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal Diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Pitt, W.; Úrbez-Torres, J.R.; Trouillas, F.P. Munkovalsaria donacina from grapevines and Desert Ash in Australia. Mycosphere 2014, 5, 656–661. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayake, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bha, T.J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/IndexFungorumPartnership.htm (accessed on 10 January 2023).
- Chaiwan, N.; Gomdola, D.; Wang, S.; Monkai, J.; Tibpromma, S.; Doilom, M.; Wanasinghe, D.N.; Mortimer, P.E.; Lumyong, S.; Hyde, K.D. An online database providing updated information of microfungi in the Greater Mekong Subregion. Mycosphere 2021, 12, 1513–1526. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; de Farias, A.R.G.; Bhunjun, C.S.; Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Herath, I.S.; Thambugala, K.M.; Manawasinghe, I.S.; et al. What is a species in fungal plant pathogens? Fungal Divers. 2021, 109, 239–266. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Molecular systematics of the Hypocreales: A teleomorph gene phylogeny and the status of their anamorphs. Can. J. Bot. 1995, 73, 816–882. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.L.; Höhna, S.; Larget, B.; Liu, L.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest, Version 2.2; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rannala, B.; Yang, Z.H. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, Version 1.4.0; University of Edinburgh: Edinburgh, UK, 2014. [Google Scholar]



| Species | Strain | ITS (504 bp) | LSU (842 bp) | SSU (971 bp) |
|---|---|---|---|---|
| M. camporesii | MFLUCC 16–1369 | 14 (2.7%) | 7 (0.8%) | 4 (0.4%) |
| M. cirsii | MFLUCC 13–0680 | 14 (2.7%) | 7 (0.8%) | 2 (0.2%) |
| M. scabiosae | MFLUCC 14–0954 | 15 (2.9%) | 7 (0.8%) | 6 (0.6%) |
| Species | Strain | ITS (504 bp) | LSU (842 bp) | SSU (971 bp) |
|---|---|---|---|---|
| M. aloes | CBS 132531 | 20 (4%) | 10 (1.2%) | not available |
| M. appendiculata | CBS 10927 | 18 (3.6%) | 16 (1.9%) | not available |
| M. chiangraiensis | MFLUCC 17–1420 | 13 (2.6%) | 18 (2.1%) | 27 (2.8%) |
| M. chromolaenae | MFLUCC 17–1435 | 14 (2.7%) | 18 (2.1%) | 32 (3.3%) |
| Species | Ascomata | Asci | Ascospores | Reference |
|---|---|---|---|---|
| M. chromolaenicola | Solitary, scattered, semi-immersed to erumpent, brown to dark brown, globose to obpyriform, 300–320 × 215–310 μm, ostiole papilla | Bitunicate, elongate-clavate, 8-spored, 80–100 × 10–13 μm, long pedicel | Broadly fusiform to ellipsoid, brown to dark brown, overlapping 1–2-seriate, 1 transverse septum, constricted at the septum, asymmetrical, 15–17 × 5–6.5 μm | Mapook et al. [8] |
| M. donacina | Ascostromata with flat bottom, gregarious, immersed to erumpent, black, globose to pyriform, 500 μm, pseudothecial ostiole | Bitunicate, clavate, 8-spored, with a long pedicel | Ellipsoid, brown, 1–2-seriate, 1 transverse septum, constricted at the septum, asymmetrical, 12–17 × 4–6.5 μm | Aptroot [13], Pitt et al. [28] |
| M. puerensis | Solitary, scattered, semi-immersed, black, globose, 300–600 × 230–380 μm, with a central ostiole | Bitunicate, elongate-clavate, 8-spored, 70–105 × 10–15 μm, long pedicel | Ellipsoid, brown to dark brown, biseriate, 1 transverse septum, constricted at the septum, asymmetrical, 10–20 × 4–7 μm | Du et al. [21] |
| M. saikhuensis | Solitary, scattered, immersed, globose, brown to dark brown, 400–450 × 400–500 μm, ostiolate | Bitunicate, elongate-clavate to short-cylindrical, 8-spored, 70–100 × 10–12μm, long pedicel | Ellipsoid, brown to blackish brown, overlapping 1–2-seriate, 1 transverse septum, unequally and strongly constricted at the septum, asymmetrical, 12–16 × 4–6 µm | Wanasinghe et al. [14] |
| M. thailandica | Solitary, scattered, immersed to erumpent, brown to dark brown, globose to obpyriform, brown to dark brown, 405–415 × 330–350 μm, ostiole papillate | Bitunicate, elongate-clavate, slightly curved, 8-spored, 80–100 × 9–15 μm, long pedicel | Broadly fusiform to ellipsoid, brown to reddish-brown, overlapping 1–2-seriate, 1 transverse septum, constricted at the septum, asymmetrical, 14–17 × 4.5–7.5 μm | Mapook et al. [8] |
| Species | Strain | ITS (504 bp) | LSU (842 bp) | SSU (971 bp) | tef1-α (840 bp) |
|---|---|---|---|---|---|
| M. chromolaenicola | MFLUCC 17–1469 | 0 | 3 | 1 | 10 |
| M. puerensis | KUMCC 20–0225 | 0 | 0 | 1 | 8 |
| M. saikhuensis | MFLUCC 16–0315 | 4 | 2 | 3 | not available |
| M. thailandica | MFLUCC 17–1508 | 3 | 2 | 3 | 2 |
| Locus | Primers | PCR Procedures | Reference | |
|---|---|---|---|---|
| Name | Sequence (5′-3′) | |||
| LSU | LR0R | ACCCGCTGAACTTAAGC | 94 °C—3 min; 94 °C—30 s; 52 °C—30 s; 72 °C—1 min; Repeat 2–4 for 35 cycles; 72 °C—8 min; 4 °C on hold | White et al. [34], Rehner and Samuels [35] |
| LR5 | TCCTGAGGGAAACTTCG | |||
| SSU | NS1 | GTAGTCATATGCTTGTCTC | White et al. [34] | |
| NS4 | CTTCCGTCAATTCCTTTAAG | |||
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | ||
| ITS4 | TCCTCCGCTTATTGATATGC | |||
| tef1-α | EF1-983F | GCYCCYGGHCAYCGTGAYTTYAT | 94 °C 2 min; 36 cycles of 66 °C–56 °C (touchdown 9 cycles), 94 °C 30 s, 56 °C 1 min, 72°C 1 min; 72 °C 10 min; 4 °C on hold | Rehner and Buckley [36] |
| EF1-2218R | ATGACACCRACRGCRACRGTYTG | |||
| Species | Strain/Voucher No. | LSU | SSU | ITS | tef-α |
|---|---|---|---|---|---|
| Bimuria omanensis | SQUCC 15280 | NG_071257 | N/A | NR_173301 | MT279046 |
| Bimuria novae-zelandiae | CBS 107.79 | AY016356 | N/A | MH861181 | DQ471087 |
| Deniquelata barringtoniae | MFLUCC 11−0422 | JX254655 | JX254656 | NR_111779 | N/A |
| Deniquelata quercina | ABRIICC 10068 | MH316157 | MH316155 | MH316153 | N/A |
| Didymocrea leucaenae | MFLUCC 17−0896 | NG_066304 | MK347826 | NR_164298 | MK360052 |
| Didymocrea sadasivanii | CBS 438.65 | DQ384103 | DQ384066 | MH870299 | N/A |
| Fuscostagonospora sasae | HHUF 29106 | NG_059395 | NG_061003 | NR_153964 | AB808524 |
| Fuscostagonospora cytisi | MFLUCC 16−0622 | KY770978 | KY770977 | N/A | KY770979 |
| Letendraea cordylinicola | MFLUCC 11−0148 | NG_059530 | NG_068362 | NR_154118 | N/A |
| Letendraea helminthicola | CBS 884.85 | AY016362 | AY016345 | MK404145 | MK404174 |
| Montagnula acaciae | MFLUCC 18−1636 | ON117298 | ON117267 | ON117280 | ON158093 |
| Montagnula acaciae | NCYUCC 19−0087 | ON117299 | ON117268 | ON117281 | ON158094 |
| Montagnula aloes | CPC 19671 | JX069847 | N/A | JX069863 | N/A |
| Montagnula aloes | CBS 132531 | NG_042676 | N/A | NR_111757 | N/A |
| Montagnula aquatica | MFLU 22−0171 | OP605986 | OP600504 | OP605992 | N/A |
| Montagnula appendiculata | CBS 109027 | AY772016 | N/A | DQ435529 | N/A |
| Montagnula bellevaliae | MFLUCC 14−0924 | KT443902 | KT443904 | KT443906 | KX949743 |
| Montagnula camporesii | MFLUCC 16−1369 | NG_070946 | NG_068418 | MN401746 | MN397908 |
| Montagnula chiangraiensis | MFLUCC 17−1420 | NG_068707 | NG_070155 | NR_168864 | N/A |
| Montagnula chromolaenae | MFLUCC 17−1435 | NG_068708 | NG_070156 | NR_168865 | N/A |
| Montagnula cirsii | MFLUCC 13−0680 | KX274249 | KX274255 | KX274242 | KX284707 |
| Montagnula cylindrospora | UTHSC DI16-208 | LN907351 | N/A | LT796834 | LT797074 |
| Montagnula donacina | HVVV01 | KJ628377 | KJ628376 | KJ628375 | N/A |
| Montagnula donacina | HFG07004 | MF183940 | N/A | MF967419 | N/A |
| Montagnula donacina | KUMCC 21−0653 | OP059052 | OP059003 | OP058961 | OP135938 |
| Montagnula donacina | KUMCC 21−0579 | OP059054 | OP059005 | OP058963 | OP135940 |
| Montagnula donacina | KUMCC 21−0631 | OP059053 | OP059004 | OP058962 | OP135939 |
| Montagnula donacina | HKAS 124552 | OP605987 | N/A | OP605991 | N/A |
| Montagnula donacina (M. chromolaenicola) | MFLUCC 17−1469 | NG_070948 | NG_070157 | NR_168866 | MT235773 |
| Montagnula donacina (M. puerensis) | KUMCC 20−0225 | MW575866 | MW575864 | MW567739 | MW573959 |
| Montagnula donacina (M. puerensis) | KUMCC 20−0331 | MW575867 | MW575865 | MW567740 | MW573960 |
| Montagnula donacina (M. saikhuensis) | MFLUCC 16−0315 | KU743210 | KU743211 | KU743209 | N/A |
| Montagnula donacina (M. thailandica) | MFLUCC 17−1508 | NG_070949 | NG_070158 | MT214352 | MT235774 |
| Montagnula donacina (M. thailandica) | MFLUCC 21−0075 | MZ538549 | N/A | MZ538515 | N/A |
| Montagnula donacina (M. thailandica) | ZHKUCC 22−0206 | OP297777 | OP297791 | OP297807 | OP321576 |
| Montagnula donacina (M. thailandica) | ZHKUCC 22−0207 | OP297778 | OP297792 | OP297808 | OP321577 |
| Montagnula graminicola | MFLUCC 13−0352 | KM658315 | KM658316 | KM658314 | N/A |
| Montagnula guiyangensis | HKAS 124556 | OP600484 | OP600500 | OP605989 | N/A |
| Montagnula guiyangensis | HGUP 22−800 | OP600485 | OP600501 | OP605990 | N/A |
| Montagnula jonesii | MFLUCC 16−1448 | KY273276 | KY313618 | KY313619 | KY313620 |
| Montagnula jonesii | MFLU 18−0084 | ON117300 | ON117269 | ON117282 | ON158095 |
| Montagnula krabiensis | MFLUCC 16−0250 | NG_068826 | NG_068385 | NR_168179 | MH412776 |
| Montagnula opulenta | CBS 16834 | NG_027581 | AF164370 | AF383966 | LT797074 |
| Montagnula scabiosae | MFLUCC 14−0954 | KT443903 | KT443905 | KT443907 | N/A |
| Neokalmusia brevispora | KT 2313 | AB524601 | AB524460 | NR_154262 | AB539113 |
| Neokalmusia kunmingensis | KUMCC 18−0120 | MK079889 | MK079887 | MK079886 | MK070172 |
| Neptunomyces aureus | CMG10A | N/A | N/A | MK912119 | MK947998 |
| Neptunomyces aureus | CMG14 | N/A | N/A | MK912123 | MK948002 |
| Paramassariosphaeria anthostomoides | CBS 615.86 | MH873693 | GU205246 | MH862005 | N/A |
| Paramassariosphaeria clematidicola | MFLU 16−0172 | KU743207 | KU743208 | KU743206 | N/A |
| Phaeodothis winteri | AFTOL-ID 1590 | DQ678073 | DQ678021 | N/A | DQ677917 |
| Phaeodothis winteri | CBS 182.58 | GU301857 | GU296183 | N/A | N/A |
| Pseudopithomyces chartarum | NCYUCC 19−0168 | MW063220 | MW079349 | MW063159 | N/A |
| Pseudopithomyces palmicola | MFLUCC 17−1506 | MT214447 | N/A | MT214353 | N/A |
| Spegazzinia deightonii | yone 212 | AB807582 | AB797292 | N/A | AB808558 |
| Spegazzinia tessarthra | SH 287 | AB807584 | N/A | JQ673429 | AB808560 |
| Tremateia arundicola | MFLU 16−1275 | KX274248 | KX274254 | KX274241 | KX284706 |
| Tremateia guiyangensis | GZAAS01 | KX274247 | KX274253 | KX274240 | KX284705 |
| Tremateia murispora | GZCC 18−2787 | MK972751 | MK972750 | NR_165916 | MK986482 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.-R.; Zhang, J.-Y.; Hyde, K.D.; Wang, Y.; Jayawardena, R.S. Morphology and Phylogeny Reveal Three Montagnula Species from China and Thailand. Plants 2023, 12, 738. https://doi.org/10.3390/plants12040738
Sun Y-R, Zhang J-Y, Hyde KD, Wang Y, Jayawardena RS. Morphology and Phylogeny Reveal Three Montagnula Species from China and Thailand. Plants. 2023; 12(4):738. https://doi.org/10.3390/plants12040738
Chicago/Turabian StyleSun, Ya-Ru, Jing-Yi Zhang, Kevin D. Hyde, Yong Wang, and Ruvishika S. Jayawardena. 2023. "Morphology and Phylogeny Reveal Three Montagnula Species from China and Thailand" Plants 12, no. 4: 738. https://doi.org/10.3390/plants12040738
APA StyleSun, Y.-R., Zhang, J.-Y., Hyde, K. D., Wang, Y., & Jayawardena, R. S. (2023). Morphology and Phylogeny Reveal Three Montagnula Species from China and Thailand. Plants, 12(4), 738. https://doi.org/10.3390/plants12040738







