Identification of a Major Locus for Lodging Resistance to Typhoons Using QTL Analysis in Rice
Abstract
1. Introduction
2. Results
2.1. Phenotype Evaluation for Lodging Resistance to Typhoons and Other Agricultural Traits
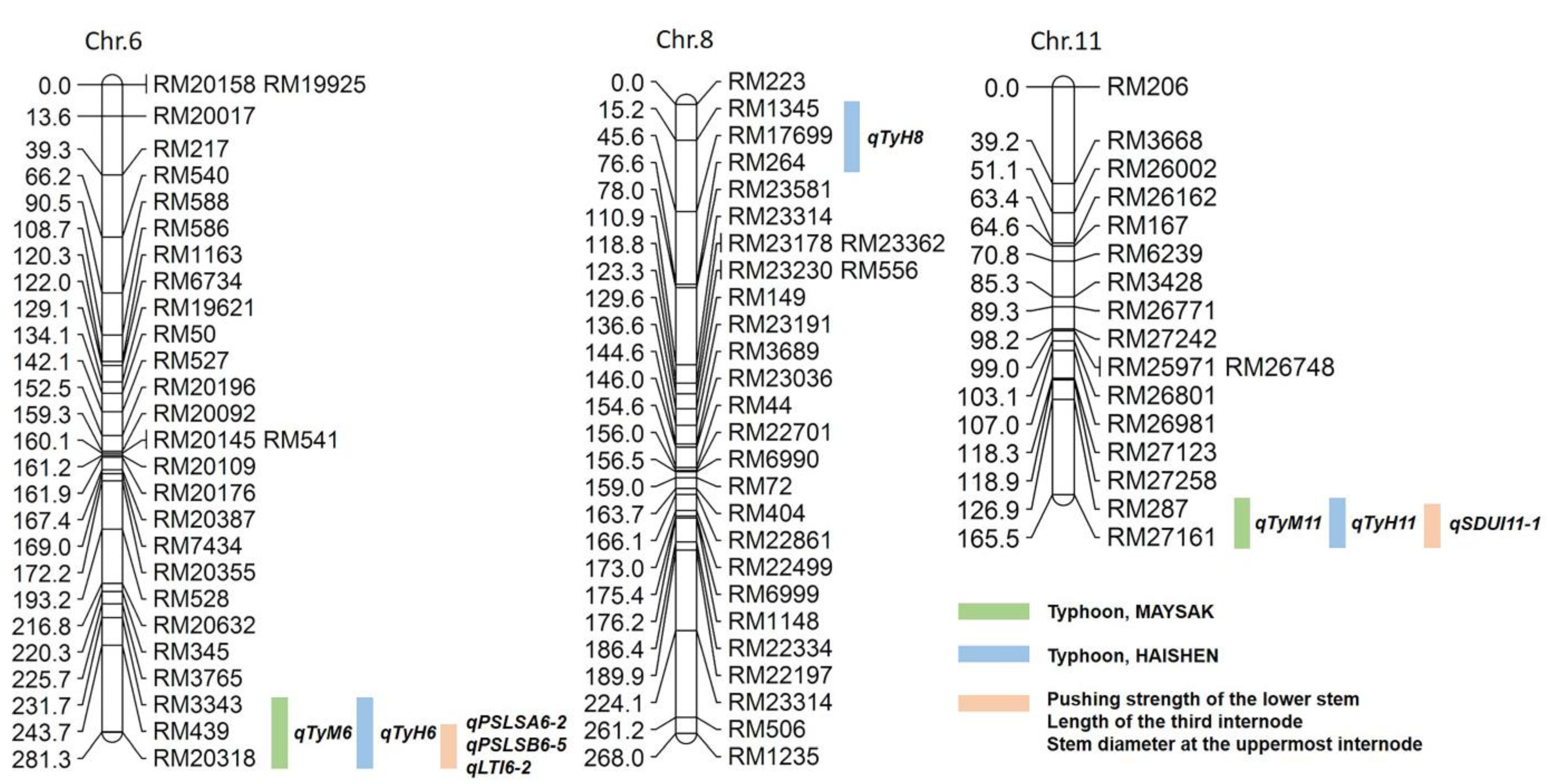
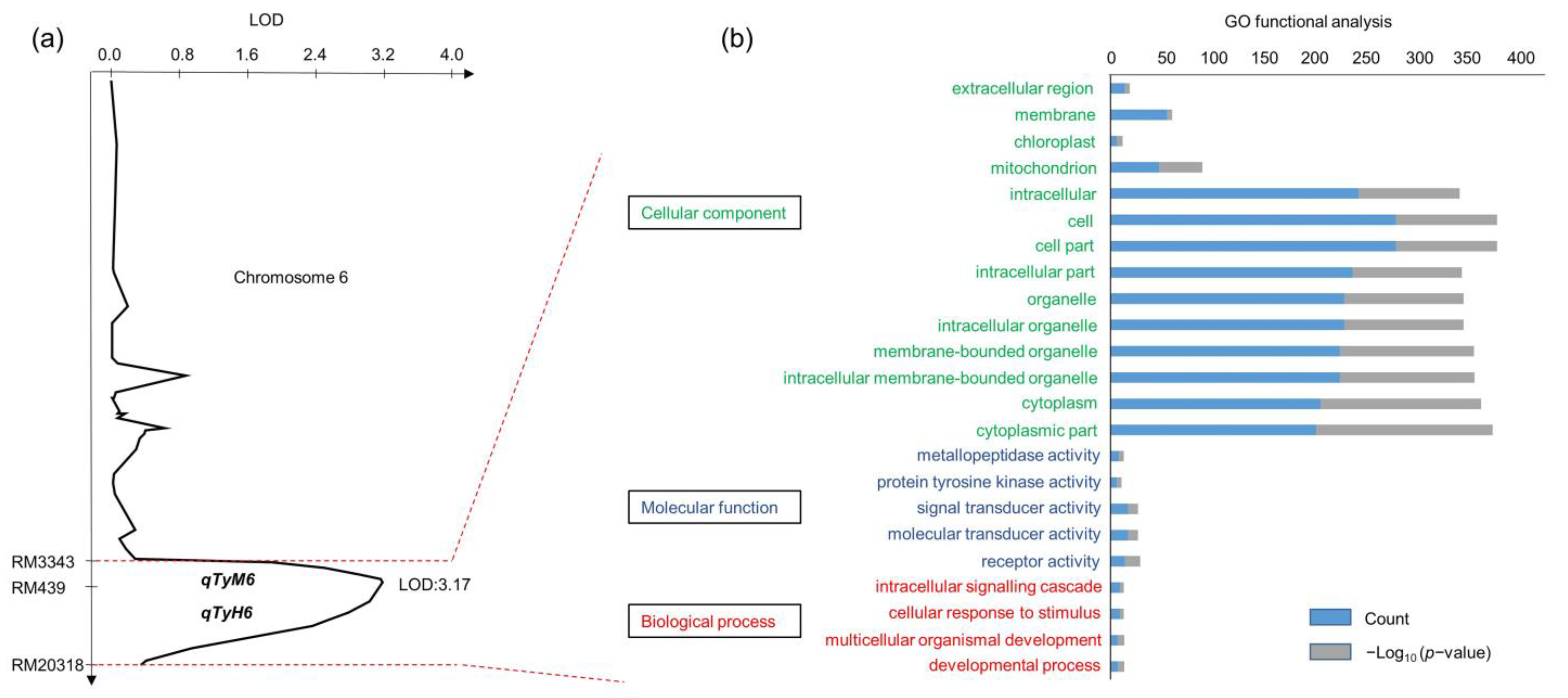
2.2. QTL Analysis Associated with Lodging Resistance to Typhoons
2.3. Gene Exploration from Target QTL Marker Interval RM3343–RM20318
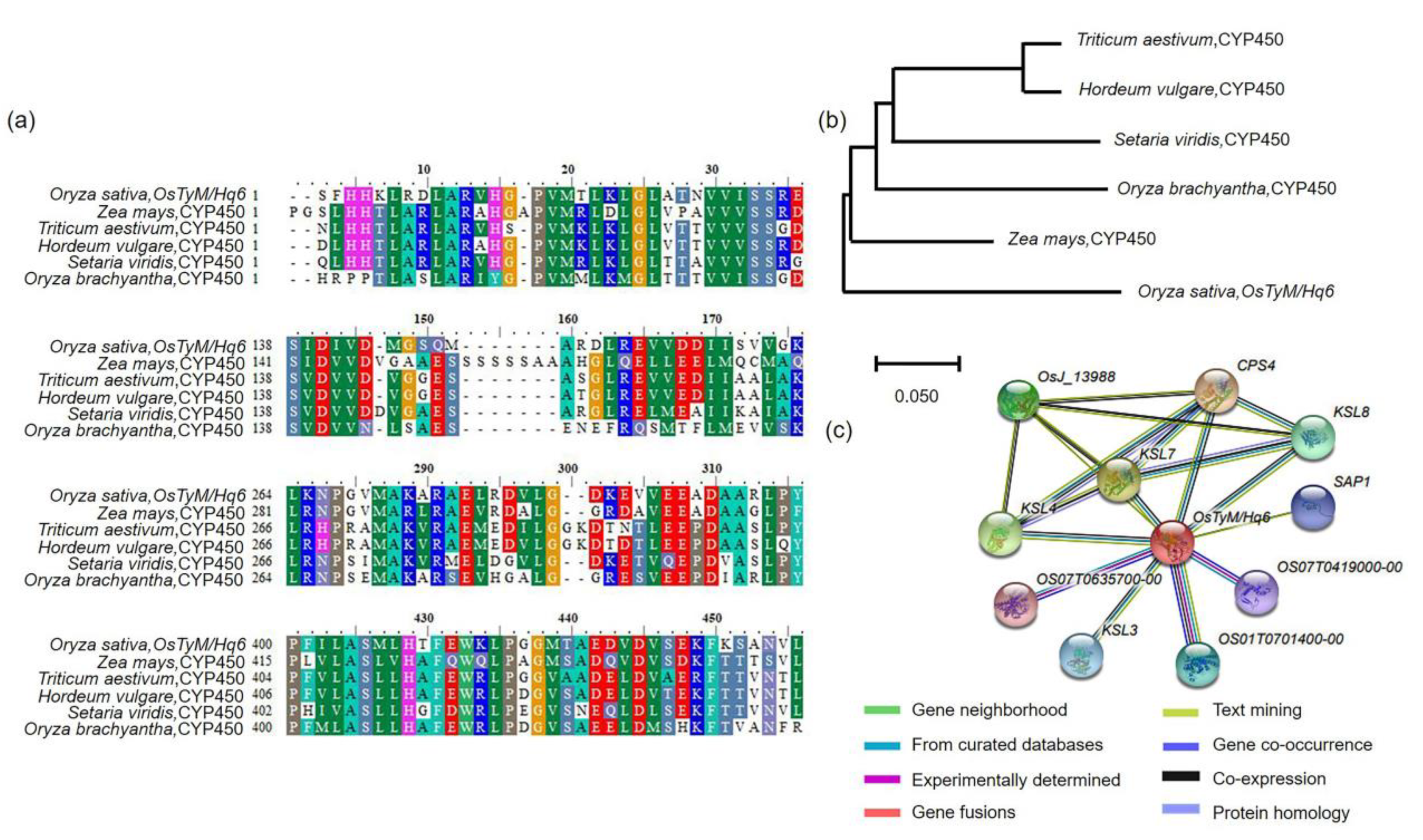
2.4. Sequence Analysis of OsTyM/Hq6
3. Discussion
4. Materials and Methods
4.1. Field Experiment Design and Plant Material
4.2. Mapping Population
4.3. Measurement of Lodging Resistance to Typhoons and Other Agricultural Traits
4.4. QTL and Statistical Analyses of Lodging Resistance to Typhoons and Other Agricultural Traits
4.5. Prediction of Related Genes and Gene Information Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Myeong, S. Impact of climate change related natural disasters on rice production in South Korea. J. Korean Soc. Hazard Mitig. 2018, 18, 53–60. [Google Scholar] [CrossRef]
- Myeong, S. Agriculture under UNFCCC and its policy implications. J. Clim. Change Res. 2014, 5, 313–321. [Google Scholar] [CrossRef]
- Shah, L.; Yahya, M.; Shah, S.M.A.; Nadeem, M.; Ali, A.; Ali, A.; Wang, J.; Riaz, M.W.; Rehman, S.; Wu, W. Improving lodging resistance: Using wheat and rice as classical examples. Int. J. Mol. Sci. 2019, 20, 4211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, G.; Song, Y.; Liu, Z.; Yang, C.; Tang, S.; Zheng, C.; Wang, S.; Ding, Y. Lodging resistance characteristics of high-yielding rice populations. Field Crop. Res. 2014, 161, 64–74. [Google Scholar] [CrossRef]
- Setter, T.; Laureles, E.; Mazaredo, A. Lodging reduces yield of rice by self-shading and reductions in canopy photosynthesis. Field Crop. Res. 1997, 49, 95–106. [Google Scholar] [CrossRef]
- Wang, J.; Li, K.; Shao, Y.; Zhang, F.; Wang, Z.; Guo, X.; Qin, Y.; Liu, X. Analysis of combining SAR and optical optimal parameters to classify typhoon-invasion lodged rice: A case study using the random forest method. Sensors 2020, 20, 7346. [Google Scholar] [CrossRef]
- Yamamoto, H.; Iwaya, K.; Suzuki, K.; Yahakawa, S.; Suzuki, Y. Agricultural disaster and salt damage in rice caused by typhoon 9918 in Kyushu and Yamaguchi districts. Jpn. J. Crop Sci. 2000, 69, 424–430. [Google Scholar] [CrossRef]
- Yamamoto, H.; Iwaya, K. Salty wind damage on rice by typhoon 0415 (MEGI) on the Sea of Japan coastal region of Tohoku and Hokuriku districts [Japan]. Jpn. J. Crop Sci. 2006, 75, 73–81. [Google Scholar] [CrossRef]
- Lai, L.-H.; Wu, P.-H. Risk analysis of rice losses caused by typhoon for Taiwan. Contemp. Manag. Res. 2010, 6, 141–158. [Google Scholar] [CrossRef]
- Bai, X.F.; Luo, L.J.; Yan, W.H.; Kovi, M.R.; Xing, Y.Z. Quantitative trait loci for rice yield-related traits using recombinant inbred lines derived from two diverse cultivars. J. Genet. 2011, 90, 209–215. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Ishimaru, K. Identification and functional analysis of a locus for improvement of lodging resistance in rice. Plant Physiol. 2004, 134, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Ookawa, T.; Hobo, T.; Yano, M.; Murata, K.; Ando, T.; Miura, H.; Asano, K.; Ochiai, Y.; Ikeda, M.; Nishitani, R. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010, 132, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Ookawa, T.; Aya, K.; Ochiai, Y.; Hirasawa, T.; Ebitani, T.; Takarada, T.; Yano, M.; Yamamoto, T.; Fukuoka, S. Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol. Plant 2015, 8, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Weng, F.; Zhang, W.; Wu, X.; Xu, X.; Ding, Y.; Li, G.; Liu, Z.; Wang, S. Impact of low-temperature, overcast and rainy weather during the reproductive growth stage on lodging resistance of rice. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Navabi, A.; Iqbal, M.; Strenzke, K.; Spaner, D. The relationship between lodging and plant height in a diverse wheat population. Can. J. Plant Sci. 2006, 86, 723–726. [Google Scholar] [CrossRef]
- Ashikari, M.; Wu, J.; Yano, M.; Sasaki, T.; Yoshimura, A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 1999, 96, 10284–10289. [Google Scholar] [CrossRef]
- Fujisawa, Y.; Kato, T.; Ohki, S.; Ishikawa, A.; Kitano, H.; Sasaki, T.; Asahi, T.; Iwasaki, Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 1999, 96, 7575–7580. [Google Scholar] [CrossRef]
- Swain, S.M.; Singh, D.P. Tall tales from sly dwarves: Novel functions of gibberellins in plant development. Trends Plant Sci. 2005, 10, 123–129. [Google Scholar] [CrossRef]
- Achard, P.; Genschik, P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 2009, 60, 1085–1092. [Google Scholar] [CrossRef]
- Ishimaru, K.; Togawa, E.; Ookawa, T.; Kashiwagi, T.; Madoka, Y.; Hirotsu, N. New target for rice lodging resistance and its effect in a typhoon. Planta 2008, 227, 601–609. [Google Scholar] [CrossRef]
- Yadav, S.; Singh, U.M.; Naik, S.M.; Venkateshwarlu, C.; Ramayya, P.J.; Raman, K.A.; Sandhu, N.; Kumar, A. Molecular mapping of QTLs associated with lodging resistance in dry direct-seeded rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1431. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Dan, D.; Yuan, Z.; Chen, Y.; Jin, J.; Yang, W.; Zhang, Z.; Li, N.; Li, S. Deciphering the genetic basis of lodging resistance in wild rice oryza longistaminata. Front. Plant Sci. 2020, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, T.; Munakata, J.; Ishimaru, K. Functional analysis of the lodging resistance QTL BSUC11 on morphological and chemical characteristics in upper culms of rice. Euphytica 2016, 210, 233–243. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Togawa, E.; Hirotsu, N.; Ishimaru, K. Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor. Appl. Genet. 2008, 117, 749–757. [Google Scholar] [CrossRef]
- Zhao, D.-D.; Son, J.H.; Lee, G.-S.; Kim, K.-M. Screening for a Novel Gene, OsPSLSq6, Using QTL Analysis for Lodging Resistance in Rice. Agronomy 2021, 11, 334. [Google Scholar] [CrossRef]
- Zhao, D.-D.; Son, J.-H.; Farooq, M.; Kim, K.-M. Identification of Candidate Gene for Internode Length in Rice to Enhance Resistance to Lodging Using QTL Analysis. Plants 2021, 10, 1369. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, M.; Ohta, D. Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 2010, 61, 291–315. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Tamiru, M.; Undan, J.R.; Takagi, H.; Abe, A.; Yoshida, K.; Undan, J.Q.; Natsume, S.; Uemura, A.; Saitoh, H.; Matsumura, H. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 85–99. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Ashikari, M. Rice growth adapting to deepwater. Curr. Opin. Plant Biol. 2011, 14, 100–105. [Google Scholar] [CrossRef]
- Schwechheimer, C. Understanding gibberellic acid signaling—Are we there yet? Curr. Opin. Plant Biol. 2008, 11, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.R.; Lu, Y.C.; Zhang, J.J.; Luo, F.; Yang, H. A collection of cytochrome P450 monooxygenase genes involved in modification and detoxification of herbicide atrazine in rice (Oryza sativa) plants. Ecotoxicol. Environ. Saf. 2015, 119, 25–34. [Google Scholar] [CrossRef]
- Luo, A.; Qian, Q.; Yin, H.; Liu, X.; Yin, C.; Lan, Y.; Tang, J.; Tang, Z.; Cao, S.; Wang, X. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 2006, 47, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Nomura, T.; Hanada, A.; Takeda-Kamiya, N.; Ohnishi, T.; Shinma, Y.; Katsumata, T.; Kawaide, H.; Kamiya, Y.; Yamaguchi, S. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Winkler, R.G.; Helentjaris, T. The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. Plant Cell 1995, 7, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Kim, K.; Manigbas, N.L.; Yi, G.; Sohn, J. Identification of quantitative trait loci for resistance to white-backed planthopper (Sogatella furcifera) in rice with Milyang 46 (Cheongcheongbyeo) background. Philipp. J. Crop. Sci. 2013, 38, 30–36. [Google Scholar]
- Yun, B.-W.; Kim, M.-G.; Handoyo, T.; Kim, K.-M. Analysis of rice grain quality-associated quantitative trait loci by using genetic mapping. Am. J. Plant Sci. 2014, 5, 1125–1132. [Google Scholar] [CrossRef]
- Lincoln, S.E.; Daly, M.J.; Lander, E.S. Constructing genetic linkage maps with MAPMAKER/EXP Version 3.0: A tutorial and reference manual, 3rd ed.; Whitehead Institute: Cambridge, MA, USA, 1993; p. 49. [Google Scholar]
- Zeng, Z.-B. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [CrossRef]
- McCough, S.R.; Doerge, R.W. QTL mapping in rice. Trends Genet. 1995, 11, 482–487. [Google Scholar] [CrossRef]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.-c.; Iwamoto, M.; Abe, T. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, 1206–1213. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit Version 7.0.0. Distributed by Author. Available online: www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 15 November 2022).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.-D.; Jang, Y.-H.; Kim, E.-G.; Park, J.-R.; Jan, R.; Lubna; Asaf, S.; Asif, S.; Farooq, M.; Chung, H.; et al. Identification of a Major Locus for Lodging Resistance to Typhoons Using QTL Analysis in Rice. Plants 2023, 12, 449. https://doi.org/10.3390/plants12030449
Zhao D-D, Jang Y-H, Kim E-G, Park J-R, Jan R, Lubna, Asaf S, Asif S, Farooq M, Chung H, et al. Identification of a Major Locus for Lodging Resistance to Typhoons Using QTL Analysis in Rice. Plants. 2023; 12(3):449. https://doi.org/10.3390/plants12030449
Chicago/Turabian StyleZhao, Dan-Dan, Yoon-Hee Jang, Eun-Gyeong Kim, Jae-Ryoung Park, Rahmatullah Jan, Lubna, Sajjad Asaf, Saleem Asif, Muhammad Farooq, Hyunjung Chung, and et al. 2023. "Identification of a Major Locus for Lodging Resistance to Typhoons Using QTL Analysis in Rice" Plants 12, no. 3: 449. https://doi.org/10.3390/plants12030449
APA StyleZhao, D.-D., Jang, Y.-H., Kim, E.-G., Park, J.-R., Jan, R., Lubna, Asaf, S., Asif, S., Farooq, M., Chung, H., Kang, D.-J., & Kim, K.-M. (2023). Identification of a Major Locus for Lodging Resistance to Typhoons Using QTL Analysis in Rice. Plants, 12(3), 449. https://doi.org/10.3390/plants12030449







