Transcriptome-Based Identification of the Optimal Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Analyses of Lingonberry Fruits throughout the Growth Cycle
Abstract
:1. Introduction
2. Results
2.1. RNA
2.2. Screening Results for the Candidate Internal Reference Genes
2.3. Gene-Specific PCR Amplification Efficiency Analysis
2.4. Analysis of Expression Stability
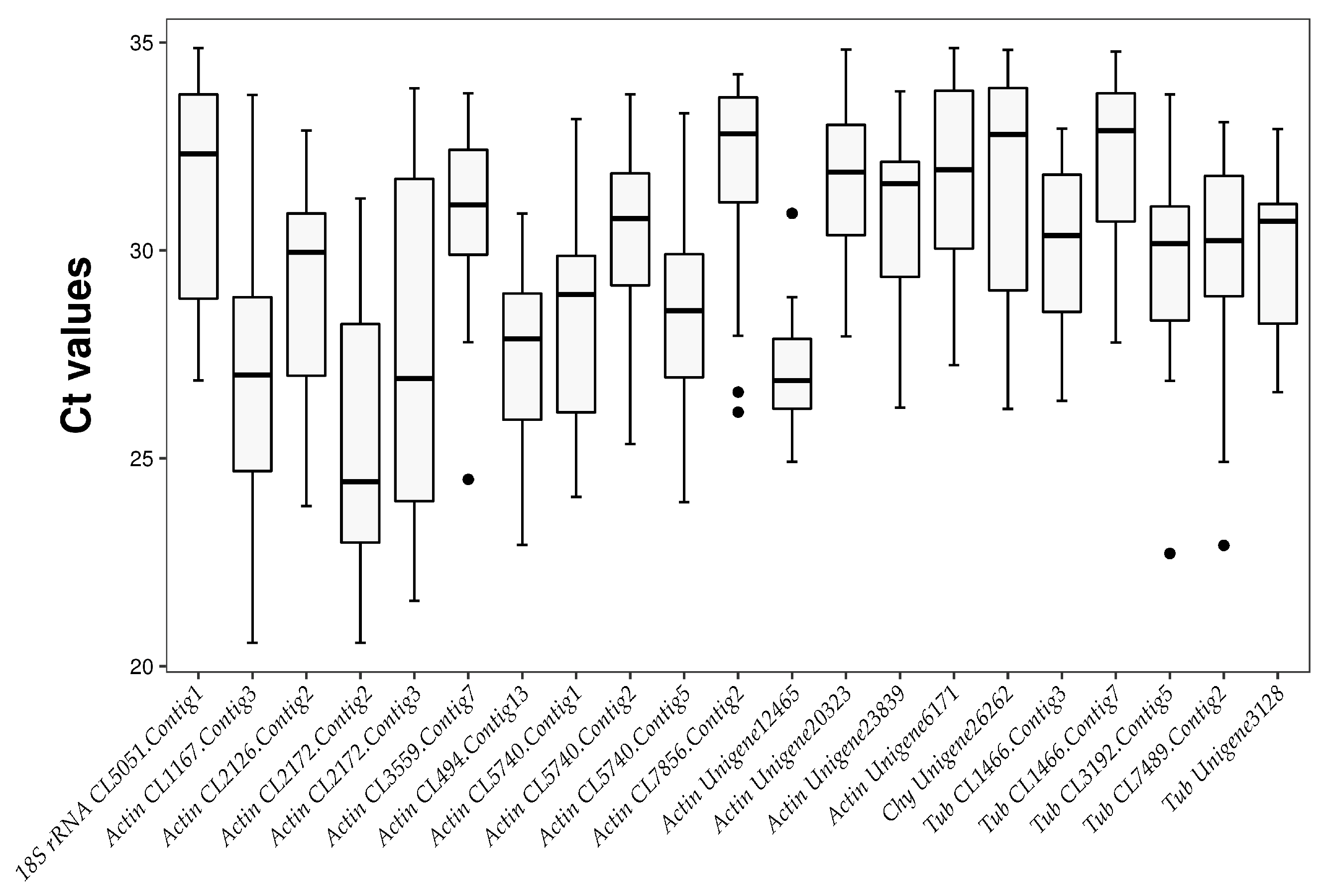
2.4.1. Analysis of Ct Values
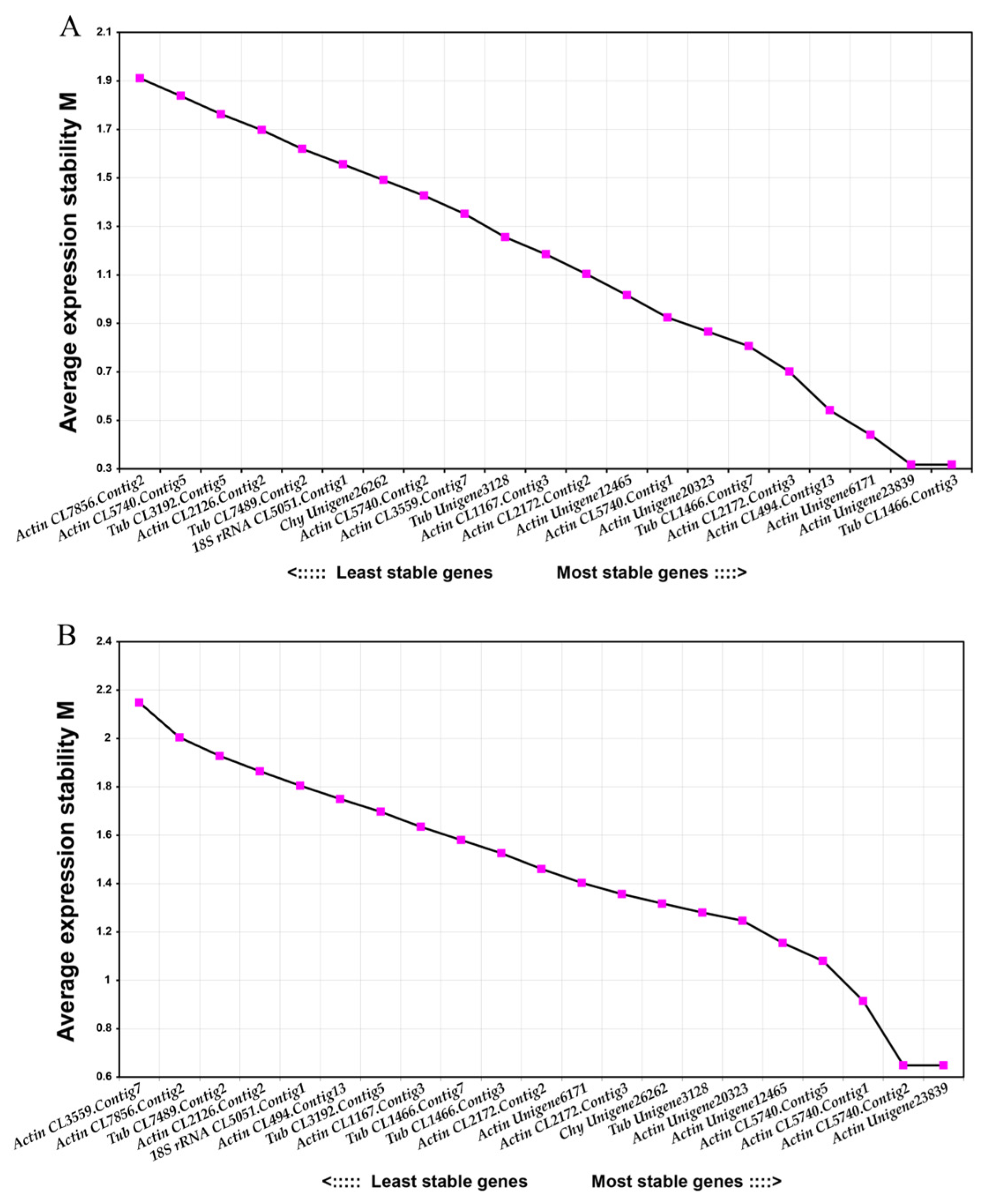
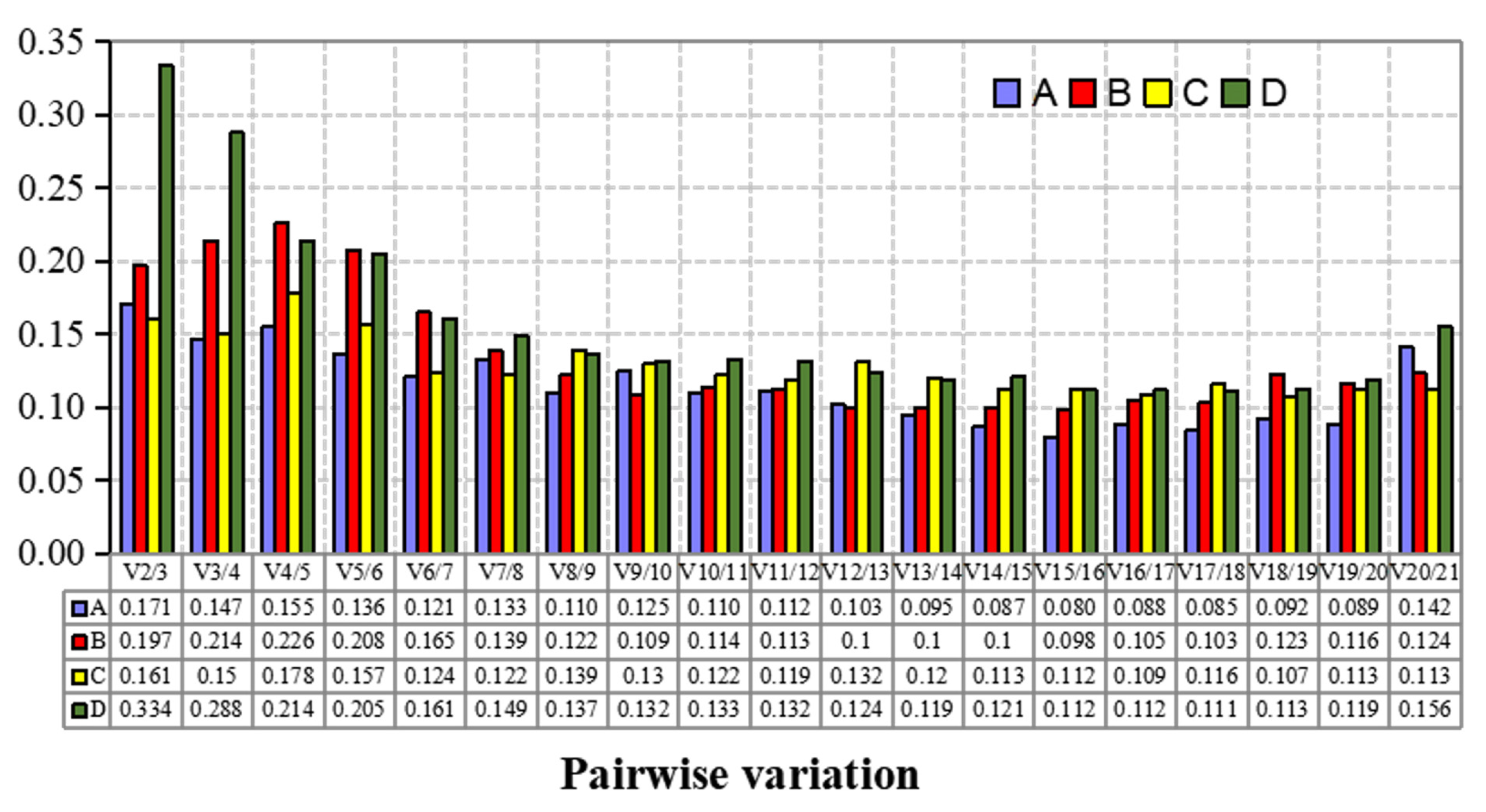
2.4.2. GeNorm Analysis
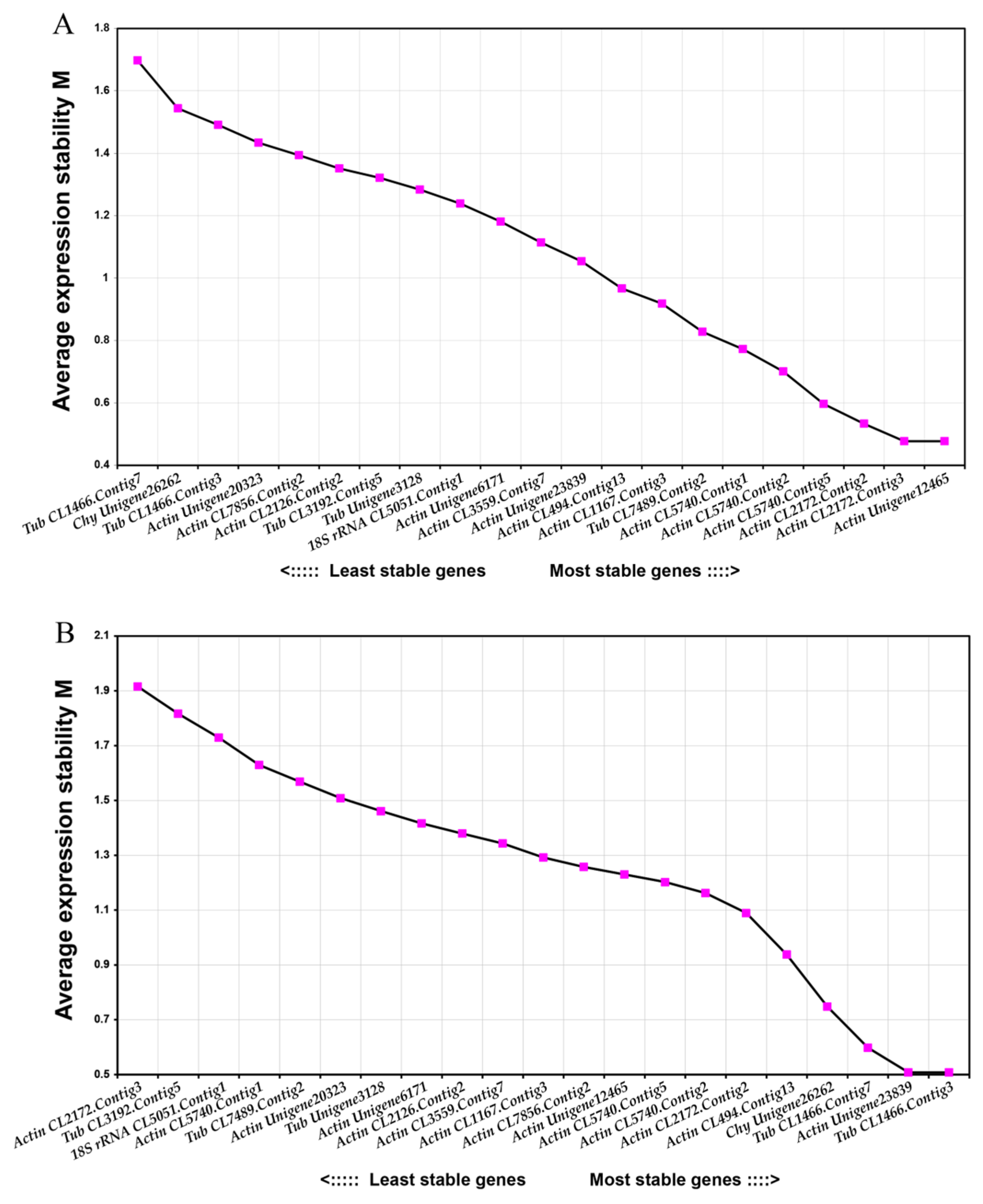
2.4.3. NormFinder Analysis
2.4.4. BestKeeper Analysis
2.4.5. Comprehensive Analysis of the Data
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Treatment, and Tissue Collection
4.2. RNA Isolation and cDNA Synthesis
4.3. Selection of Candidate Reference Genes
4.4. Primer Design and Analysis of the Amplification Efficiency for qRT-PCR
4.5. Determination and Validation of Reference Gene Expression Stability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brennan, R. Blueberries. By JB Retamales and JF Hancock. Wallingford, UK: CABI (2012), pp. 336, £ 45.00. ISBN 978-1-84593-826-0. Exp. Agric. 2012, 48, 598. [Google Scholar] [CrossRef]
- Brown, P.N.; Turi, C.E.; Shipley, P.R.; Murch, S.J. Comparisons of large (Vaccinium macrocarpon Ait.) and small (Vaccinium oxycoccos L., Vaccinium vitis-idaea L.) cranberry in British Columbia by phytochemical determination, antioxidant potential, and metabolomic profiling with chemometric analysis. Planta Med. 2012, 78, 630–640. [Google Scholar] [CrossRef]
- Karppinen, K.; Zoratti, L.; Nguyenquynh, N.; Häggman, H.; Jaakola, L. On the developmental and environmental regulation of secondary metabolism in Vaccinium spp. Berries. Front. Plant Sci. 2016, 7, 655. [Google Scholar] [CrossRef] [PubMed]
- Vanguilder, H.D.; Vrana, K.E.; Freeman, W.M.; VanGuilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. BioTechniques 2008, 44, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fu, Y.; Ban, L.; Wang, Z.; Feng, G.; Li, J.; Gao, H. Selection of reliable reference genes for quantitative real-time RT-PCR in alfalfa. Genes Genet. Syst. 2014, 90, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pan, Y.; Yang, L.; Cai, S.; Huang, X. Validation of internal reference genes for qRT-PCR normalization in ‘Guanxi Sweet Pummelo’ (Citrus grandis). J. Fruit Sci. 2013, 30, 48–54. [Google Scholar]
- Goidin, D.; Mamessier, A.; Staquet, M.J.; Schmitt, D.; Berthier-Vergnes, O. Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and beta-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and noninvasive human melanoma cell subpopulations. Anal. Biochem. 2001, 295, 17–21. [Google Scholar] [CrossRef]
- Wang, M.; Lu, S. Validation of suitable reference genes for quantitative gene expression analysis in Panax ginseng. Front. Plant Sci. 2016, 6, 1259. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.S.; Albert, P.S.; Gillespie, J.W.; Rodriguez-Canales, J.; Marston Linehan, W.; Pinto, P.A.; Chuaqui, R.F.; Emmert-Buck, M.R. Quantitative RT-PCR gene expression analysis of laser microdissected tissue samples. Nat. Protoc. 2009, 4, 902–922. [Google Scholar] [CrossRef]
- Wang, H.L.; Chen, J.H.; Tian, Q.Q.; Xia, X.L.; Yin, W.L. Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol. Plant. 2014, 152, 529–545. [Google Scholar] [CrossRef]
- Wang, H.-L.; Li, L.; Tang, S.; Yuan, C.; Tian, Q.; Su, Y.; Li, H.-G.; Zhao, L.; Yin, W.; Zhao, R.; et al. Evaluation of appropriate reference genes for reverse Transcription-Quantitative PCR studies in different tissues of a desert poplar via comparision of different algorithms. Int. J. Mol. Sci. 2015, 16, 20468–20491. [Google Scholar] [CrossRef]
- Li, C.; Xu, J.; Deng, Y.; Sun, H.; Li, Y. Selection of reference genes for normalization of cranberry (Vaccinium macrocarpon Ait.) gene expression under different experimental conditions. PLoS ONE 2019, 14, e0224798. [Google Scholar] [CrossRef] [PubMed]
- Vashisth, T.; Johnson, L.K.; Malladi, A. An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep. 2011, 30, 2167–2176. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, J.; Hua, Q.; Tel-Zur, N.; Qin, Y. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Z.; Tang, M.Y.; Zhong, M.Y.; Wang, R.J.; Chen, J.Y.; Ye, Y.T.; Zhang, X.Q.; Nie, G. Optimum reference gene selection in miscanthus sinensis root tissue with various abiotic stress. J. Sichuan Agric. Univ. 2020, 38, 699–707. [Google Scholar]
- Deng, Y.; Li, Y.; Sun, H. Selection of reference genes for RT-qPCR normalization in blueberry (Vaccinium corymbosum × angustifolium) under various abiotic stresses. FEBS Open Bio 2020, 10, 1418–1435. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, Z.; Ma, H.; Gu, Y.; Li, Y.; Sun, H. Comparative transcriptome analysis of lingonberry (Vaccinium vitis-idaea) provides insights into genes associated with flavonoids metabolism during fruit development. Biotechnol. Biotechnol. Equip. 2020, 34, 1252–1264. [Google Scholar] [CrossRef]
- Royeen, C.B. The boxplot: A screening test for research data. Am. J. Occup. Ther. 1986, 40, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Li, Q.; Wang, G.X.; Ma, Q.H.; Zhu, L.Q. Reference genes selection and system establishment for Real-Time qPCR analysis in Ping’ou Hybrid Hazelnut (C. heterophylla Fisch. × C. avellana L.). Sci. Agric. Sin. 2017, 50, 2399–2410. [Google Scholar]
- Deng, L.T. Transcriptome Based Reference Genes Selection and the Expression Analyses of Genes Involved in the Biosyn-Thesis of Three Metabolic Pathway. Ph.D. Thesis, South China Agricultural University, Guangzhou, China, 2016. [Google Scholar]
- Sang, J. Validation of Reference Genes and Analysis of HSF Gene Family Based on RNA-Seq in Hyper-Accumulating Se-dum alfredii Hance. Ph.D. Thesis, Chinese Academy of Forestry, Beijing, China, 2014. [Google Scholar]
- Mallona, I.; Lischewski, S.; Weiss, J.; Hause, B.; Egea-Cortines, M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 2010, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Stürzenbaum, S.R.; Kille, P. Control genes in quantitative molecular biological techniques: The variability of invariance. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2001, 130, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wang, Y.; Yang, D.; Wei, C.; Gao, L.; Xia, T.; Shan, Y.; Luo, Y. Reference genes for real-time fluorescence quantitative PCR in Camellia sinensis. Chin. Bull. Bot. 2010, 45, 579. [Google Scholar]
- Zhang, J.Y.; Huang, S.N.; Wang, T.; Pan, D.L.; Zhai, M.; Guo, Z.R. Screening of reference genes for reverse transcription quantitative real-time PCR in Actinidida deliciosa. Acta Agric. Shanghai 2018, 34, 5. [Google Scholar]
- Thellin, O.; Zorzi, W.; Lakaye, B.; Borman, B.D.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef]
- Egert, A.; Keller, F.; Peters, S. Abiotic stress-induced accumulation of raffinose in Arabidopsis leaves is mediated by a single raffinose synthase (RS5, At5g40390). BMC Plant Biol. 2013, 13, 218. [Google Scholar] [CrossRef]
- Gong, X.; Liu, M.; Zhang, L.; Ruan, Y.; Ding, R.; Ji, Y.; Zhang, N.; Zhang, S.; Farmer, J.; Wang, C. Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiol. Plant 2015, 153, 119–136. [Google Scholar] [CrossRef]
- Liu, Y.; Ji, X.; Nie, X.; Qu, M.; Zheng, L.; Tan, Z.; Zhao, H.; Huo, L.; Liu, S.; Zhang, B.; et al. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Kisseleva, L.; Sawa, S.; Furukawa, T.; Komatsu, S.; Koshiba, T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 2004, 45, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Ashrafi, M.; Azimi Moqadam, M.R.; Moradi, P.; Mohsenifard, E.; Shekari, F. Evaluation and validation of housekeeping genes in two contrast species of thyme plant to drought stress using real-time PCR. Plant Physiol. Biochem. 2018, 132, 54–60. [Google Scholar] [CrossRef]
- Joseph, J.T.; Poolakkalody, N.J.; Shah, J.M. Plant reference genes for development and stress response studies. J. Biosci. 2018, 43, 173–187. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, N.; Si, H.; Calderón-Urrea, A. Selection and validation of reference genes for RT-qPCR analysis in potato under abiotic stress. Plant Methods 2017, 13, 85. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Brilhante, O.; Grossi-de-Sa, M.F.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef]
- Diretto, G.; Welsch, R.; Tavazza, R.; Mourgues, F.; Pizzichini, D.; Beyer, P.; Giuliano, G. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007, 7, 11. [Google Scholar] [CrossRef]
- Libault, M.; Thibivilliers, S.; Bilgin, D.; Radwan, O.; Benitez, M.; Clough, S.; Stacey, G. Identification of four soybean reference genes for gene expression normalization. In The Plant Genome; Wiley: Hoboken, NJ, USA, 2008; Volume 1. [Google Scholar]
- Thellin, O.; ElMoualij, B.; Heinen, E.; Zorzi, W. A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol. Adv. 2009, 27, 323–333. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yang, L.; Jin, Y.; Lin, J.; Liu, F. Generation, annotation, and analysis of a large-scale expressed sequence tag library from Arabidopsis pumila to explore salt-responsive genes. Front. Plant Sci. 2017, 8, 955. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| A | Gene ID | Primer Sequences | Amplicfication Efficency (%) | Amplicon Length (bp) | TM (°C) |
|---|---|---|---|---|---|
| Actin | CL1167.Contig3_All | F: GCCAAATCATCGCCGTGTT | 92.482 | 80 | 82.56 |
| R: CCTCTCCTGTCACTGCTTTAATCTC | |||||
| Actin | CL2126.Contig2_All | F: GCCTTCAACCAACCAGACTTACG | 97.940 | 82 | 85.21 |
| R: GAACTAGAATCCCAGAGGCAAATG | |||||
| Actin | CL2172.Contig2_All | F: CCGACTGAAATGGATCTCGAA | 97.220 | 80 | 83.27 |
| R: GCGATGCGAAAACCCTCTATAC | |||||
| Actin | CL2172.Contig3_All | F: CCGACTGAAATGGATCTCGAA | 104.69 | 80 | 81.86 |
| R: GCGATGCGAAAACCCTCTATAC | |||||
| Actin | CL3559.Contig7_All | F: CAGAAGCGCCTCTCAATCCA | 90.482 | 82 | 79.24 |
| R: CATAGCAGGAGCGTTGAACGT | |||||
| Actin | CL494.Contig13_All | F: GTCGGCTCTAAATCCAGAATCCT | 95.123 | 80 | 82.63 |
| R: TTCGGAGAGAAGCTGAGAAGCA | |||||
| Actin | CL5740.Contig1_All | F: ACCTTTTGGATTGTGGGCTAGA | 91.052 | 80 | 83.56 |
| R: AGCTCCACTTGCACTTTTCCTT | |||||
| Actin | CL5740.Contig2_All | F: ACCTTTTGGATTGTGGGCTAGA | 98.549 | 81 | 80.23 |
| R: AGCTCCACTTGCACTTTTCCTT | |||||
| Actin | CL5740.Contig5_All | F: CTGGACAAAAGGCCGGAATT | 94.082 | 80 | 82.64 |
| R: TTTGCCATGTGCAGACTTTGG | |||||
| Actin | CL7856.Contig2_All | F: GCTCCTGCTTGCCTTCTTGT | 98.792 | 80 | 82.36 |
| R: CCCTGATAGCAGGATCTCAAGTTT | |||||
| Actin | Unigene12465_All | F: CTGGTAGCAAAACCCCACTCTGA | 96.517 | 80 | 83.49 |
| R: ATCCCACCTCCTTGGCCATAT | |||||
| Actin | Unigene20323_All | F: TCGCAGCCTCAACTCCAAAT | 95.378 | 80 | 84.91 |
| R: CCATTAACGGTGGCAAATCTC | |||||
| Actin | Unigene23839_All | F: TACTGACACTGCCCTTTGCTTTG | 95.836 | 80 | 78.66 |
| R: ACTTGCGACCAAGCATTTCC | |||||
| Actin | Unigene6171_All | F: ACGCCTGGGAAAAGACAAAA | 94.019 | 80 | 81.84 |
| R: AGAACCGACGACACCATTGAC | |||||
| Chy | Unigene26262_All | F: CATTGTGATGGCTGCGGTAT | 103.26 | 83 | 82.36 |
| R: GGCCTAAGCTAATCGAGATGCTT | |||||
| 18S rRNA | CL5051.Contig1_All | F: CAACCTCTCCCGCCAAATCT | 96.502 | 80 | 85.42 |
| R: GCAGTGGTGGTGATGCCATT | |||||
| Tub | CL1466.Contig3_All | F: ACGTCCAAGGTGGCCAATGT | 96.525 | 80 | 84.25 |
| R: TGGGTCTATGCCGTGTTCATC | |||||
| Tub | CL1466.Contig7_All | F: ACTCAGCACCCCATCCTTTG | 97.366 | 80 | 79.25 |
| R: GGAATCGCAAGCAGCAAGTC | |||||
| Tub | CL3192.Contig5_All | F: TTGGACCGCATTCGTAAGC | 99.78 | 80 | 83.45 |
| R: GTACCCCCACCAACAGCATT | |||||
| Tub | CL7489.Contig2_All | F: ATCGACCTTGCAGGCCTGTT | 100.677 | 81 | 81.32 |
| R: CTCCCGACAAGCTTCGGATATC | |||||
| Tub | Unigene3128_All | F: CGGAAGCGATTTACTGAGGAA | 100.745 | 80 | 80.54 |
| R: TGTATGTTGTGCCGCTCACA |
| Gene | Different Cultivars Samples in Lingonberry | |||||||
|---|---|---|---|---|---|---|---|---|
| GeNorm | NormFinder | BestKeeper | Com. | |||||
| M | Rank | S | Rank | SD | CV (%) | Rank | Rank | |
| 18S rRNA CL5051.Contig1 | 1.56 | 16 | 1.262 | 16 | 0.44 | 1.55 | 1 | 11 |
| Actin CL1167.Contig3 | 1.18 | 11 | 0.804 | 9 | 1.21 | 4.45 | 7 | 9 |
| Actin CL2126.Contig2 | 1.70 | 18 | 1.434 | 18 | 1.49 | 5.32 | 10 | 15 |
| Actin CL2172.Contig2 | 1.10 | 10 | 0.871 | 11 | 1.42 | 6.31 | 8 | 10 |
| Actin CL2172.Contig3 | 0.70 | 5 | 0.532 | 6 | 1.10 | 4.67 | 4 | 2 |
| Actin CL3559.Contig7 | 1.35 | 13 | 1.101 | 13 | 1.59 | 5.75 | 13 | 14 |
| Actin CL494.Contig13 | 0.54 | 4 | 0.449 | 4 | 1.80 | 6.57 | 15 | 8 |
| Actin CL5740.Contig1 | 0.92 | 8 | 0.641 | 8 | 2.05 | 7.52 | 17 | 11 |
| Actin CL5740.Contig2 | 1.43 | 14 | 1.125 | 14 | 2.29 | 7.79 | 18 | 15 |
| Actin CL5740.Contig5 | 1.84 | 20 | 1.594 | 20 | 2.92 | 10.48 | 20 | 21 |
| Actin CL7856.Contig2 | 1.91 | 21 | 1.630 | 21 | 1.10 | 3.44 | 4 | 15 |
| Actin Unigene12465 | 1.02 | 9 | 0.852 | 10 | 0.54 | 2.04 | 2 | 7 |
| Actin Unigene20323 | 0.87 | 7 | 0.469 | 5 | 1.19 | 3.87 | 6 | 6 |
| Actin Unigene23839 | 0.32 | 1 | 0.110 | 1 | 1.54 | 5.20 | 12 | 1 |
| Actin Unigene6171 | 0.44 | 3 | 0.319 | 3 | 1.50 | 5.16 | 11 | 4 |
| Chy Unigene26262 | 1.49 | 15 | 1.201 | 15 | 2.00 | 6.48 | 16 | 15 |
| Tub CL1466.Contig3 | 0.32 | 1 | 0.309 | 2 | 1.61 | 5.54 | 14 | 4 |
| Tub CL1466.Contig7 | 0.81 | 6 | 0.586 | 7 | 1.02 | 3.22 | 3 | 3 |
| Tub CL3192.Contig5 | 1.76 | 19 | 1.448 | 19 | 2.32 | 8.48 | 19 | 20 |
| Tub CL7489.Contig2 | 1.62 | 17 | 1.293 | 17 | 3.47 | 12.69 | 21 | 19 |
| Tub Unigene3128 | 1.23 | 12 | 1.006 | 12 | 1.43 | 5.09 | 9 | 11 |
| Best genes | Actin Unigene23839/ | Actin Unigene23839 | 18S rRNA CL5051.Contig1 | Actin Unigene 23839 | ||||
| Tub CL1466.Contig3 | ||||||||
| Worst genes | Actin CL7856.Contig2 | Actin CL7856.Contig2 | Tub CL7489.Contig2 | Actin CL5740 Contig5 | ||||
| Gene | Different Tissues Samples in Lingonberry | |||||||
|---|---|---|---|---|---|---|---|---|
| GeNorm | NormFinder | BestKeeper | Com. | |||||
| M | Rank | S | Rank | SD | CV (%) | Rank | Rank | |
| 18S rRNA CL5051.Contig1 | 1.80 | 17 | 1.341 | 17 | 1.44 | 4.95 | 10 | 16 |
| Actin CL1167.Contig3 | 1.63 | 14 | 1.162 | 14 | 1.73 | 6.41 | 15 | 15 |
| Actin CL2126.Contig2 | 1.86 | 18 | 1.414 | 18 | 1.90 | 6.88 | 18 | 18 |
| Actin CL2172.Contig2 | 1.46 | 11 | 0.779 | 9 | 1.09 | 4.89 | 4 | 8 |
| Actin CL2172.Contig3 | 1.36 | 9 | 0.658 | 7 | 0.91 | 3.90 | 3 | 5 |
| Actin CL3559.Contig7 | 2.15 | 21 | 2.253 | 21 | 1.63 | 5.18 | 12 | 18 |
| Actin CL494.Contig13 | 1.75 | 16 | 1.176 | 15 | 1.37 | 5.34 | 8 | 14 |
| Actin CL5740.Contig1 | 0.91 | 3 | 0.615 | 3 | 1.68 | 6.34 | 14 | 7 |
| Actin CL5740.Contig2 | 0.65 | 1 | 0.567 | 2 | 1.24 | 4.32 | 5 | 1 |
| Actin CL5740.Contig5 | 1.08 | 4 | 0.624 | 4 | 1.38 | 5.21 | 9 | 3 |
| Actin CL7856.Contig2 | 2.00 | 20 | 1.657 | 20 | 2.71 | 9.02 | 21 | 21 |
| Actin Unigene12465 | 1.15 | 5 | 0.514 | 1 | 0.86 | 3.19 | 2 | 1 |
| Actin Unigene20323 | 1.25 | 6 | 0.642 | 6 | 1.31 | 4.33 | 6 | 4 |
| Actin Unigene23839 | 0.65 | 1 | 0.636 | 5 | 1.65 | 5.68 | 13 | 5 |
| Actin Unigene6171 | 1.40 | 10 | 0.964 | 10 | 1.80 | 5.94 | 17 | 13 |
| Chy Unigene26262 | 1.32 | 8 | 1.003 | 11 | 1.75 | 6.01 | 16 | 12 |
| Tub CL1466.Contig3 | 1.53 | 12 | 1.091 | 13 | 1.93 | 6.63 | 19 | 16 |
| Tub CL1466.Contig7 | 1.58 | 13 | 1.073 | 12 | 1.31 | 4.26 | 6 | 10 |
| Tub CL3192.Contig5 | 1.70 | 15 | 1.191 | 16 | 0.82 | 2.69 | 1 | 11 |
| Tub CL7489.Contig2 | 1.93 | 19 | 1.524 | 19 | 2.38 | 8.21 | 20 | 20 |
| Tub Unigene3128 | 1.28 | 7 | 0.712 | 8 | 1.53 | 5.18 | 11 | 9 |
| Best genes | Actin CL5740.Contig2/ | Actin Unigene12465 | Tub CL3192.Contig5 | Actin CL5740.Contig2/ | ||||
| Actin Unigene23839 | Actin Unigene 12465 | |||||||
| Worst genes | Actin CL3559.Contig7 | Actin CL3559.Contig7 | Actin CL7856.Contig2 | Tub CL7489.Contig2 | ||||
| Gene | Lingonberries Treated by Alkali Stress | |||||||
|---|---|---|---|---|---|---|---|---|
| GeNorm | NormFinder | BestKeeper | Com. | |||||
| M | Rank | S | Rank | SD | CV (%) | Rank | Rank | |
| 18S rRNA CL5051.Contig1 | 1.24 | 13 | 0.942 | 16 | 1.62 | 5.28 | 21 | 18 |
| Actin CL1167.Contig3 | 0.92 | 8 | 0.847 | 15 | 1.55 | 5.08 | 18 | 14 |
| Actin CL2126.Contig2 | 1.35 | 16 | 0.842 | 13 | 1.25 | 4.08 | 15 | 16 |
| Actin CL2172.Contig2 | 0.53 | 3 | 0.551 | 4 | 0.79 | 3.24 | 6 | 2 |
| Actin CL2172.Contig3 | 0.48 | 1 | 0.642 | 7 | 1.16 | 4.46 | 13 | 5 |
| Actin CL3559.Contig7 | 1.11 | 11 | 0.745 | 9 | 0.45 | 1.40 | 1 | 5 |
| Actin CL494.Contig13 | 0.97 | 9 | 0.535 | 3 | 1.16 | 4.03 | 13 | 10 |
| Actin CL5740.Contig1 | 0.77 | 6 | 0.361 | 2 | 0.60 | 1.98 | 3 | 1 |
| Actin CL5740.Contig2 | 0.70 | 5 | 0.359 | 1 | 0.95 | 2.98 | 8 | 3 |
| Actin CL5740.Contig5 | 0.60 | 4 | 0.634 | 6 | 1.02 | 3.37 | 10 | 4 |
| Actin CL7856.Contig2 | 1.39 | 17 | 0.945 | 17 | 0.51 | 1.53 | 2 | 13 |
| Actin Unigene12465 | 0.48 | 1 | 0.779 | 11 | 1.15 | 4.16 | 12 | 9 |
| Actin Unigene20323 | 1.43 | 18 | 1.072 | 18 | 0.62 | 1.88 | 5 | 14 |
| Actin Unigene23839 | 1.05 | 10 | 0.560 | 5 | 0.94 | 2.89 | 7 | 7 |
| Actin Unigene6171 | 1.18 | 12 | 0.657 | 8 | 0.60 | 1.79 | 3 | 8 |
| Chy Unigene26262 | 1.54 | 20 | 1.236 | 20 | 1.55 | 4.69 | 18 | 20 |
| Tub CL1466.Contig3 | 1.49 | 19 | 1.141 | 19 | 1.47 | 4.80 | 17 | 19 |
| Tub CL1466.Contig7 | 1.70 | 21 | 2.058 | 21 | 1.61 | 4.93 | 20 | 21 |
| Tub CL3192.Contig5 | 1.32 | 15 | 0.843 | 14 | 1.33 | 4.45 | 16 | 17 |
| Tub CL7489.Contig2 | 0.83 | 7 | 0.753 | 10 | 1.10 | 3.59 | 11 | 11 |
| Tub Unigene3128 | 1.28 | 14 | 0.808 | 12 | 0.95 | 3.08 | 8 | 12 |
| Best genes | Actin CL2172.Contig3/ | Actin CL5740.Contig2 | Actin CL3559.Contig7 | Actin CL5740.Contig1 | ||||
| Actin Unigene12465 | ||||||||
| Worst genes | Tub CL1466.Contig7 | Tub CL1466.Contig7 | 18S rRNA CL5051.Contig1 | Tub CL1466.Contig7 | ||||
| Gene | Lingonberries Treated by PEG-Simulated Drought Stress | |||||||
|---|---|---|---|---|---|---|---|---|
| GeNorm | NormFinder | BestKeeper | Com. | |||||
| M | Rank | S | Rank | SD | CV (%) | Rank | Rank | |
| 18S rRNA CL5051.Contig1 | 1.73 | 19 | 1.510 | 19 | 1.96 | 6.14 | 19 | 19 |
| Actin CL1167.Contig3 | 1.29 | 11 | 0.820 | 12 | 1.27 | 4.22 | 16 | 14 |
| Actin CL2126.Contig2 | 1.38 | 13 | 0.649 | 4 | 0.99 | 3.31 | 8 | 9 |
| Actin CL2172.Contig2 | 1.09 | 6 | 0.439 | 1 | 0.58 | 2.45 | 4 | 1 |
| Actin CL2172.Contig3 | 1.91 | 21 | 1.795 | 21 | 2.44 | 8.79 | 21 | 21 |
| Actin CL3559.Contig7 | 1.34 | 12 | 0.661 | 5 | 0.72 | 2.32 | 7 | 8 |
| Actin CL494.Contig13 | 0.94 | 5 | 0.482 | 2 | 0.69 | 2.40 | 6 | 3 |
| Actin CL5740.Contig1 | 1.63 | 18 | 1.302 | 18 | 1.61 | 5.45 | 18 | 18 |
| Actin CL5740.Contig2 | 1.16 | 7 | 0.697 | 6 | 1.01 | 3.23 | 12 | 9 |
| Actin CL5740.Contig5 | 1.20 | 8 | 0.764 | 9 | 1.15 | 3.93 | 13 | 11 |
| Actin CL7856.Contig2 | 1.26 | 10 | 1.071 | 15 | 0.99 | 3.04 | 8 | 12 |
| Actin Unigene12465 | 1.23 | 9 | 0.606 | 3 | 1.00 | 3.71 | 11 | 7 |
| Actin Unigene20323 | 1.51 | 16 | 1.240 | 17 | 1.19 | 3.62 | 14 | 16 |
| Actin Unigene23839 | 0.51 | 1 | 0.766 | 10 | 0.55 | 1.71 | 3 | 4 |
| Actin Unigene6171 | 1.42 | 14 | 0.982 | 14 | 0.99 | 2.96 | 8 | 13 |
| Chy Unigene26262 | 0.75 | 4 | 0.746 | 8 | 0.36 | 1.07 | 2 | 4 |
| Tub CL1466.Contig3 | 0.51 | 1 | 0.814 | 11 | 0.65 | 2.10 | 5 | 6 |
| Tub CL1466.Contig7 | 0.60 | 3 | 0.745 | 7 | 0.28 | 0.82 | 1 | 1 |
| Tub CL3192.Contig5 | 1.82 | 20 | 1.523 | 20 | 2.26 | 7.40 | 20 | 20 |
| Tub CL7489.Contig2 | 1.57 | 17 | 1.180 | 16 | 1.53 | 4.94 | 17 | 17 |
| Tub Unigene3128 | 1.46 | 15 | 0.847 | 13 | 1.20 | 3.91 | 15 | 15 |
| Best genes | Actin Unigene23839/ | Actin CL2172.Contig2 | Tub CL1466.Contig7 | 18S rRNA CL5051. Contig1 | ||||
| Tub CL1466.Contig3 | ||||||||
| Worst genes | Actin CL2172.Contig3 | Actin CL2172.Contig3 | Actin CL2172.Contig3 | Actin CL2172.Contig3 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Xu, J.; Wang, Q.; Li, J.; Li, Y.; Dong, M.; Sun, H. Transcriptome-Based Identification of the Optimal Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Analyses of Lingonberry Fruits throughout the Growth Cycle. Plants 2023, 12, 4180. https://doi.org/10.3390/plants12244180
Zhang W, Xu J, Wang Q, Li J, Li Y, Dong M, Sun H. Transcriptome-Based Identification of the Optimal Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Analyses of Lingonberry Fruits throughout the Growth Cycle. Plants. 2023; 12(24):4180. https://doi.org/10.3390/plants12244180
Chicago/Turabian StyleZhang, Wanchen, Jian Xu, Qiang Wang, Jing Li, Yadong Li, Mei Dong, and Haiyue Sun. 2023. "Transcriptome-Based Identification of the Optimal Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Analyses of Lingonberry Fruits throughout the Growth Cycle" Plants 12, no. 24: 4180. https://doi.org/10.3390/plants12244180
APA StyleZhang, W., Xu, J., Wang, Q., Li, J., Li, Y., Dong, M., & Sun, H. (2023). Transcriptome-Based Identification of the Optimal Reference Genes for Quantitative Real-Time Polymerase Chain Reaction Analyses of Lingonberry Fruits throughout the Growth Cycle. Plants, 12(24), 4180. https://doi.org/10.3390/plants12244180






