Effects of Plant Regulators on the Seed Germination and Antioxidant Enzyme Activity of Cotton under Compound Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Seed Germination
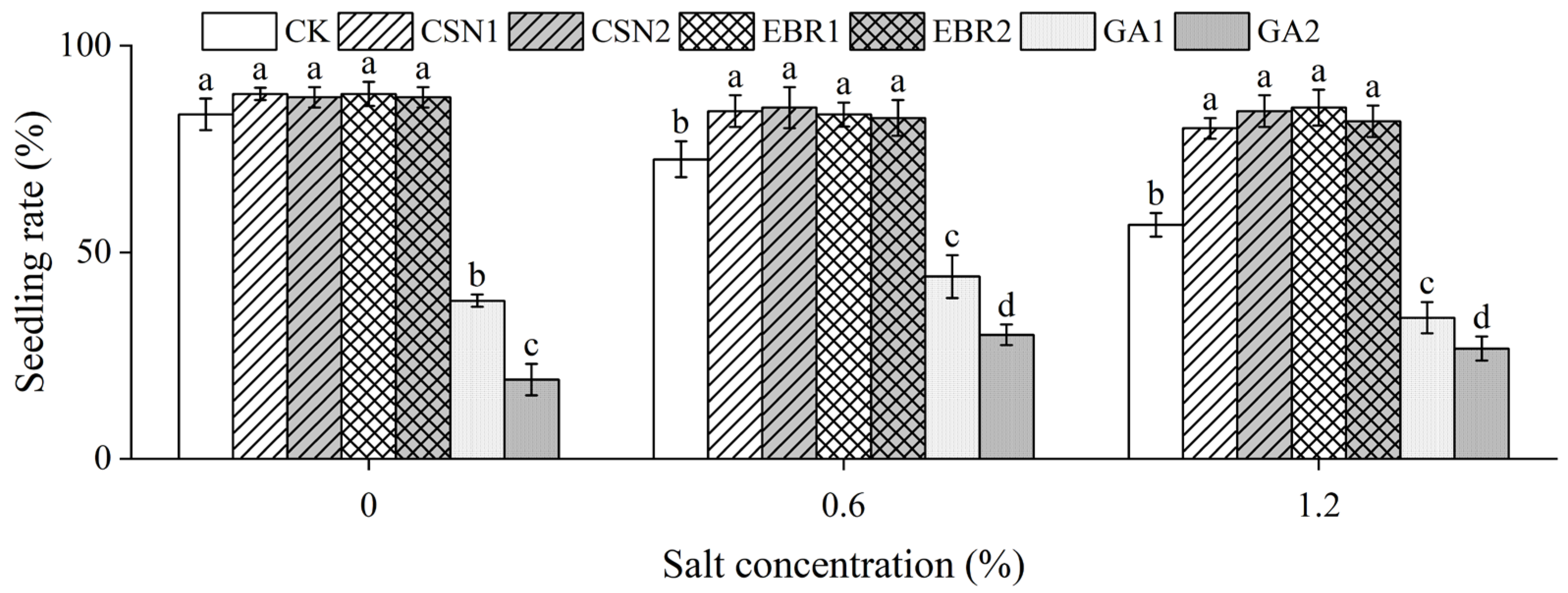
2.2. Seedling Rate
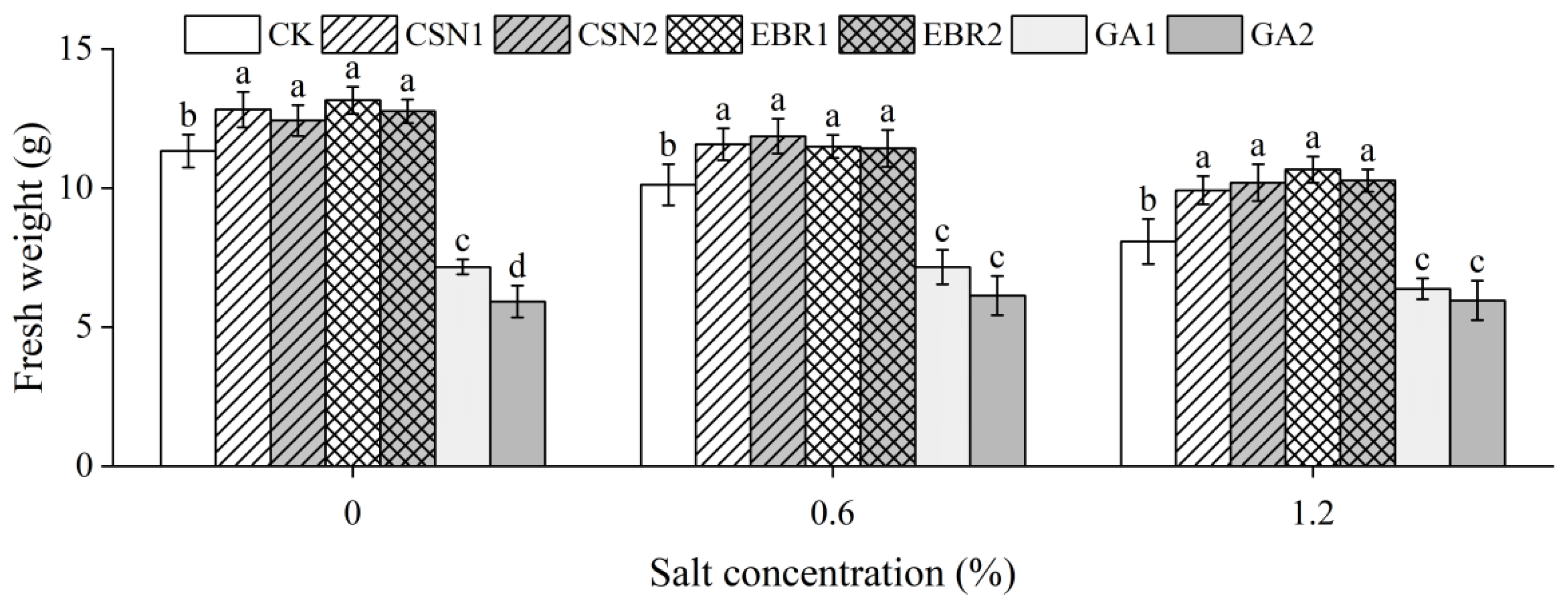
2.3. Fresh Weight
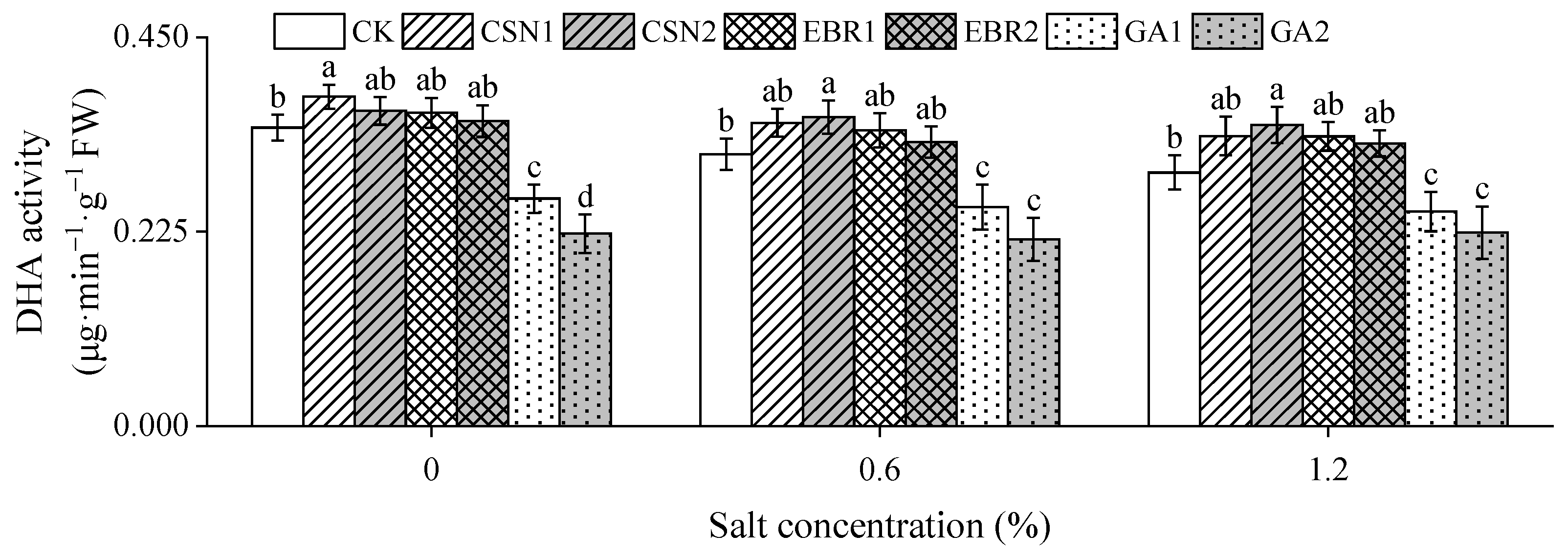
2.4. Dehydrogenase Activity
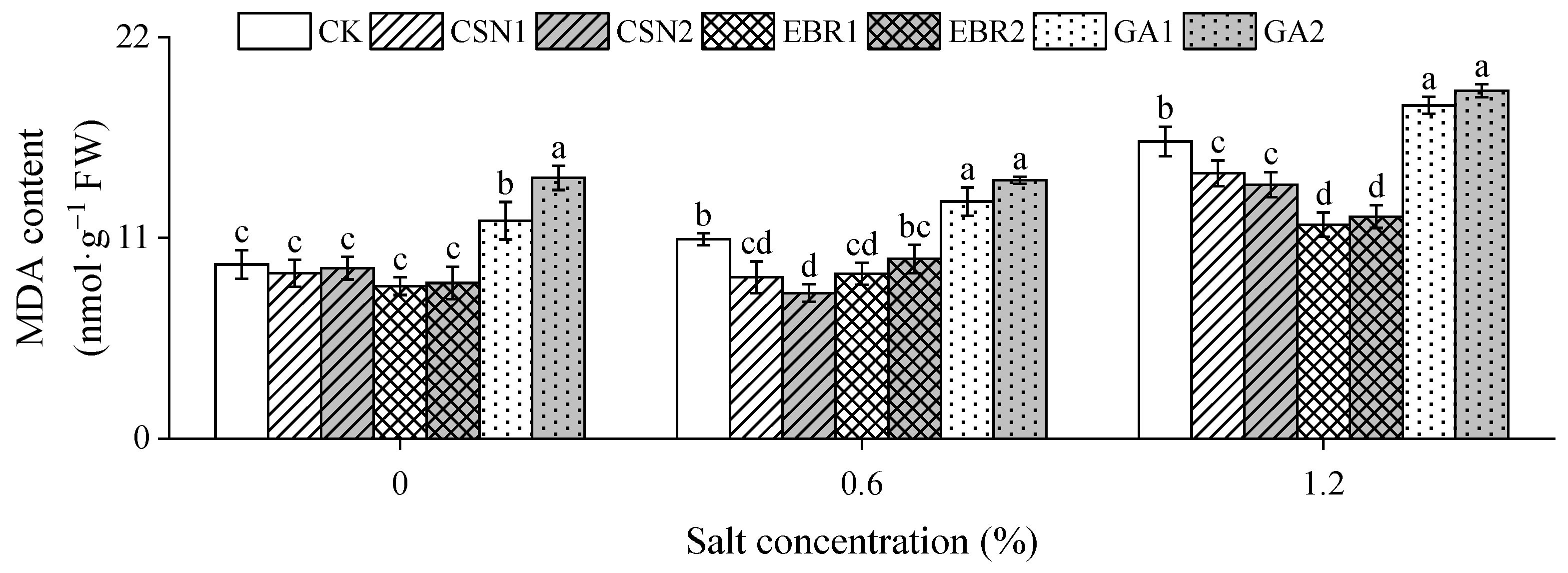
2.5. Malondialdehyde Content
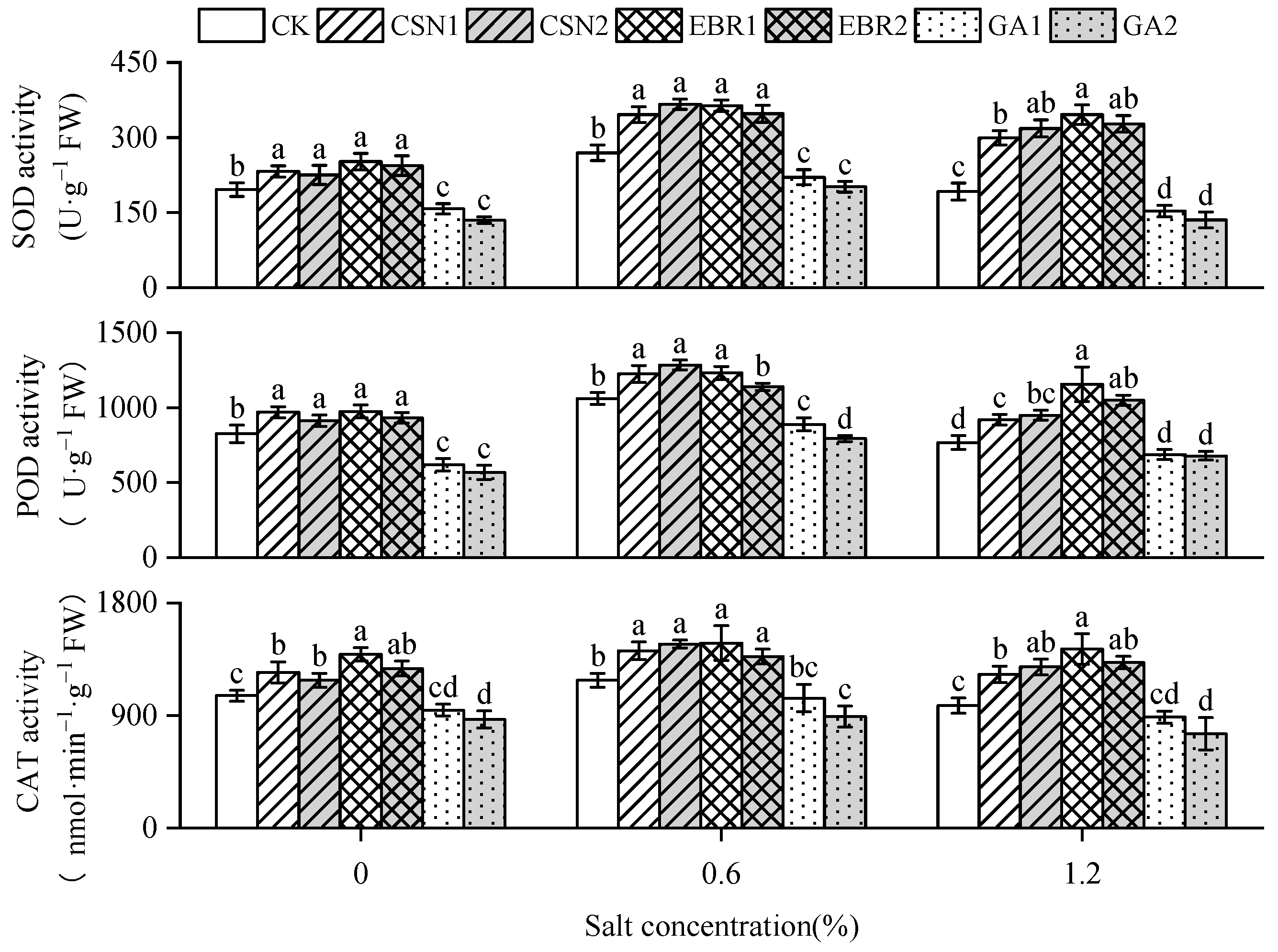
2.6. Antioxidant Enzyme Activity
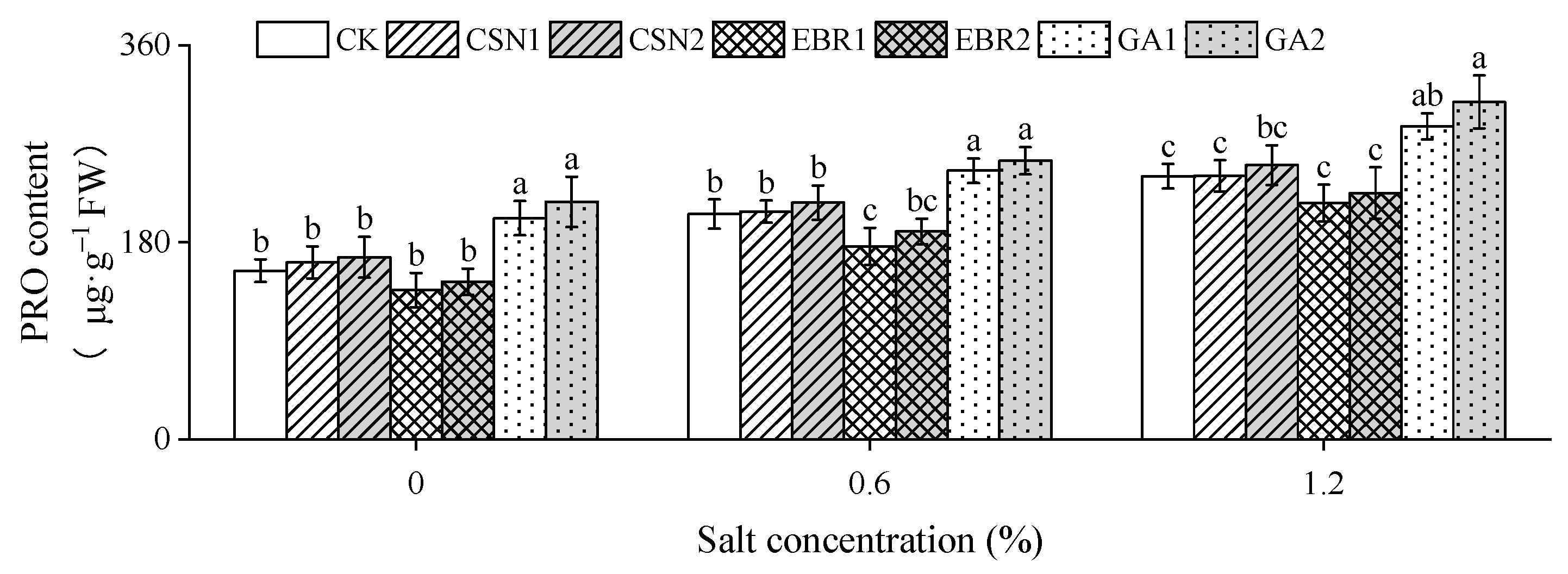
2.7. Proline Content
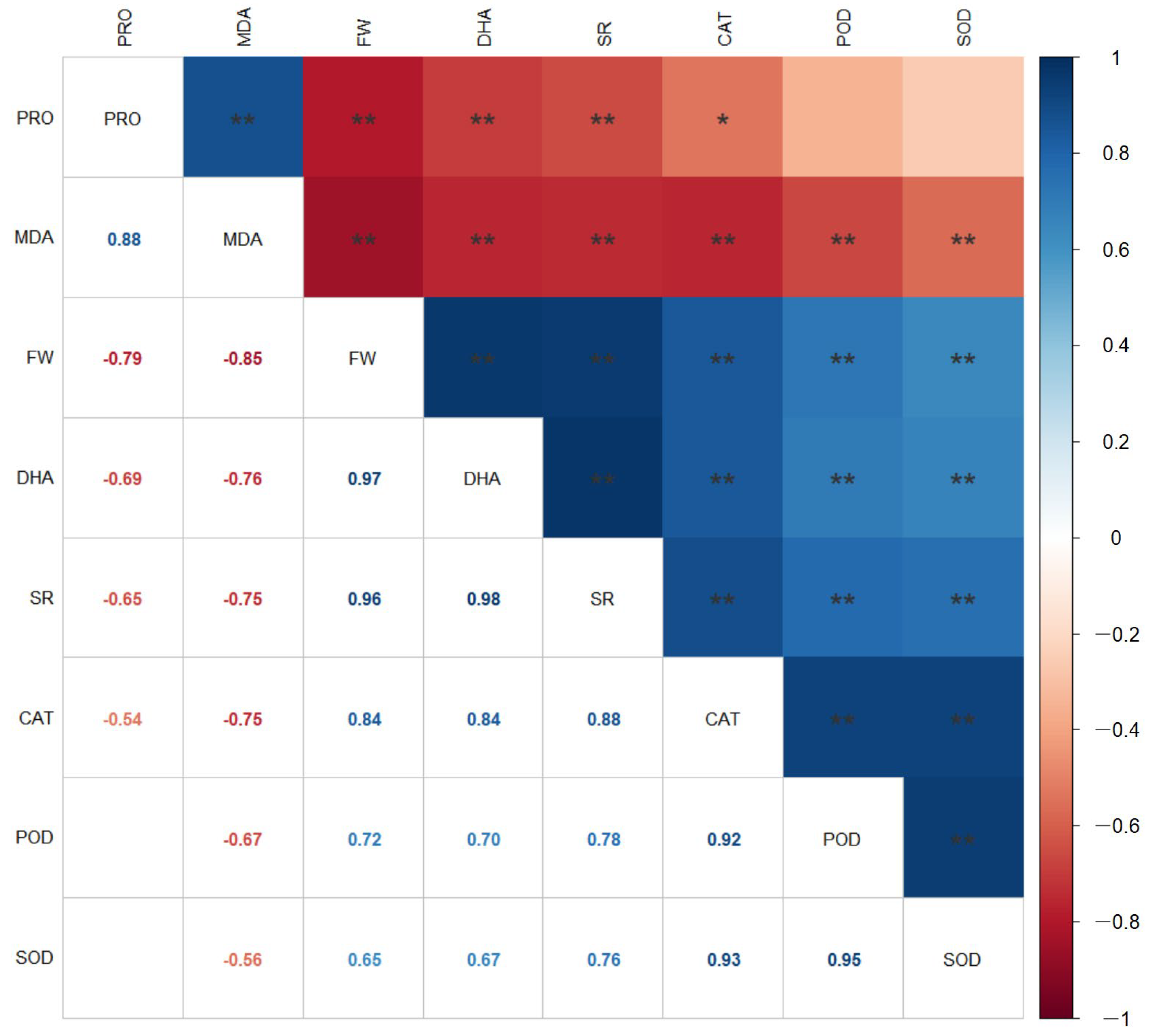
2.8. Correlation Analysis
2.9. Comprehensive Evaluation
3. Discussion
4. Materials and Methods
4.1. Experimental Location and Materials
4.2. Cotton Seed Treatment
4.3. Experimental Design
4.3.1. Seed Germination Test
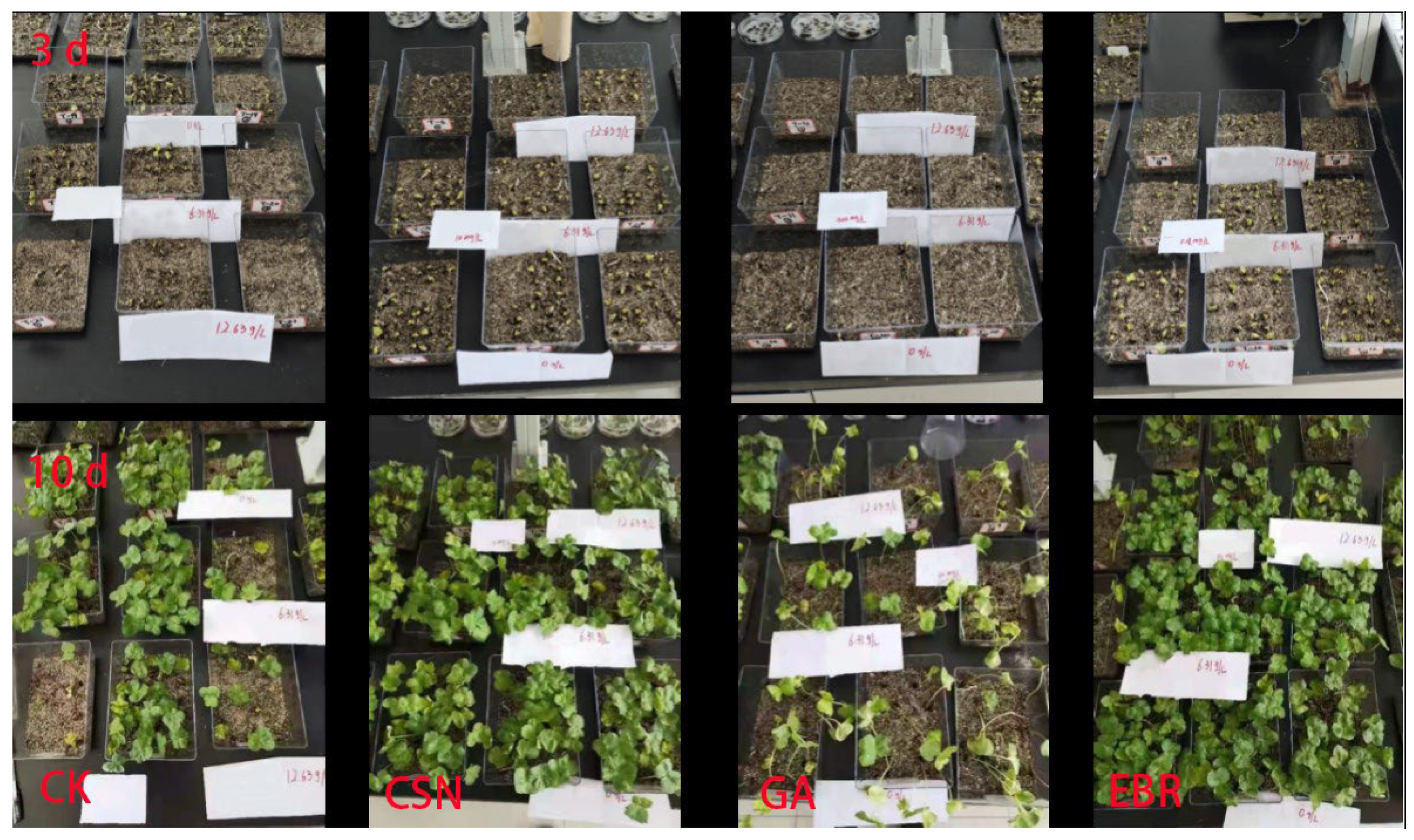
4.3.2. Seedling Growth Test
4.4. Estimation of Plant Growth Parameters
4.5. Estimation of Root Viability
4.6. Estimation of Lipid Peroxidation
4.7. Antioxidant Enzyme Activity Determination
4.8. Estimation of Proline Content
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, W.J.; Jiang, X.H.; Fu, Y.Y.; Shen, X.J.; Gao, Y.; Wang, X.P. The Effects of Salt Stress on Chlorophyll Fluorescence of Cotton Seedling Leaves. J. Irrig. Drain. 2021, 40, 23–28. [Google Scholar] [CrossRef]
- Liu, Y. Cotton sowing area in the region exceeds 1.4 million acres. Xinjiang Daily, 10 April 2023, Version 001; Available online: https://xjrb.ts.cn/xjrb/20230410/207597.html (accessed on 1 September 2023).
- Khorsandi, F.; Anagholi, A. Reproductive compensation of cotton after salt stress relief at different growth stages. J. Agron. Crop Sci. 2009, 195, 278–283. [Google Scholar] [CrossRef]
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gómez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the physiological and biochemical impacts of salt stress on plant growth and development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- Johnson, R.; Puthur, J.T. Seed priming as a cost effective technique for developing plants with cross tolerance to salinity stress. Plant Physiol. Biochem. 2021, 162, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Abido, W.; Allem, A.; Zsombic, L.; Attila, N. Effect of gibberellic acid on germination of six wheat cultivars under salinity stress levels. Asian J. Biol. Sci. 2019, 12, 51–60. [Google Scholar] [CrossRef]
- Sedghi, M.; Nemati, A.; Esmaielpour, B. Effect of seed priming on germination and seedling growth of two medicinal plants under salinity. Emir. J. Food Agric. 2010, 22, 130–139. [Google Scholar] [CrossRef]
- Xia, J.; Hao, X.Z.; Wang, T.G.; Li, H.Q.; Shi, X.J.; Liu, Y.C.; Luo, H.H. Seed Priming with Gibberellin Regulates the Germination of Cotton Seeds Under Low-Temperature Conditions. Plant Growth Regul. 2023, 42, 319–334. [Google Scholar] [CrossRef]
- Swaefy, H.M.; El-Ziat, R.A. Calendula Response to Salinity Stress. New Perspect. Agric. Crop Sci. 2020, 3, 978. [Google Scholar] [CrossRef]
- Huang, B.; Li, W.K.; Sun, M.T.; Yan, Y.; Wang, J.; He, C.X.; Yu, X.C.; Li, Y.S. Effect of sodium nitrophenol on seed germination and cold tolerance of seedlings at low temperature. J. Agron. Crop Sci. 2022, 36, 845–855. [Google Scholar] [CrossRef]
- Przybysz, A.; Gawrońska, H.; Gajc-Wolska, J. Biological mode of action of a nitrophenolates-based biostimulant: Case study. Front. Plant Sci. 2014, 5, 713. [Google Scholar] [CrossRef] [PubMed]
- Batool, Z.; Ishfaq, M.; Akbar, N.; Zulfiqar, U.; Anjum, S.A.; Shafiq, M.; Nazir, S.; Aziz, A. Exogenous application of Atonik (sodium nitrophenolate) under skip irrigation regimes modulated the physiology, growth and productivity of Zea mays L. Arch. Agron. Soil Sci. 2023, 69, 2325–2339. [Google Scholar] [CrossRef]
- Rattan, A.; Kapoor, D.; Kapoor, N.; Bhardwaj, R.; Sharma, A. Brassinosteroids regulate functional components of antioxidative defense system in salt stressed maize seedlings. J. Plant Growth Regul. 2020, 39, 1465–1475. [Google Scholar] [CrossRef]
- Semida, W.M.; Rady, M.M. Pre-soaking in 24-epibrassinolide or salicylic acid improves seed germination, seedling growth, and anti-oxidant capacity in Phaseolus vulgaris L. grown under NaCl stress. J. Hortic. Sci. Biotechnol. 2014, 89, 338–344. [Google Scholar] [CrossRef]
- Guo, H.J.; Hu, Z.Q.; Zhang, H.M.; Min, W.; Hou, Z.N. Comparative Effects of Salt and Alkali Stress on Antioxidant System in Cotton (Gossypium hirsutum L.) Leaves. Open Chem. 2019, 17, 1352–1360. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Wei, Z.H.; Liu, J.; Liu, X.Z.; Liu, F. Growth and physiological responses of cotton plants to salt stress. J. Agron. Crop Sci. 2021, 207, 565–576. [Google Scholar] [CrossRef]
- Gao, R.R.; Wei, X.Y.; He, Z.; Zhao, R.H.; Wang, K.; Yang, X.J.; Walck, J.L. Soil salt and NaCl have different effects on seed germination of the halophyte Suaeda salsa. J. Plant Nutr. Soil Sci. 2018, 181, 488–497. [Google Scholar] [CrossRef]
- Khan, M.O.; Irfan, M.; Muhammad, A.; Ullah, I.; Nawaz, S.; Khalil, M.K.; Ahmad, M. A practical and economical strategy to mitigate salinity stress through seed priming. Front. Environ. Sci. 2022, 10, 1–14. [Google Scholar] [CrossRef]
- Saradadevi, G.P.; Das, D.; Mangrauthia, S.K.; Mohapatra, S.; Chikkaputtaiah, C.; Roorkiwal, M.; Solanki, M.; Sundaram, R.M.; Chirravuri, N.N.; Sakhare, A.S. Genetic, Epigenetic, Genomic and Microbial Approaches to Enhance Salt Tolerance of Plants: A comprehensive review. Biology 2021, 10, 1255. [Google Scholar] [CrossRef]
- Alkharabsheh, H.M.; Seleiman, M.F.; Hewedy, O.A.; Battaglia, M.L.; Jalal, R.S.; Alhammad, B.A.; Schillaci, C.; Ali, N.; Al-Doss, A. Field Crop Responses and Management Strategies to Mitigate Soil Salinity in Modern Agriculture: A Review. Agronomy 2021, 11, 2299. [Google Scholar] [CrossRef]
- Tao, R.; Liu, T.; Chu, G.X. Effects of Nitrapyrin Soaking on Cotton Germination Rate and Its Salt Resistant Physiological Characteristics in Salt Stress Condition. Cotton Sci. 2013, 25, 426–431. [Google Scholar]
- Li, H.P.; Sun, H.C.; Ping, W.C.; Liu, L.T.; Zhang, Y.J.; Zhang, K.; Bai, Z.Y.; Li, A.C.; Zhu, J.J.; Li, C.D. Exogenous Ethylene Promotes the Germination of Cotton Seeds Under Salt Stress. J. Plant Growth Regul. 2023, 42, 3923–3933. [Google Scholar] [CrossRef]
- Niedergerke, R.; Lüttgau, H.C. Calcium and the Contraction of the Heart: Antagonism between Calcium and Sodium Ions. Nature 1957, 179, 1066–1067. [Google Scholar] [CrossRef] [PubMed]
- Appenroth, K.J.; Gabrys, H.; Scheuerlein, R.W. Ion antagonism in phytochrome-mediated calcium-dependent germination of turions of Spirodela polyrhiza (L.) Schleiden. Planta 1999, 208, 583–587. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Zhou, X.P.; Tao, M.; Yuan, F.; Liu, L.L.; Wu, F.H.; Wu, X.M.; Xiang, Y.; Niu, Y.; Liu, F. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nat. Aust. 2019, 572, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Chakma, S.P.; Chileshe, S.M.; Thomas, R.; Krishna, P. Cotton seed priming with brassinosteroid promotes germination and seedling growth. Agronomy 2021, 11, 566. [Google Scholar] [CrossRef]
- Chang, D.; Yang, Y.; Wang, Y.; Zhang, X.Y.; Zhang, F.C.; LI, F.G. Effect of 24-Epi Brassinolide on Seed Germination under Stresses of Salt and PEG in Cotton. Acta Agric. Boreali-Occident. Sin. 2015, 24, 96–101. [Google Scholar] [CrossRef]
- Qu, X.; Jiao, Y.S.; Wang, R.H.; Liu, S.Y.; Li, Y.J. Effects of gibberellin and sodium nitrophenolate on seed germination and seedling vigor of pepper. China Cucurbits Veg. 2019, 32, 59–63. [Google Scholar] [CrossRef]
- Huang, Y.T.; Cai, S.Y.; Ruan, X.L.; Xu, J.; Cao, D.D. CSN improves seed vigor of aged sunflower seeds by regulating the fatty acid, glycometabolism, and abscisic acid metabolism. J. Adv. Res. 2021, 33, 1–13. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Han, C.; Liu, Z.M.; Yu, F.Y.; Wu, Q.K. 24-Epibrassinolide promotes fatty acid accumulation and the expression of related genes in Styrax tonkinensis seeds. Int. J. Mol. Sci. 2022, 23, 8897. [Google Scholar] [CrossRef]
- Wang, N.; Gao, J.; Huang, W.J.; Li, B.; Tang, Z.S.; Song, Z.X.; Liu, A.P. Time-concentration effects of GA3 on germination of Astragalus membranaceus under severe drought and salt stresses. Chin. J. Ecol. 2018, 37, 2339–2344. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Wei, Q.H.; Wang, R.M.; Zhang, X. Effect of exogenous gibberellin on foxtail millet seed germination under salt stress. Crops 2012, 06, 139–141. [Google Scholar] [CrossRef]
- Xue, Z.Z.; Wu, X.H. Effect of Different GA3 on the Germination of Tomato Seed Under Salt Stress. North. Hortic. 2011, 15, 59–61. [Google Scholar]
- Su, W.X.; Xu, L.X.; Jiang, W.T.; Liu, Y.L.; Wang, F.; Lv, Y.R.; Yan, J.X. Effects of Different Exogenous Substances on Seed Germination and Seedling Growth and Physiology of Perilla frutescens under Saline-alkali Stress. Acta Agrestia Sin. 2022, 30, 2415–2422. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Taylor, N.L.; Millar, A.H. The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci. 2011, 16, 614–623. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.F.; Hill, C.B.; Zhang, X.Q.; Li, C.D. Genome-Wide Association Study of Salinity Tolerance During Germination in Barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Teakle, N.L.; Tyerman, S.D. Mechanisms of Cl-transport contributing to salt tolerance. Plant Cell Environ. Behav. 2010, 33, 566–589. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Zhang, S.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Brassinosteroid improves seed germination and early development of tomato seedling under phenanthrene stress. Plant Growth Regul. 2012, 68, 87–96. [Google Scholar] [CrossRef]
- Tanveer, M.; Shahzad, B.; Sharma, A.; Biju, S.; Bhardwaj, R. 24-Epibrassinolide; an active brassinolide and its role in salt stress tolerance in plants: A review. Plant Physiol. Biochem. 2018, 130, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Seth, C.S. 24-Epibrassinolide Regulates Functional Components of Nitric Oxide Signalling and Antioxidant Defense Pathways to Alleviate Salinity Stress in Brassica juncea L. cv. Varuna. J. Plant Growth Regul. 2023, 42, 4207–4222. [Google Scholar] [CrossRef]
- Fang, S.; Hou, X.; Liang, X.L. Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 2021, 12, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Munawar, W.; Hameed, A.; Khan, M.K.R. Differential Morphophysiological and Biochemical Responses of Cotton Genotypes Under Various Salinity Stress Levels During Early Growth Stage. Front. Plant Sci. 2021, 12, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.S.; An, M.Y.; Han, L.B.; Yin, S.X. Foliar Application of 24-Epibrassinolide Improved Salt Stress Tolerance of Perennial Ryegrass. HortSci. Horts 2015, 50, 1518–1523. [Google Scholar] [CrossRef]
- Sousa, V.Q.; Messias, W.F.S.; Pereira, Y.C.; da Silva, B.R.S.; Lobato, E.M.S.G.; Alyemeni, M.N.; Ahmad, P.; Lobato, A.K.d.S. Pretreatment with 24-Epibrassinolide Synergistically Protects Root Structures and Chloroplastic Pigments and Upregulates Antioxidant Enzymes and Biomass in Na+-Stressed Tomato Plants. J. Plant Growth Regul. 2021, 41, 2869–2885. [Google Scholar] [CrossRef]
- Bao, S.D. Soil Agrochemical Analysis, 3rd ed.; China Agricultural Press: Beijing China, 2000; p. 187. [Google Scholar]
- Li, C.; Lu, B.; Liu, L.T.; Duan, W.J.; Dan, J.; Li, J.; Zhang, K.; Sun, H.C.; Zhang, Y.J.; Li, C.D. Melatonin promotes seed germination under salt stress by regulating ABA and GA3 in cotton (Gossypium hirsutum L.). Plant Physiol. Biochem. 2021, 162, 506–516. [Google Scholar] [CrossRef]
- Hu, G.F.; Zhang, X.Y.; Mo, H.T.; Xiao, C.X.; Meng, Q.S. The Effects of Resistance Salt-alkaline Preparation on the Characters of Physiological and Biochemical Index of Cotton Seedling. Acta Agric. Boreali-Occident. Sin 2009, 18, 169–172. [Google Scholar] [CrossRef]
- Su, N.N.; Wu, Q.; Chen, J.H.; Shabala, L.; Mithöfer, A.; Wang, H.Y.; Qu, M.; Yu, M.; Cui, J.; Shabala, S. GABA operates upstream of H+-ATPase and improves salinity tolerance in Arabidopsis by enabling cytosolic K+ retention and Na+ exclusion. JExB 2019, 70, 6349–6361. [Google Scholar] [CrossRef]
- Mahmud, J.A.; Hasanuzzaman, M.; Khan, M.I.R.; Nahar, K.; Fujita, M. β-Aminobutyric Acid Pretreatment Confers Salt Stress Tolerance in Brassica napus L. by Modulating Reactive Oxygen Species Metabolism and Methylglyoxal Detoxification. Plants 2020, 9, 241. [Google Scholar] [CrossRef] [PubMed]
| Salt Concentration (%) | Treatment | Germination Potential (%) | Germination Rate (%) |
|---|---|---|---|
| 0 | CK | 88.33 ± 2.89 b | 96.67 ± 2.89 a |
| CSN1 | 95 ± 5 a | 98.33 ± 2.89 a | |
| CSN2 | 93.33 ± 2.89 ab | 98.33 ± 2.89 a | |
| EBR1 | 96.67 ± 2.89 a | 100 ± 0 a | |
| EBR2 | 93.33 ± 2.89 ab | 98.33 ± 2.89 a | |
| GA1 | 48.33 ± 2.89 c | 63.33 ± 2.89 b | |
| GA2 | 23.33 ± 2.89 d | 31.67 ± 2.89 c | |
| 0.6 | CK | 66.67 ± 2.89 b | 78.33 ± 2.89 b |
| CSN1 | 78.33 ± 2.89 a | 90 ± 0 a | |
| CSN2 | 83.33 ± 2.89 a | 91.67 ± 2.89 a | |
| EBR1 | 78.33 ± 2.89 a | 90 ± 0 a | |
| EBR2 | 78.33 ± 2.89 a | 88.33 ± 2.89 a | |
| GA1 | 53.33 ± 2.89 c | 66.67 ± 2.89 c | |
| GA2 | 33.33 ± 2.89 d | 53.33 ± 2.89 d | |
| 1.2 | CK | 36.67 ± 2.89 c | 55 ± 5 b |
| CSN1 | 53.33 ± 2.89 b | 71.67 ± 2.89 a | |
| CSN2 | 58.33 ± 2.89 ab | 73.33 ± 2.89 a | |
| EBR1 | 60 ± 5 a | 76.67 ± 2.89 a | |
| EBR2 | 56.67 ± 2.89 ab | 75 ± 5 a | |
| GA1 | 31.67 ± 2.89 cd | 48.33 ± 2.89 bc | |
| GA2 | 26.67 ± 2.89 d | 43.33 ± 2.89 c |
| Salt Concentration | Treatment | Membership Function | D Value | Rank | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GP | GR | SR | FW | DHA | MDA | SOD | POD | CAT | ||||
| 0 | CK | 0.89 | 0.95 | 0.93 | 0.75 | 0.77 | 0.80 | 0.52 | 0.63 | 0.36 | 0.73 | 5 |
| CSN1 | 0.98 | 0.98 | 1.00 | 0.95 | 1.00 | 0.88 | 0.83 | 0.99 | 0.72 | 0.93 | 2 | |
| CSN2 | 0.95 | 0.98 | 0.99 | 0.90 | 0.90 | 0.84 | 0.77 | 0.85 | 0.60 | 0.86 | 4 | |
| EBR1 | 1.00 | 1.00 | 1.00 | 1.00 | 0.88 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1 | |
| EBR2 | 0.95 | 0.98 | 0.99 | 0.95 | 0.82 | 0.97 | 0.93 | 0.90 | 0.78 | 0.92 | 3 | |
| GA1 | 0.34 | 0.46 | 0.28 | 0.17 | 0.26 | 0.40 | 0.20 | 0.13 | 0.14 | 0.26 | 6 | |
| GA2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 7 | |
| 0.6 | CK | 0.67 | 0.65 | 0.77 | 0.70 | 0.70 | 0.52 | 0.41 | 0.54 | 0.49 | 0.61 | 5 |
| CSN1 | 0.90 | 0.96 | 0.98 | 0.95 | 0.95 | 0.86 | 0.87 | 0.88 | 0.90 | 0.92 | 3 | |
| CSN2 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1 | |
| EBR1 | 0.90 | 0.96 | 0.97 | 0.93 | 0.91 | 0.83 | 0.98 | 0.89 | 1.00 | 0.93 | 2 | |
| EBR2 | 0.90 | 0.91 | 0.95 | 0.92 | 0.81 | 0.70 | 0.89 | 0.70 | 0.82 | 0.84 | 4 | |
| GA1 | 0.40 | 0.35 | 0.26 | 0.18 | 0.33 | 0.19 | 0.12 | 0.19 | 0.25 | 0.25 | 6 | |
| GA2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.19 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 7 | |
| 1.2 | CK | 0.30 | 0.35 | 0.51 | 0.45 | 0.56 | 0.38 | 0.27 | 0.19 | 0.33 | 0.37 | 5 |
| CSN1 | 0.80 | 0.85 | 0.91 | 0.84 | 0.90 | 0.62 | 0.78 | 0.50 | 0.70 | 0.77 | 4 | |
| CSN2 | 0.95 | 0.90 | 0.99 | 0.90 | 1.00 | 0.70 | 0.87 | 0.57 | 0.79 | 0.85 | 3 | |
| EBR1 | 1.00 | 1.00 | 1.00 | 1.00 | 0.89 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1 | |
| EBR2 | 0.90 | 0.95 | 0.94 | 0.92 | 0.83 | 0.94 | 0.91 | 0.78 | 0.84 | 0.89 | 2 | |
| GA1 | 0.15 | 0.15 | 0.13 | 0.09 | 0.20 | 0.11 | 0.08 | 0.02 | 0.20 | 0.12 | 6 | |
| GA2 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 7 | |
| Title | Unit | Low-Salt (0.6%) | High-Salt (1.2%) |
|---|---|---|---|
| pH | — | 7.25 | 7.19 |
| Salinity | g·L−1 | 6.2 | 12.4 |
| electrical conductivity | S·m−1 | 1.041 | 1.785 |
| HCO3− | g·kg−1 | 0.2348 | 0.1981 |
| Cl− | g·kg−1 | 2.0046 | 3.7661 |
| SO42− | g·kg−1 | 3.0816 | 6.3686 |
| Ca2+ | g·kg−1 | 0.1284 | 0.3082 |
| Mg2+ | g·kg−1 | 0.0783 | 0.0731 |
| K+ | g·kg−1 | 0.0337 | 0.0455 |
| Na+ | g·kg−1 | 2.547 | 4.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Huang, J.; Qi, T.; Fu, Q.; Meng, A.; Fu, Y. Effects of Plant Regulators on the Seed Germination and Antioxidant Enzyme Activity of Cotton under Compound Salt Stress. Plants 2023, 12, 4112. https://doi.org/10.3390/plants12244112
Dong Z, Huang J, Qi T, Fu Q, Meng A, Fu Y. Effects of Plant Regulators on the Seed Germination and Antioxidant Enzyme Activity of Cotton under Compound Salt Stress. Plants. 2023; 12(24):4112. https://doi.org/10.3390/plants12244112
Chicago/Turabian StyleDong, Zhiduo, Jian Huang, Tong Qi, Qiuping Fu, Ajing Meng, and Yanbo Fu. 2023. "Effects of Plant Regulators on the Seed Germination and Antioxidant Enzyme Activity of Cotton under Compound Salt Stress" Plants 12, no. 24: 4112. https://doi.org/10.3390/plants12244112
APA StyleDong, Z., Huang, J., Qi, T., Fu, Q., Meng, A., & Fu, Y. (2023). Effects of Plant Regulators on the Seed Germination and Antioxidant Enzyme Activity of Cotton under Compound Salt Stress. Plants, 12(24), 4112. https://doi.org/10.3390/plants12244112





