In Vitro Studies of Endophytic Bacteria Isolated from Ginger (Zingiber officinale) as Potential Plant-Growth-Promoting and Biocontrol Agents against Botrytis cinerea and Colletotrichum acutatum
Abstract
:1. Introduction
2. Results
2.1. Endophytic Bacteria Isolation and Identification
2.2. Detection of Genes Involved in the Synthesis of Lipopeptides
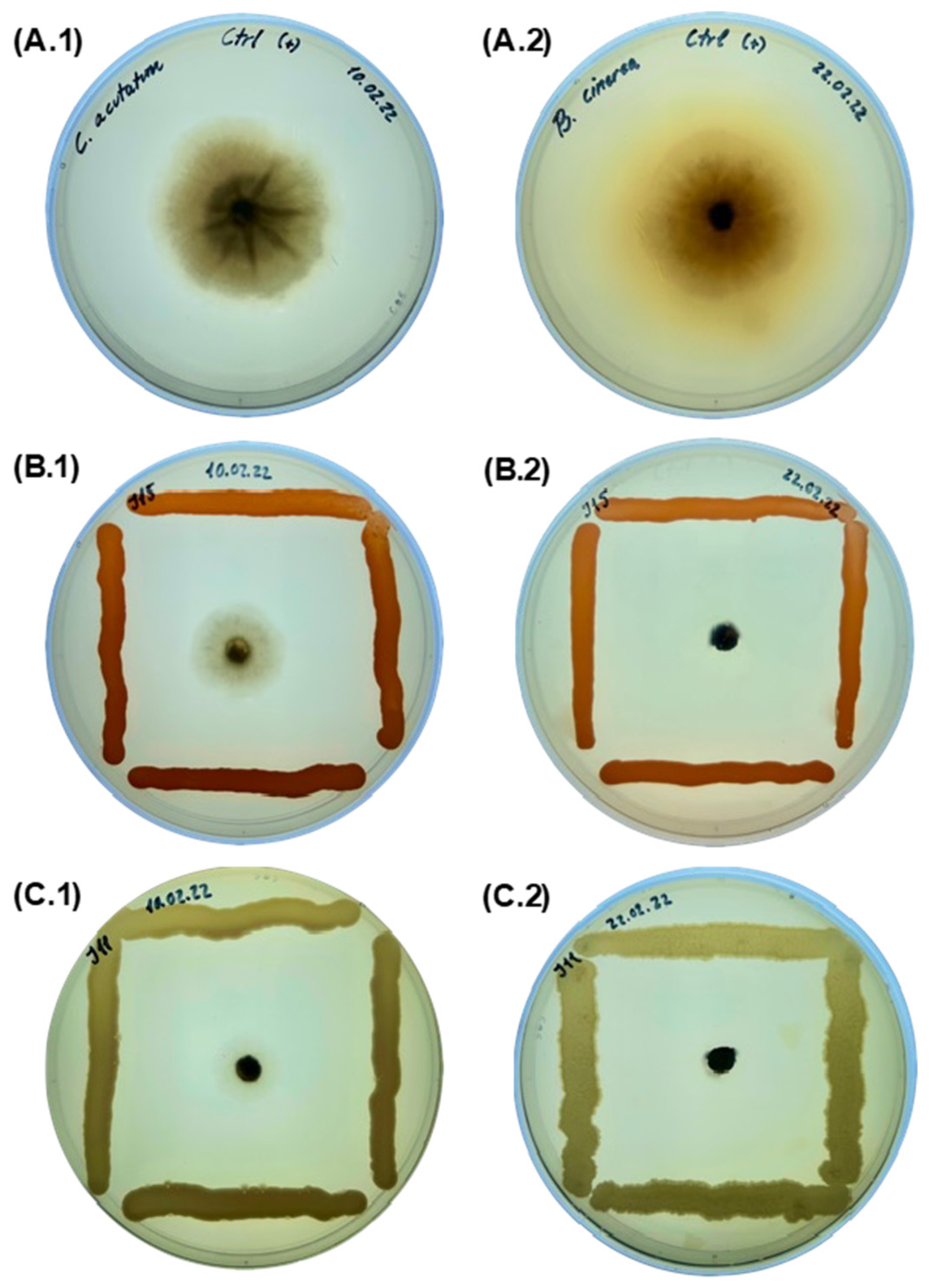
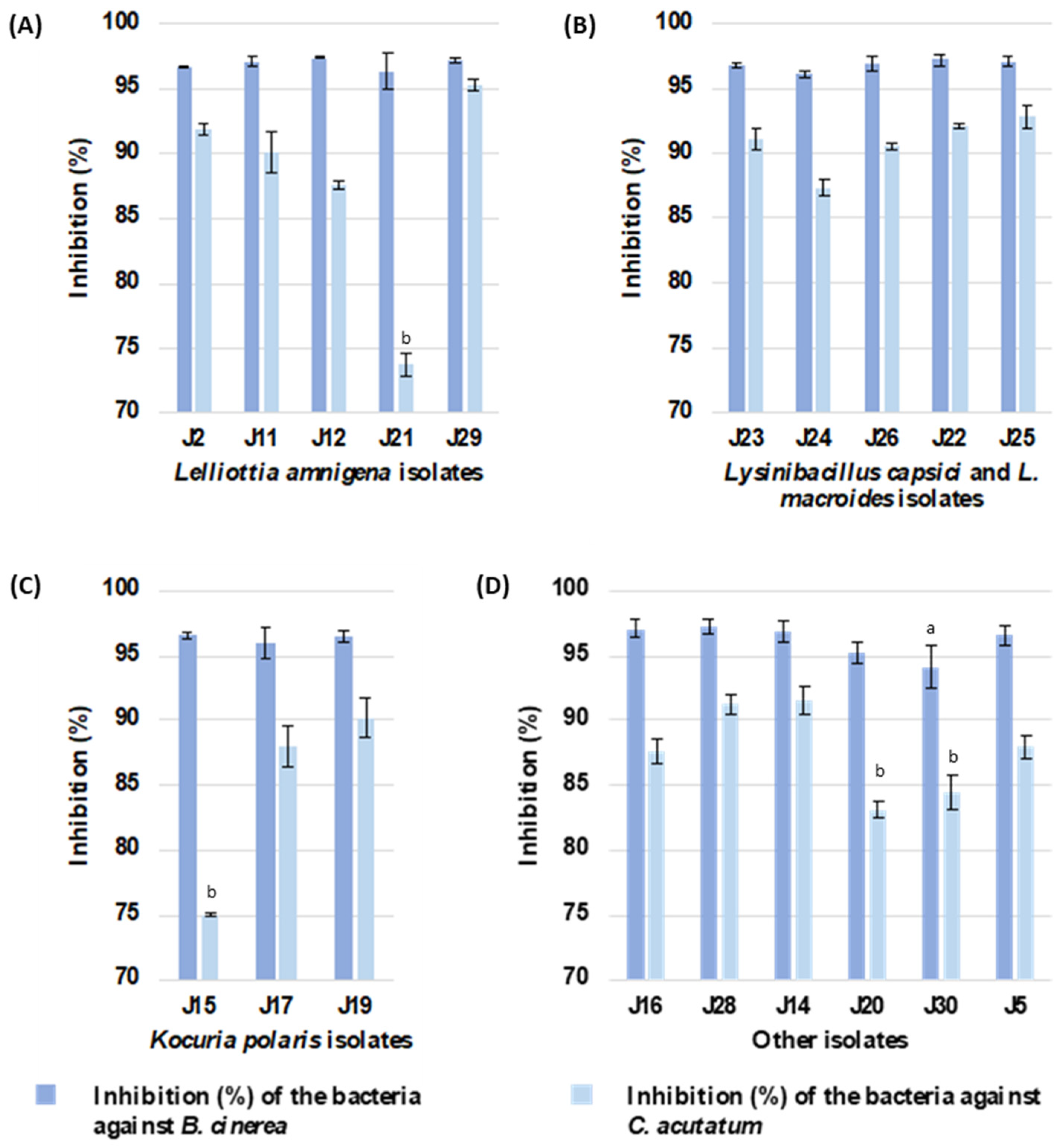
2.3. In Vitro Antagonistic Activity Assay against Phytopathogenic Fungi
2.4. In Vitro Characterization of Bacteria for Plant-Growth-Promoting Potential
2.4.1. Extracellular Enzyme Production Assays
2.4.2. Atmospheric Nitrogen Fixation, Potassium, and Phosphate Solubilization
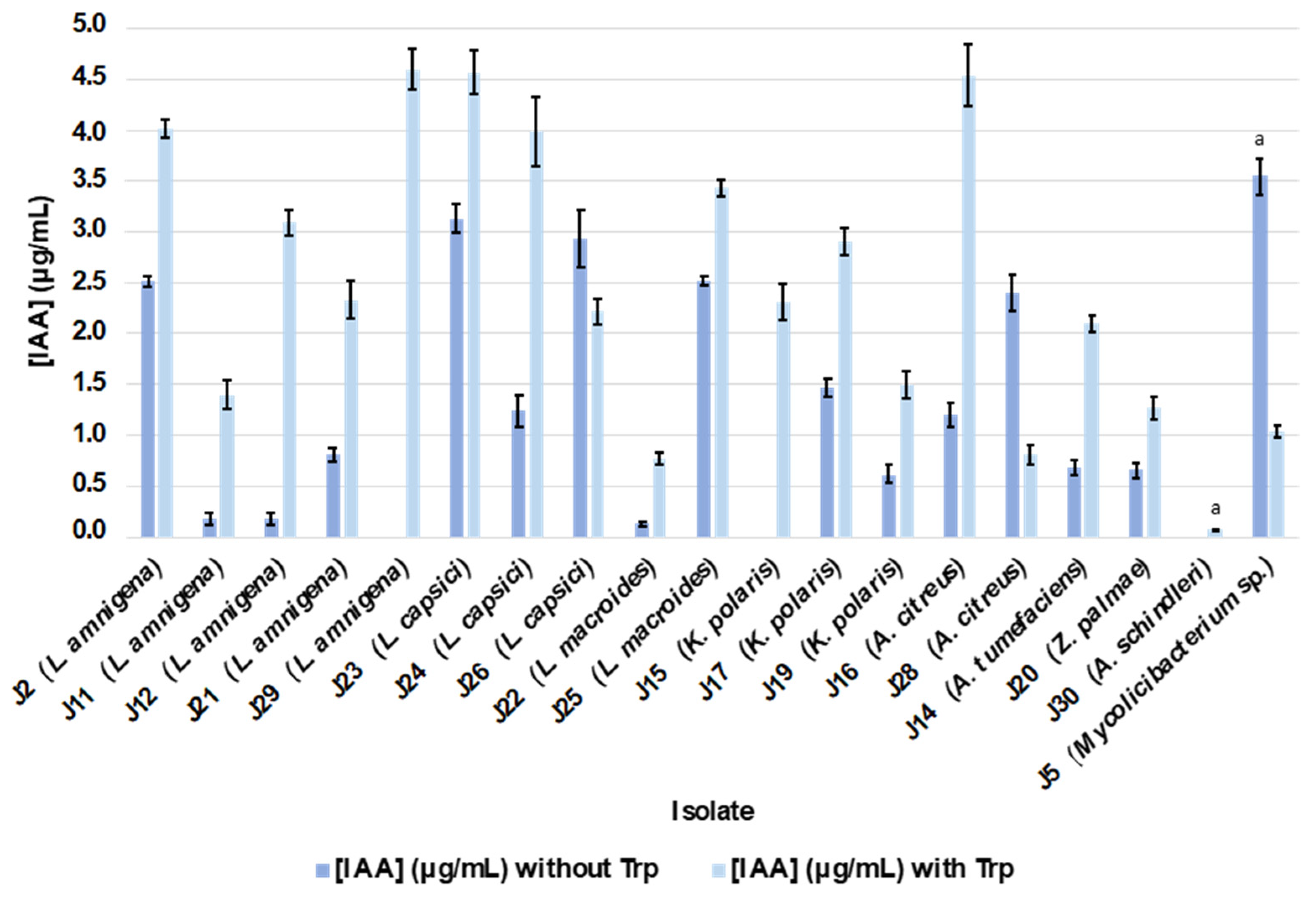
2.4.3. Siderophore and IAA Production
2.4.4. Biofilm Production
3. Discussion
4. Materials and Methods
4.1. Isolation of Endophytic Bacteria from Ginger
4.2. Molecular Identification of Ginger Isolates
4.3. Detection of Genes Involved in the Synthesis of Lipopeptides
4.4. In Vitro Antagonistic Activity Assay against Phytopathogenic Fungi
4.5. In Vitro Characterization of Bacteria for Plant Growth Promoting Potential
4.5.1. Extracellular Enzyme Production Assays
4.5.2. Atmospheric Nitrogen Fixation
4.5.3. Potassium Solubilization
4.5.4. Phosphate Solubilization
4.5.5. Siderophore Production
4.5.6. Indole Acetic Acid (IAA) Production
4.5.7. Biofilm Production
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carvalho, F.P. Pesticides, Environment, and Food Safety. Food Energy Secur. 2017, 6, 48–60. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kabir, E.; Jahan, S.A. Exposure to Pesticides and the Associated Human Health Effects. Sci. Total Environ. 2017, 575, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Khatri, N.; Tyagi, S. Influences of Natural and Anthropogenic Factors on Surface and Groundwater Quality in Rural and Urban Areas. Front. Life Sci. 2015, 8, 23–39. [Google Scholar] [CrossRef]
- Alengebawy, A.; Abdelkhalek, S.T.; Qureshi, S.R.; Wang, M.-Q. Heavy Metals and Pesticides Toxicity in Agricultural Soil and Plants: Ecological Risks and Human Health Implications. Toxics 2021, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Tudi, M.; Daniel Ruan, H.; Wang, L.; Lyu, J.; Sadler, R.; Connell, D.; Chu, C.; Phung, D.T. Agriculture Development, Pesticide Application and Its Impact on the Environment. Int. J. Environ. Res. Public Health 2021, 18, 1112. [Google Scholar] [CrossRef] [PubMed]
- Ashok Kumar, A.K.; Priyanka Singh, P.S.; Dubey, N.K. Botanicals in Agricultural Pest Management. In Management of Fungal Plant Pathogens; CABI: Wallingford, CT, USA, 2010; pp. 14–27. [Google Scholar]
- Garrido, C.; Carbú, M.; Fernández-Acero, F.J.; González-Rodríguez, V.E.; Cantoral, J.M. New Insights in the Study of Strawberry Fungal Pathogens. Genes Genomes Genom. 2011, 5, 24–39. [Google Scholar]
- Guo, Z.; Luo, C.-X.; Wu, H.-J.; Peng, B.; Kang, B.-S.; Liu, L.-M.; Zhang, M.; Gu, Q.-S. Colletotrichum Species Associated with Anthracnose Disease of Watermelon (Citrullus Lanatus) in China. J. Fungi 2022, 8, 790. [Google Scholar] [CrossRef] [PubMed]
- Sarven, M.S.; Hao, Q.; Deng, J.; Yang, F.; Wang, G.; Xiao, Y.; Xiao, X. Biological Control of Tomato Gray Mold Caused by Botrytis cinerea with the Entomopathogenic Fungus Metarhizium anisopliae. Pathogens 2020, 9, 213. [Google Scholar] [CrossRef]
- Elad, Y.; Stewart, A. Microbial Control of Botrytis spp. In Botrytis: Biology, Pathology and Control; Springer: Dordrecht, The Netherlands, 2007; pp. 223–241. [Google Scholar]
- Cosseboom, S.D.; Hu, M. Ontogenic Susceptibility of Grapevine Clusters to Ripe Rot, Caused by the Colletotrichum acutatum and C. gloeosporioides Species Complexes. Phytopathology 2022, 112, 1956–1964. [Google Scholar] [CrossRef]
- Rupp, S.; Weber, R.W.S.; Rieger, D.; Detzel, P.; Hahn, M. Spread of Botrytis cinerea Strains with Multiple Fungicide Resistance in German Horticulture. Front. Microbiol. 2017, 7, 2075. [Google Scholar] [CrossRef]
- Hahn, M. The Rising Threat of Fungicide Resistance in Plant Pathogenic Fungi: Botrytis as a Case Study. J. Chem. Biol. 2014, 7, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Heydari, A.; Pessarakli, M. A Review on Biological Control of Fungal Plant Pathogens Using Microbial Antagonists. J. Biol. Sci. 2010, 10, 273–290. [Google Scholar] [CrossRef]
- Nath Yadav, A. Plant Growth Promoting Bacteria: Biodiversity and Multifunctional Attributes for Sustainable Agriculture. Adv. Biotechnol. Microbiol. 2017, 5, 555671. [Google Scholar] [CrossRef]
- Doty, S.L. Nitrogen-Fixing Endophytic Bacteria for Improved Plant Growth. In Bacteria in Agrobiology: Plant Growth Responses; Springer: Berlin/Heidelberg, Germany, 2011; pp. 183–199. [Google Scholar]
- Bonaterra, A.; Badosa, E.; Daranas, N.; Francés, J.; Roselló, G.; Montesinos, E. Bacteria as Biological Control Agents of Plant Diseases. Microorganisms 2022, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- McGuinness, M.; Dowling, D. Plant-Associated Bacterial Degradation of Toxic Organic Compounds in Soil. Int. J. Environ. Res. Public Health 2009, 6, 2226–2247. [Google Scholar] [CrossRef]
- Mekuriya, W.; Mekibib, B. Review on the Medicinal Values of Ginger for Human and Animal Ailments. J. Vet. Sci. Technol. 2018, 9, 1000519. [Google Scholar] [CrossRef]
- Chen, T.; Chen, Z.; Ma, G.H.; Du, B.H.; Shen, B.; Ding, Y.Q.; Xu, K. Diversity and Potential Application of Endophytic Bacteria in Ginger. Genet. Mol. Res. 2014, 13, 4918–4931. [Google Scholar] [CrossRef]
- Jasim, B.; Joseph, A.A.; John, C.J.; Mathew, J.; Radhakrishnan, E.K. Isolation and Characterization of Plant Growth Promoting Endophytic Bacteria from the Rhizome of Zingiber officinale. 3 Biotech 2014, 4, 197–204. [Google Scholar] [CrossRef]
- Rajan, P.P.; Gupta, S.R.; Sarma, Y.R.; Jackson, G.V.H. Endophytic Bacteria: Its Disease Suppressive and Growth Promotive Activities in Ginger (Zingiber officinale Rosc.). In Spices and Aromatic Plants. Challenges and Opportunities in the New Century; Indian Society for Spices: Calicut, Kerala, India, 2000; pp. 254–258. [Google Scholar]
- Rohini, S.; Aswani, R.; Kannan, M.; Sylas, V.P.; Radhakrishnan, E.K. Culturable Endophytic Bacteria of Ginger Rhizome and Their Remarkable Multi-Trait Plant Growth-Promoting Features. Curr. Microbiol. 2018, 75, 505–511. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A Modified Microtiter-Plate Test for Quantification of Staphylococcal Biofilm Formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Darwis, W.; Supriyanto, A.P.; Wibowo, R.H.; Sipriyadi, S.; Supriati, R. Endophytic Bacteria Identification of Red Ginger (Zingiber Officinale Var. Rubrum) From Enggano Island. Elkawnie 2022, 8, 119. [Google Scholar] [CrossRef]
- Zhang, C.; Kong, F. Isolation and Identification of Potassium-Solubilizing Bacteria from Tobacco Rhizospheric Soil and Their Effect on Tobacco Plants. Appl. Soil Ecol. 2014, 82, 18–25. [Google Scholar] [CrossRef]
- Parashar, M.; Dhar, S.K.; Kaur, J.; Chauhan, A.; Tamang, J.; Singh, G.B.; Lyudmila, A.; Perveen, K.; Khan, F.; Bukhari, N.A.; et al. Two Novel Plant-Growth-Promoting Lelliottia amnigena Isolates from Euphorbia prostrata Aiton Enhance the Overall Productivity of Wheat and Tomato. Plants 2023, 12, 3081. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, E.; Ramirez-Bahena, M.H.; Peix, A.; Tejedor, C.; Mulas, R.; González-Andrés, F.; Velázquez, E. Analysis of Cultivable Endophytic Bacteria in Roots of Maize in a Soil from León Province in Mainland Spain. In Biological Nitrogen Fixation and Beneficial Plant-Microbe Interaction; Springer International Publishing: Cham, Switzerland, 2016; pp. 45–53. [Google Scholar]
- Wang, L.L.; Wang, E.T.; Liu, J.; Li, Y.; Chen, W.X. Endophytic Occupation of Root Nodules and Roots of Melilotus dentatus by Agrobacterium tumefaciens. Microb. Ecol. 2006, 52, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Gillani, R.A.; Shenaz, N.; Matloob, S.; Haq, F.; Wan Ngah, W.S.; Nasim, W.; Munis, M.F.H.; Rehman, A.; Chaudhary, H.J. Biosorption of Cr(III) and Pb(II) by Endophytic Agrobacterium tumefaciens 12b3: Equilibrium and Kinetic Studies. Desalination Water Treat. 2017, 67, 206–214. [Google Scholar] [CrossRef]
- Forni, C.; Riov, J.; Grilli Caiola, M.; Tel-Or, E. Indole-3-Acetic Acid (IAA) Production by Arthrobacter Species Isolated from Azolla. J. Gen. Microbiol. 1992, 138, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Munaganti, R.K.; Muvva, V.; Konda, S.; Naragani, K.; Mangamuri, U.K.; Dorigondla, K.R.; Akkewar, D. Antimicrobial Profile of Arthrobacter kerguelensis VL-RK_09 Isolated from Mango Orchards. Braz. J. Microbiol. 2016, 47, 1030–1038. [Google Scholar] [CrossRef]
- Tavarideh, F.; Pourahmad, F.; Nemati, M. Diversity and Antibacterial Activity of Endophytic Bacteria Associated with Medicinal Plant, Scrophularia striata. Vet. Res. Forum 2022, 13, 409. [Google Scholar]
- Choopan Vamerzani, M.; Pordeli, H.; Moghbeli, M. Isolation and Molecular Identification of Lysinibacillus macroides Isolated from Wheat Roots and Investigate Their Antifungal Activity against Trichoderma Spp. J. Microb. World 2016, 8, 290–298. [Google Scholar]
- Liu, T.; Zhai, C.; Zhang, J.; Coulter, J.A. Genetic Diversity and Promotion Plant Growth of Culturable Endophytic Diazotrophs Associated with Seashore Paspalum Cultivars. N. Z. J. Crop. Hortic. Sci. 2021, 49, 243–257. [Google Scholar] [CrossRef]
- Okamoto, T.; Taguchi, H.; Nakamura, K.; Ikenaga, H.; Kuraishi, H.; Yamasato, K. Zymobacter palmae Gen. Nov., Sp. Nov., a New Ethanol-Fermenting Peritrichous Bacterium Isolated from Palm Sap. Arch. Microbiol. 1993, 160, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.-S.; Deng, T.; Gao, Y.; Jiang, W.; Ou, X.; Wang, Y.; Guo, L.; Zhou, T. Genome Resource for Acinetobacter schindleri H4-3-C1: An Endophyte of Pseudostellaria heterophylla with Degradation Activity to Toxins Produced by Fungal Pathogens. Mol. Plant-Microbe Interact. 2022, 35, 1124–1126. [Google Scholar] [CrossRef] [PubMed]
- Alagawadi, A.R.; Ammanna, S.; Doddagoudar, C.K. Isolation and Molecular Characterization of Endorhizospheric Diazotrophs of Western Ghats. J. Pure Appl. Microbiol. 2014, 8, 3273–3278. [Google Scholar]
- del Barrio-Duque, A.; Ley, J.; Samad, A.; Antonielli, L.; Sessitsch, A.; Compant, S. Beneficial Endophytic Bacteria-Serendipita indica Interaction for Crop Enhancement and Resistance to Phytopathogens. Front. Microbiol. 2019, 10, 2888. [Google Scholar] [CrossRef]
- Duong, B.; Nguyen, H.X.; Phan, H.V.; Colella, S.; Trinh, P.Q.; Hoang, G.T.; Nguyen, T.T.; Marraccini, P.; Lebrun, M.; Duponnois, R. Identification and Characterization of Vietnamese Coffee Bacterial Endophytes Displaying in Vitro Antifungal and Nematicidal Activities. Microbiol. Res. 2021, 242, 126613. [Google Scholar] [CrossRef] [PubMed]
- Baldan, E.; Nigris, S.; Populin, F.; Zottini, M.; Squartini, A.; Baldan, B. Identification of Culturable Bacterial Endophyte Community Isolated from Tissues of Vitis vinifera “Glera”. Plant Biosyst.—Int. J. Deal. All Asp. Plant Biol. 2014, 148, 508–516. [Google Scholar] [CrossRef]
- Fillinger, S.; Walker, A.S. Chemical Control and Resistance Management of Botrytis Diseases. In Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Fillinger, S., Elad, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 189–216. ISBN 978-3-319-23370-3. [Google Scholar]
- Bhattacharyya, P.N.; Jha, D.K. Plant Growth-Promoting Rhizobacteria (PGPR): Emergence in Agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Escribano-Viana, R.; Portu, J.; Garijo, P.; Gutiérrez, A.R.; Santamaría, P.; López-Alfaro, I.; López, R.; González-Arenzana, L. Evaluating a Preventive Biological Control Agent Applied on Grapevines against Botrytis cinerea and Its Influence on Winemaking. J. Sci. Food Agric. 2018, 98, 4517–4526. [Google Scholar] [CrossRef]
- Gholami, M.; Khakvar, R.; AliasgarZad, N. Application of Endophytic Bacteria for Controlling Anthracnose Disease (Colletotrichum lindemuthianum) on Bean Plants. Arch. Phytopathol. Plant Prot. 2013, 46, 1831–1838. [Google Scholar] [CrossRef]
- Murtado, A.; Mubarik, N.R.; Tjahjoleksono, A. Isolation and Characterization Endophytic Bacteria as Biological Control of Fungus Colletotrichum Sp. on Onion Plants (Allium cepa L.). IOP Conf. Ser. Earth Envion. Sci. 2020, 457, 012043. [Google Scholar] [CrossRef]
- Prasad, M.; Srinivasan, R.; Chaudhary, M.; Mahawer, S.K.; Jat, L.K. Endophytic Bacteria: Role in Sustainable Agriculture. In Microbial Endophytes; Elsevier: Amsterdam, The Netherlands, 2020; pp. 37–60. [Google Scholar]
- Geisseler, D.; Horwath, W.R.; Joergensen, R.G.; Ludwig, B. Pathways of Nitrogen Utilization by Soil Microorganisms—A Review. Soil Biol. Biochem. 2010, 42, 2058–2067. [Google Scholar] [CrossRef]
- Gupta, A.; Gupta, R.; Singh, R.L. Microbes and Environment. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Singapore, 2017; pp. 43–84. [Google Scholar]
- Narayanan, M.; Kandasamy, S.; He, Z.; Kumarasamy, S. Ecological Impacts of Pesticides on Soil and Water Ecosystems and Its Natural Degradation Process. In Pesticides in the Natural Environment; Elsevier: Amsterdam, The Netherlands, 2022; pp. 23–49. [Google Scholar]
- Woźniak, M.; Tyśkiewicz, R.; Siebielec, S.; Gałązka, A.; Jaroszuk-Ściseł, J. Metabolic Profiling of Endophytic Bacteria in Relation to Their Potential Application as Components of Multi-Task Biopreparations. Microb. Ecol. 2023, 86, 2527–2540. [Google Scholar] [CrossRef] [PubMed]
- Carro, L.; Menéndez, E. Knock, Knock-Let the Bacteria in: Enzymatic Potential of Plant Associated Bacteria. In Molecular Aspects of Plant Beneficial Microbes in Agriculture; Elsevier: Amsterdam, The Netherlands, 2020; pp. 169–178. [Google Scholar]
- Coorevits, A.; Dinsdale, A.E.; Heyrman, J.; Schumann, P.; Van Landschoot, A.; Logan, N.A.; De Vos, P. Lysinibacillus macroides Sp. Nov., Nom. Rev. Int. J. Syst. Evol. Microbiol. 2012, 62, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.L.E.; Kilany, A.H.A.M.; Hassan, E.A. Antifungal Activity of Lysinibacillus macroides against Toxigenic Aspergillus flavus and Fusarium proliferatum and Analysis of Its Mycotoxin Minimization Potential. BMC Microbiol. 2023, 23, 269. [Google Scholar] [CrossRef] [PubMed]
- Sinha, D.; Tandon, P.K. An Overview of Nitrogen, Phosphorus and Potassium: Key Players of Nutrition Process in Plants. In Sustainable Solutions for Elemental Deficiency and Excess in Crop Plants; Springer: Singapore, 2020; pp. 85–117. [Google Scholar]
- Wang, Y.; Chen, Y.; Wu, W. Potassium and Phosphorus Transport and Signaling in Plants. J. Integr. Plant Biol. 2021, 63, 34–52. [Google Scholar] [CrossRef] [PubMed]
- Marschner, P. Rhizosphere Biology. In Marschner’s Mineral Nutrition of Higher Plants; Elsevier: Amsterdam, The Netherlands, 2012; pp. 369–388. [Google Scholar]
- Wang, Y.; Mopper, S.; Hasenstein, K.H. Effects of Salinity on Endogenous ABA, IAA, JA, AND SA in Iris hexagona. J. Chem. Ecol. 2001, 27, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A Role for Flavin Monooxygenase-Like Enzymes in Auxin Biosynthesis. Science (1979) 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Woodward, A.W. Auxin: Regulation, Action, and Interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin Biosynthesis. Arab. Book 2014, 12, e0173. [Google Scholar] [CrossRef]
- Jiang, J.-J.; Wang, J.-F.; Yang, P.; Xu, Z.-M.; He, T.; Gao, Q.; Wang, L.-L.; Li, Q.-S. Interactive Effects between Cadmium Stabilized by Palygorskite and Mobilized by Siderophores from Pseudomonas fluorescens. Ecotoxicol. Environ. Saf. 2019, 181, 265–273. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial Features of Plant Growth-Promoting Rhizobacteria for Improving Plant Growth and Health in Challenging Conditions: A Methodical Review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef] [PubMed]
- Niehus, R.; Picot, A.; Oliveira, N.M.; Mitri, S.; Foster, K.R. The Evolution of Siderophore Production as a Competitive Trait. Evolution 2017, 71, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Trapet, P.L.; Stringlis, I.A.; Kruijs, S.; Bakker, P.A.H.M.; Pieterse, C.M.J. Iron and Immunity. Annu. Rev. Phytopathol. 2017, 55, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Ricci, E.C. Investigating the Role of Pseudomonas sp. and Bacillus sp. Biofilms as Plant Growth Promoting Inoculants; McGill University: Montreal, QC, Canada, 2016. [Google Scholar]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus Subtilis against Infection of Arabidopsis Roots by Pseudomonas syringae Is Facilitated by Biofilm Formation and Surfactin Production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yan, F.; Chai, Y.; Liu, H.; Kolter, R.; Losick, R.; Guo, J. Biocontrol of Tomato Wilt Disease Bacillus subtilis Isolates from Natural Environments Depends on Conserved Genes Mediating Biofilm Formation. Environ. Microbiol. 2013, 15, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghearachchi, H.S.; Seneviratne, G. A Bradyrhizobial-Penicillium Spp. Biofilm with Nitrogenase Activity Improves N2 Fixing Symbiosis of Soybean. Biol. Fertil Soils 2004, 40, 432–434. [Google Scholar] [CrossRef]
- Khan, M.S.; Ahmad, I.; Cameotra, S.S.; Botha, F. Sub-MICs of Carum copticum and Thymus vulgaris Influence Virulence Factors and Biofilm Formation in Candida Spp. BMC Complement. Altern. Med. 2014, 14, 337. [Google Scholar] [CrossRef]
- Triveni, S.; Prasanna, R.; Saxena, A.K. Optimization of Conditions for in Vitro Development of Trichoderma viride-Based Biofilms as Potential Inoculants. Folia Microbiol. 2012, 57, 431–437. [Google Scholar] [CrossRef]
- Maksimov, I.V.; Singh, B.P.; Cherepanova, E.A.; Burkhanova, G.F.; Khairullin, R.M. Prospects and Applications of Lipopeptide-Producing Bacteria for Plant Protection (Review). Appl. Biochem. Microbiol. 2020, 56, 15–28. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus Lipopeptides: Versatile Weapons for Plant Disease Biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Mora Pons, I.; Cabrefiga Olamendi, J.; Montesinos Seguí, E. Antimicrobial Peptide Genes in Bacillus Strains from Plant Environments. Int. Microbiol. 2011, 14, 213–223. [Google Scholar]
- Farace, G.; Fernandez, O.; Jacquens, L.; Coutte, F.; Krier, F.; Jacques, P.; Clément, C.; Barka, E.A.; Jacquard, C.; Dorey, S. Cyclic Lipopeptides from Bacillus subtilis Activate Distinct Patterns of Defence Responses in Grapevine. Mol. Plant Pathol. 2015, 16, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Bolivar-Anillo, H.J.; González-Rodríguez, V.E.; Cantoral, J.M.; García-Sánchez, D.; Collado, I.G.; Garrido, C. Endophytic Bacteria Bacillus subtilis, Isolated from Zea mays, as Potential Biocontrol Agent against Botrytis cinerea. Biololy 2021, 10, 492. [Google Scholar] [CrossRef] [PubMed]
- Fickers, P.; Guez, J.-S.; Damblon, C.; Leclère, V.; Béchet, M.; Jacques, P.; Joris, B. High-Level Biosynthesis of the Anteiso-C17 Isoform of the Antibiotic Mycosubtilin in Bacillus subtilis and Characterization of Its Candidacidal Activity. Appl. Environ. Microbiol. 2009, 75, 4636–4640. [Google Scholar] [CrossRef] [PubMed]
- González-Rodríguez, V.E.; Garrido, C.; Cantoral, J.M.; Schumacher, J. The F-Actin Capping Protein Is Required for Hyphal Growth and Full Virulence but Is Dispensable for Septum Formation in Botrytis cinerea. Fungal Biol. 2016, 120, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Scarpellini, M.; Franzetti, L.; Galli, A. Development of PCR Assay to Identify Pseudomonas fluorescens and Its Biotype. FEMS Microbiol. Lett. 2004, 236, 257–260. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, H.; Wang, F.; Yang, Y.; Wang, X.; Chen, B.; Stampfli, M.R.; Zhou, H.; Shu, W.; Brightling, C.E.; et al. A Refined View of Airway Microbiome in Chronic Obstructive Pulmonary Disease at Species and Strain-Levels. Front. Microbiol. 2020, 11, 1758. [Google Scholar] [CrossRef]
- Tenorio-Salgado, S.; Tinoco, R.; Vazquez-Duhalt, R.; Caballero-Mellado, J.; Perez-Rueda, E. Identification of Volatile Compounds Produced by the Bacterium Burkholderia tropica That Inhibit the Growth of Fungal Pathogens. Bioengineered 2013, 4, 236–243. [Google Scholar] [CrossRef]
- Castro, R.A.; Quecine, M.C.; Lacava, P.T.; Batista, B.D.; Luvizotto, D.M.; Marcon, J.; Ferreira, A.; Melo, I.S.; Azevedo, J.L. Isolation and Enzyme Bioprospection of Endophytic Bacteria Associated with Plants of Brazilian Mangrove Ecosystem. Springerplus 2014, 3, 382. [Google Scholar] [CrossRef]
- Baldani, J.I.; Reis, V.M.; Videira, S.S.; Boddey, L.H.; Baldani, V.L.D. The Art of Isolating Nitrogen-Fixing Bacteria from Non-Leguminous Plants Using N-Free Semi-Solid Media: A Practical Guide for Microbiologists. Plant Soil 2014, 384, 413–431. [Google Scholar] [CrossRef]
- Shahid, M.; Hameed, S.; Tariq, M.; Zafar, M.; Ali, A.; Ahmad, N. Characterization of Mineral Phosphate-Solubilizing Bacteria for Enhanced Sunflower Growth and Yield-Attributing Traits. Ann. Microbiol. 2015, 65, 1525–1536. [Google Scholar] [CrossRef]
- Pérez-Miranda, S.; Cabirol, N.; George-Téllez, R.; Zamudio-Rivera, L.S.; Fernández, F.J. O-CAS, a Fast and Universal Method for Siderophore Detection. J. Microbiol. Methods 2007, 70, 127–131. [Google Scholar] [CrossRef]
- Merritt, J.H.; Kadouri, D.E.; O’Toole, G.A. Growing and Analyzing Static Biofilms. Curr. Protoc. Microbiol. 2011, 22, 1B.1.1–1B.1.18. [Google Scholar] [CrossRef]

| Isolate | Identification | GenBank Acc. N. | |
|---|---|---|---|
| 16S | rpoB | ||
| J2 | Lelliottia amnigena | OR840641 | OR879032 |
| J5 | Mycolicibacterium sp. | OR840642 | OR879039 |
| J11 | Lelliottia amnigena | OR840643 | OR879034 |
| J12 | Lelliottia amnigena | OR840644 | OR879035 |
| J14 | Agrobacterium tumefaciens | OR840645 | OR879040 |
| J15 | Kocuria polaris | OR840646 | OR879029 |
| J16 | Agrococcus citreus | OR840647 | OR879037 |
| J17 | Kocuria polaris | OR840648 | OR879030 |
| J19 | Kocuria polaris | OR840649 | OR879031 |
| J20 | Zymobacter palmae | OR840650 | OR879041 |
| J21 | Lelliottia amnigena | OR840651 | OR879033 |
| J22 | Lysinibacillus macroides | OR840652 | OR879027 |
| J23 | Lysinibacillus capsici | OR840653 | OR879024 |
| J24 | Lysinibacillus capsici | OR840654 | OR879025 |
| J25 | Lysinibacillus macroides | OR840655 | OR879028 |
| J26 | Lysinibacillus capsici | OR840656 | OR879026 |
| J28 | Agrococcus citreus | OR840657 | OR879038 |
| J29 | Lelliottia amnigena | OR840658 | OR879036 |
| J30 | Acinetobacter schindleri | OR840659 | OR879042 |
| Isolate | Identification | Bacylisin | Bacillomycin | Fengycin | Iturin | Subtilin | Surfactin |
|---|---|---|---|---|---|---|---|
| J2 | Lelliottia amnigena | - | + | + | + | - | + |
| J11 | - | - | - | - | - | - | |
| J12 | - | - | - | - | + | + | |
| J21 | - | - | - | - | + | + | |
| J29 | - | - | - | - | + | + | |
| J23 | Lysinibacillus capsici | - | - | - | - | + | + |
| J24 | - | - | - | - | - | + | |
| J26 | - | - | - | - | + | + | |
| J22 | Lysinibacillus macroides | + | - | - | + | - | + |
| J25 | - | - | - | - | - | + | |
| J15 | Kocuria polaris | - | - | - | - | - | - |
| J17 | - | - | - | - | - | + | |
| J19 | - | - | - | - | - | + | |
| J16 | Agrococcus citreus | - | - | - | - | - | - |
| J28 | - | - | - | - | + | + | |
| J14 | Agrobacterium tumefaciens | - | + | + | + | - | + |
| J20 | Zymobacter palmae | - | + | - | - | + | + |
| J30 | Acinetobacter schindleri | - | - | - | - | + | - |
| J5 | Mycolicibacterium sp. | - | - | - | - | + | + |
| Isolate | Identification | Amylase | Cellulase | Esterase | Lipase | Protease |
|---|---|---|---|---|---|---|
| J2 | Lelliottia amnigena | - | - | - | - | + |
| J11 | - | - | - | - | - | |
| J12 | - | - | - | - | + | |
| J21 | - | - | + | + | + | |
| J29 | - | - | + | + | - | |
| J23 | Lysinibacillus capsici | - | - | + | + | - |
| J24 | - | - | - | - | + | |
| J26 | - | - | + | + | - | |
| J22 | Lysinibacillus macroides | - | - | + | + | + |
| J25 | - | - | - | - | + | |
| J15 | Kocuria polaris | - | - | + | + | - |
| J17 | - | - | - | - | + | |
| J19 | - | - | - | - | + | |
| J16 | Agrococcus citreus | - | - | - | - | - |
| J28 | - | - | + | + | - | |
| J14 | Agrobacterium tumefaciens | - | - | + | + | - |
| J20 | Zymobacter palmae | - | - | - | + | + |
| J30 | Acinetobacter schindleri | - | - | - | + | - |
| J5 | Mycolicibacterium sp. | - | - | - | + | + |
| Isolate | Identification | Fixation of Atmospheric Nitrogen | Solubilization of Potassium | Solubilization of Phosphate | Production of Siderophores | Production of IAA (without Trp/with Trp) | Production of Biofilm |
|---|---|---|---|---|---|---|---|
| J2 | Lelliottia amnigena | + | - | - | - | +/+ | + |
| J11 | - | - | - | - | +/+ | + | |
| J12 | - | - | - | + | +/+ | + | |
| J21 | - | - | - | - | +/+ | + | |
| J29 | - | + | + | - | -/+ | + | |
| J23 | Lysinibacillus capsici | - | - | - | - | +/+ | - |
| J24 | + | - | - | - | +/+ | + | |
| J26 | + | - | - | + | +/+ | + | |
| J22 | Lysinibacillus macroides | - | - | - | - | +/+ | - |
| J25 | + | - | - | - | +/+ | + | |
| J15 | Kocuria polaris | - | - | - | - | -/+ | - |
| J17 | + | - | - | - | +/+ | + | |
| J19 | + | - | - | - | +/+ | + | |
| J16 | Agrococcus citreus | + | - | - | - | +/+ | + |
| J28 | + | - | - | + | +/+ | + | |
| J14 | Agrobacterium tumefaciens | - | - | - | - | +/+ | + |
| J20 | Zymobacter palmae | - | - | - | - | +/+ | - |
| J30 | Acinetobacter schindleri | - | - | - | - | -/+ | - |
| J5 | Mycolicibacterium sp. | + | - | - | + | +/+ | + |
| Strongly Adherent | Moderately Adherent | Weakly Adherent | Not Adherent |
|---|---|---|---|
| Mycolicibacterium sp. J5 K. polaris J19 L. capsici J26 A. citreus J28 | L. amnigena J2, 12 K. polaris J17 L. macroides J25 | L. amnigena J11, J21, J29 A. tumefaciens J14 A. citreus J16 L. capsici J24 | L. macroides J22 L. capsici J23 K. polaris J15 Z. palmae J20 A. schindleri J30 |
| Primer | Sequence (5′ → 3′) | Target |
|---|---|---|
| 16S_F | GAAGAGTTTGATCATGGCTC | 16S rRNA gene [81] |
| 16S_R | AAGGAGGTGATCCAGCCGCA | |
| Univ_rpoβ_F_deg | GGYTWYGAAGTNCGHGACGTDCA | rpoβ gene [82] |
| Univ_rpoβ_R_deg | TGACGYTGCATGTTBGMRCATMA |
| Primer | Sequence (5′ → 3′) | Product Size (bp) | Used for |
|---|---|---|---|
| BACF | CAGCTCATGGGAATGCTTTT | 500 | Detection of bacA gene (bacylisin) |
| BACR | CTCGGTCCTGAAGGGACAAG | ||
| BMYBF | GAATCCCGTTGTTCTCCAAA | 370 | Detection of bmyB gene (bacillomycin) |
| BMYBR | GCGGGTATTGAATGCTTGTT | ||
| FENDF | GGCCCGTTCTCTAAATCCAT | 270 | Detection of fenD gene (fengycin) |
| FENDR | GTCATGCTGACGAGAGCAAA | ||
| ITUCF | GGCTGCTGCAGATGCTTTAT | 423 | Detection of ituC gene (iturin) |
| ITUCR | TCGCAGATAATCGCAGTGAG | ||
| SPASF | GGTTTGTTGGATGGAGCTGT | 375 | Detection of spaS gene (subtilin) |
| SPASR | GCAAGGAGTCAGAGCAAGGT | ||
| SRFAF | TCGGGACAGGAAGACATCAT | 200 | Detection of sfrAA gene (surfactin) |
| SRFAR | CCACTCAAACGGATAATCCTGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bódalo, A.; Borrego, R.; Garrido, C.; Bolivar-Anillo, H.J.; Cantoral, J.M.; Vela-Delgado, M.D.; González-Rodríguez, V.E.; Carbú, M. In Vitro Studies of Endophytic Bacteria Isolated from Ginger (Zingiber officinale) as Potential Plant-Growth-Promoting and Biocontrol Agents against Botrytis cinerea and Colletotrichum acutatum. Plants 2023, 12, 4032. https://doi.org/10.3390/plants12234032
Bódalo A, Borrego R, Garrido C, Bolivar-Anillo HJ, Cantoral JM, Vela-Delgado MD, González-Rodríguez VE, Carbú M. In Vitro Studies of Endophytic Bacteria Isolated from Ginger (Zingiber officinale) as Potential Plant-Growth-Promoting and Biocontrol Agents against Botrytis cinerea and Colletotrichum acutatum. Plants. 2023; 12(23):4032. https://doi.org/10.3390/plants12234032
Chicago/Turabian StyleBódalo, Alejandro, Rogelio Borrego, Carlos Garrido, Hernando José Bolivar-Anillo, Jesús M. Cantoral, María Dolores Vela-Delgado, Victoria E. González-Rodríguez, and María Carbú. 2023. "In Vitro Studies of Endophytic Bacteria Isolated from Ginger (Zingiber officinale) as Potential Plant-Growth-Promoting and Biocontrol Agents against Botrytis cinerea and Colletotrichum acutatum" Plants 12, no. 23: 4032. https://doi.org/10.3390/plants12234032
APA StyleBódalo, A., Borrego, R., Garrido, C., Bolivar-Anillo, H. J., Cantoral, J. M., Vela-Delgado, M. D., González-Rodríguez, V. E., & Carbú, M. (2023). In Vitro Studies of Endophytic Bacteria Isolated from Ginger (Zingiber officinale) as Potential Plant-Growth-Promoting and Biocontrol Agents against Botrytis cinerea and Colletotrichum acutatum. Plants, 12(23), 4032. https://doi.org/10.3390/plants12234032








