Genetic Engineering and Genome Editing Advances to Enhance Floral Attributes in Ornamental Plants: An Update
Abstract
:1. Introduction
2. General Mechanism of Gene Editing and Genetic Engineering
3. Importance of Improving Ornamental Attributes
4. Applications of Genetic Engineering and Genome Editing to Improve Floral Traits in Ornamentals
4.1. Floral Color
4.2. Floral Scent
4.3. Flower Longevity
4.4. Floral Anatomy
4.5. Flowering Time and Development
5. Conclusions and Future Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lecomte, C.; Alabouvette, C.; Edel-Hermann, V.; Robert, F.; Steinberg, C. Biological control of ornamental plant diseases caused by Fusarium oxysporum: A review. Biol. Cont. 2016, 101, 17–30. [Google Scholar] [CrossRef]
- Jin, C.; Dong, L.; Chang, W.; Wani, M.A.; Yang, C.; Li, S.; Li, F. Creating novel ornamentals via new strategies in the era of genome editing. Front. Plant Sci. 2023, 14, 1142866. [Google Scholar] [CrossRef]
- Giovannini, A.; Laura, M.; Nesi, B.; Savona, M.; Cardi, T. Genes and genome editing tools for breeding desirable phenotypes in ornamentals. Plant Cell Rep. 2021, 40, 461–478. [Google Scholar] [CrossRef]
- Mekapogu, M.; Kwon, O.K.; Song, H.Y.; Jung, J.A. Towards the improvement of ornamental attributes in chrysanthemum: Recent progress in biotechnological advances. Int. J. Mol. Sci. 2022, 23, 12284. [Google Scholar] [CrossRef] [PubMed]
- Royal FloraHolland in Facts and Figures. 2022 Annual Report. Available online: https://www.royalfloraholland.com/en (accessed on 18 September 2023).
- Mekapogu, M.; Vasamsetti, B.M.K.; Kwon, O.K.; Ahn, M.S.; Lim, S.H.; Jung, J.A. Anthocyanins in floral colors: Biosynthesis and regulation in chrysanthemum flowers. Int. J. Mol. Sci. 2020, 21, 6537. [Google Scholar] [CrossRef] [PubMed]
- Kuligowska, K.; Lutken, H.; Muller, R. Towards development of new ornamental plants: Status and progress in wide hybridization. Planta 2016, 244, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef]
- Mba, C. Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy 2013, 3, 200–231. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Deng, J.; Khalid, N.; Sanaullah, T.; Shuilin, H. Biotechnological advancements for improving floral attributes in ornamental plants. Front. Plant Sci. 2017, 8, 530. [Google Scholar] [CrossRef] [PubMed]
- Raman, R. The impact of genetically modified (GM) crops in modern agriculture: A review. GM Crops Food 2017, 8, 195–208. [Google Scholar] [CrossRef]
- Mekapogu, M.; Jung, J.A.; Kwon, O.K.; Ahn, M.S.; Song, H.Y.; Jang, S. Recent progress in enhancing fungal disease resistance in ornamental plants. Int. J. Mol. Sci. 2021, 22, 7956. [Google Scholar] [CrossRef]
- Boutigny, A.L.; Dohin, N.; Pornin, D.; Rolland, M. Overview and detectability of the genetic modifications in ornamental plants. Hortic. Res. 2020, 7, 11. [Google Scholar] [CrossRef]
- Bruetschy, C. The EU regulatory framework on genetically modified organisms (GMOs). Trans. Res. 2019, 28, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.S.; Ding, J.; Li, Y. Genome-editing technologies and their potential application in horticultural crop breeding. Hortic. Res. 2015, 2, 15019. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Caroll, D. Genome engineering with zinc-finger nucleases. Genetics 2011, 188, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Ritcher, C.; Chang, J.T.; Fineran, P.C. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) systems. Viruses 2012, 4, 2291–2311. [Google Scholar]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Hsu, P.D.; Zhang, F. Dissecting natural function using targeted genome engineering technologies. ACS Chem. Neurosci. 2012, 3, 603–610. [Google Scholar] [CrossRef]
- Hahne, G.; Tomlinson, L.; Nogue, F. Precision genetic engineering tools for the next-generation plant breeding. Plant Cell Rep. 2019, 38, 435–436. [Google Scholar] [CrossRef] [PubMed]
- Songstad, D.D.; Petolino, J.F.; Voytas, D.F.; Reichert, N.A. Genome editing of plants. Crit. Rev. Plant Sci. 2017, 36, 1–3. [Google Scholar] [CrossRef]
- Marton, I.; Zuker, A.; Shklarman, E.; Zeevi, V.; Tovkach, A.; Roffe, S.; Ovadis, M.; Tzfira, T.; Vainstein, A. Non-transgenic genome modification in plant cells. Plant Physiol. 2010, 154, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Ramirez, C.L.; Joung, J.K.; Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 2011, 8, 765–770. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Cong, L.; Zhou, Y.; Cunniff, M.M.; Feng, G.P.; Zhang, F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012, 7, 171–192. [Google Scholar] [CrossRef] [PubMed]
- Morbitzer, R.; Romer, P.; Boch, J.; Lahaye, T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl. Acad Sci. USA 2010, 107, 21617–21622. [Google Scholar] [CrossRef]
- Puchta, H.; Fauser, F. Synthetic nucleases for genome engineering in plants: Prospects for a bright future. Plant J. 2014, 78, 727–741. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; He, A.L. CRISPR-Cas9: Tool for qualitative and quantitative plant genome editing. Front. Plant Sci. 2016, 7, 1740. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Durest, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Gasiunas, G.; Siksnys, V. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: Holy grail of genome editing? Trends Microbio. 2013, 21, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.G.; Doudna, J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Lemos, B.R.; Kaplan, A.C.; Bae, J.E.; Ferrazzoli, A.E.; Kuo, J.; Anand, R.P.; Waterman, D.P.; Haber, J.E. CRISPR/Cas9 cleavages in budding yeast reveal template insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. USA 2018, 115, E2040–E2047. [Google Scholar] [CrossRef]
- Danner, E.; Bashir, S.; Yumlu, S.; Wurst, W.; Wefers, B.; Kuhn, R. Control of gene editing by manipulation of DNA repair mechanisms. Mamm. Genome 2017, 28, 262–274. [Google Scholar] [CrossRef]
- Hoshi, Y.; Kondo, M.; Mori, S.; Adachi, Y.; Nakano, M.; Kobayashi, H. Production of transgenic lily plants by Agrobacterium-mediated transformation. Plant Cell Rep. 2004, 22, 359–364. [Google Scholar] [CrossRef]
- Chin, D.P.; Mishiba, K.I.; Mii, M. Agrobacterium-mediated transformation of protocorm-like bodies in cymbidium. Plant Cell Rep. 2007, 26, 735–743. [Google Scholar] [CrossRef]
- Fang, F.; Oliva, M.; Ehi-Eromosele, S.; Zaccai, M.; Arazi, T.; Oren-Shamir, M. Successful floral-dipping transformation of post-anthesis lisianthus (Eustoma grandiflorum) flowers. Plant J. 2018, 96, 869–879. [Google Scholar] [CrossRef]
- Cheng, X.; Huang, C.; Zhang, X.; Lyu, Y. Establishment of transgenic marigold using the floral dip method. Acta Physiol. Plantarum 2019, 41, 147. [Google Scholar] [CrossRef]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing crop transformation in the era of genome editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef]
- Zhao, H.; Wolt, J.D. Risk associated with off-target plant genome editing and methods for its limitation. Emerg. Top. Life Sci. 2017, 1, 231–240. [Google Scholar]
- Tycko, J.; Wainberg, M.; Marinov, G.K.; Ursu, O.; Hess, G.T.; Ego, B.K.; Aradhana; Li, A.; Trevino, A.E.; Spees, K.; et al. Mitigation of off-target toxicity in CRISPR-Cas9 screens for essential non-coding elements. Nat. Commun. 2019, 10, 4063. [Google Scholar] [CrossRef]
- Kausch, A.P.; Nelson-Vasilchik, K.; Hague, J.; Mookkan, M.; Quemada, H.; Dellaporta, S.; Fragoso, C.; Zhang, Z.Y.J. Edit at will: Genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci. 2019, 281, 186–205. [Google Scholar] [CrossRef]
- Mao, Y.F.; Botella, J.R.; Liu, Y.G.; Zhu, J.K. Gene editing in plants: Progress and challenges. Natl. Sci. Rev. 2019, 6, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Nontaswatsri, C.; Fukai, S. Genetic transformation of carnation (Dianthus caryophyllus L.). In Protocols for In Vitro Propagation of Ornamental Plants; Mohan, S.M., Ochatt, S.J., Eds.; Springer: Amsterdam, The Netherland, 2010; pp. 87–96. [Google Scholar]
- Du, F.; Wu, Y.; Zhang, L.; Li, X.W.; Zhao, X.Y.; Wang, W.H.; Gao, Z.; Xia, Y. De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium Oriental hybrid-Sorbonne. Plant Mol. Biol. Rep. 2015, 33, 281–293. [Google Scholar] [CrossRef]
- Azadi, P.; Bagheri, H.; Nalousi, A.M.; Nazari, F.; Chandler, S.F. Current status and biotechnological advances in genetic engineering of ornamental plants. Biotechnol. Adv. 2016, 34, 1073–1090. [Google Scholar] [CrossRef] [PubMed]
- Ibitoye, D.; Akin-Idowu, P.E. Marker-assisted-selection (MAS): A fast track to increase genetic gain in horticultural crop breeding. J. Afr. J. Biotechnol. 2011, 10, 11333–11339. [Google Scholar]
- Su, J.; Jiang, J.; Zhang, F.; Liu, Y.; Ding, L.; Chen, S.; Chen, F. Current achievements and future prospects in the genetic breeding of chrysanthemum: A review. Hortic. Res. 2019, 6, 109. [Google Scholar] [CrossRef]
- Kamthan, A.; Chaudari, A.; Kamthan, M.; Datta, A. Genetically modified (GM) crops: Milestones and new advances in crop improvement. Theo. Appl. Genet. 2016, 29, 1639–1655. [Google Scholar] [CrossRef]
- Tang, J.; Ye, J.; Liu, P.; Wang, S.; Chen, F.; Song, A. Ornamental plant gene editing: Past, present and future. Ornamental Plant Res. 2023, 3, 6. [Google Scholar] [CrossRef]
- Samantha, A.; Das, G.; Das, S.K. Roles of flavonoids in plants. Carbon 2011, 100, 12–35. [Google Scholar]
- Kong, J.M.; Chia, L.S.; Goh, N.K.; Chia, T.F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochem. Rev. 2003, 64, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower color modification by engineering of the flavonoid biosynthetic pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P.; Heidmann, I.; Forkmann, G.; Saedler, H. A new petunia flower color generated by transformation of a mutant with a maize gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Koseki, M.; Goto, K.; Masuta, C.; Kanazawa, A. The star-type color pattern in Petunia hybrid ‘Red Star’ flowers is induced by sequence-specific degradation of Chalcone synthase RNA. Plant Cell Physiol. 2005, 46, 1879–1883. [Google Scholar] [CrossRef]
- Rosati, C.; Simoneau, P. Metabolite engineering of flower color in ornamental plants. J. Crop. Improv. 2006, 18, 301–324. [Google Scholar] [CrossRef]
- Nishihara, M.; Nakatsuka, T.; Yamamura, S. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett. 2005, 579, 6074–6078. [Google Scholar] [CrossRef]
- Boase, M.R.; Lewis, D.H.; Davies, K.M.; Marshall, G.B.; Patel, D.; Schwinn, K.E.; Deroles, S.C. Isolation and antisense suppression of flavonoid 3’5’-hydroxylase modifies flower pigments and color in cyclamen. BMC Plant Biol. 2010, 10, 107. [Google Scholar] [CrossRef]
- Seitz, C.; Vitten, M.; Steinbach, P.; Hartl, S.; Hirsche, J.; Rathje, W.; Treutter, D.; Forkmann, G. Redirection of anthocyanin synthesis in Osteospermum hybrida by a two-enzyme manipulation strategy. Phytochem. 2007, 68, 824–833. [Google Scholar] [CrossRef]
- Katsumoto, Y.; Fukuchi-Mizutani, M.; Fukui, Y.; Brugliera, F.; Holton, T.A.; Karan, M.; Nakamura, N.; Sakakibara, K.Y.; Togami, J.; Pigeaire, A.; et al. Engineering of the Rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007, 48, 1589–1600. [Google Scholar] [CrossRef]
- Laitinen, R.A.; Ainasoja, M.; Broholm, S.K.; Teeri, T.H.; Elomaa, P. Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida. J. Exp. Bot. 2008, 59, 3691–3703. [Google Scholar] [CrossRef]
- Qi, Y.; Lou, Q.; Quan, Y.; Liu, Y.; Wang, Y. Flower-specific expression of the Phalaenopsis flavonoid 3′, 5′-hydroxylase modifies flower color pigmentation in Petunia and Lilium. Plant Cell Tissue Organ Cult. 2013, 115, 263–273. [Google Scholar] [CrossRef]
- He, H.; Ke, H.; Keting, H.; Qiaoyan, X.; Silan, D. Flower colour modification of chrysanthemum by suppression of F3’H and overexpression of the exogenous Senecio cruentus F3’5’H gene. PLoS ONE 2013, 8, e74395. [Google Scholar] [CrossRef]
- Brugliera, F.; Tao, G.Q.; Tems, U.; Kalc, G.; Mouradova, E.; Price, K.; Stevenson, K.; Nakamura, N.; Stacey, I.; Katsumoto, Y.; et al. Violet/blue chrysanthemums- Metabolic engineering of the anthocyanin biosynthetic pathway results in novel petal colors. Plant Cell Physiol. 2013, 54, 1696–1710. [Google Scholar] [CrossRef]
- Noda, N.; Aida, R.; Kishimoto, S.; Ishiguro, K.; Fukuchi-Mizutani, M.; Tanaka, Y.; Ohmiya, A. Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol. 2013, 54, 1684–1695. [Google Scholar] [CrossRef]
- Noda, N.; Yoshioka, S.; Kishimoto, S.; Nakayama, M.; Douzono, M.; Tanaka, Y.; Aida, R. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci. Adv. 2017, 3, e1602785. [Google Scholar] [CrossRef]
- Rajabi, A.; Fahmideh, L.; Keykhasaber, M.; Omran, V.G. Genetic engineering of novel yellow color African violet (Saintpaulia ionantha) produced by accumulation of Aureusidin 6-O-glucoside. Biological Procedures Onlnie 2022, 24, 3. [Google Scholar] [CrossRef]
- Lou, Y.; Zhang, Q.; Xu, Q.; Yu, X.; Wang, W.; Gai, R.; Ming, F. PhCHS5 and PhF3’5’H genes overexpression in petunia (Petunia hybrida) and Phalaenopsis (Phalaenopsis Aphrodite) regulate flower color and branch number. Plants 2023, 12, 2204. [Google Scholar] [CrossRef]
- He, G.; Zhang, R.; Jiang, S.; Wang, H.; Ming, F. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis. Hortic. Res. 2023, 10, uhad080. [Google Scholar] [CrossRef]
- Xu, J.; Shin, J.Y.; Park, P.M.; Ahn, H.R.; Kim, Y.J.; Kim, S.J.; Lee, S.H. Flower color modification through co-overexpression of the VtF3’5’H and RhNHX genes in Rosa hybrida. Plant Cell Tis. Org. Cult. 2023, 153, 403–416. [Google Scholar] [CrossRef]
- Watanabe, K.; Kobayashi, A.; Endo, M.; Sage-Ono, K.; Toki, S.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomea (Pharbitis) nil. Sci. Rep. 2017, 7, 10028. [Google Scholar] [CrossRef]
- Watanabe, K.; Oda-Yamamizo, C.; Sage-Ono, K.; Ohmiya, A.; Ono, M. Alteration of flower color in Ipomea nil through CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 4. Transgenic Res. 2018, 27, 25–38. [Google Scholar] [CrossRef]
- Nishihara, M.; Higuchi, A.; Watanabe, A.; Tasaki, K. Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 2018, 18, 331. [Google Scholar] [CrossRef]
- Yan, R.; Wang, Z.; Ren, Y.; Li, H.; Liu, N.; Sun, H. Establishment of efficient genetic transformation systems and application of CRISPR/Cas9 genome editing technology in Lilium pumilum DC. Fisch. And Lilium longiflorum White Heaven. Int. J. Mol. Sci. 2019, 20, 2920. [Google Scholar] [CrossRef]
- Tasaki, K.; Yoshida, M.; Nakajima, M.; Higuchi, A.; Watanabe, A.; Nishihara, M. Molecular characterization of an anthocyanin-related glutathione S-transferase gene in Japanese gentian with the CRISPR/Cas9 system. BMC Plant Biol. 2020, 20, 370. [Google Scholar] [CrossRef]
- Yu, J.; Tu, L.; Subburaj, S.; Bae, S.; Lee, G.J. Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Reports 2021, 40, 1037–1045. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, X.; Huang, R.; Yang, S.; Li, M.; Guo, Y. CRISPR/Cas9-mediated targeted mutation reveals a role for AN4 rather than DPL in regulating venation formation in the corolla tube of Petunia hybrida. Hortic. Res. 2021, 8, 116. [Google Scholar] [CrossRef]
- Dudareva, N.; Pichersky, E. Biochemical and molecular genetic aspects of floral scents. Plant Physiol. 2000, 122, 627–633. [Google Scholar] [CrossRef]
- Aranovich, D.; Lewinsohn, E.; Zaccai, M. Post-harvest enhancement of aroma in transgenic lisianthus (Eustoma grandiflorum) using the Clarika breweri benzyl alcohol acetyl transferase (BEAT) gene. Postharvets Biol. Biotechnol. 2007, 43, 255–260. [Google Scholar]
- Lavy, M.; Zukea, A.; Lewinsohn, E.; Larkov, O.; Ravid, U.; Vainstein, A.; Weiss, D. Linalool and linalool oxide production in transgenic carnation flowers expressing the Clarkia breweri linalool synthase gene. Mol. Breed. 2002, 9, 103–111. [Google Scholar] [CrossRef]
- Ben Zvi, M.M.; Negre-Zakharov, F.; Masci, T.; Ovadis, M.; Shklarman, E.; Ben-Meir, H.; Tzfira, T.; Dudareva, N.; Vainstein, A. Interlinking showy traits: Co-engineering of scent and co-engineering of scent and color biosynthesis in flowers. Plant Biotech. J. 2008, 6, 403–415. [Google Scholar]
- Ben Zvi, M.M.; Shkalarman, E.; Masci, T.; Kalev, H.; Debenar, T.; Shafir, S.; Ovadis, M.; Vainstein, A. Pap1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. N. Phytol. 2012, 195, 335–345. [Google Scholar]
- Yang, T.; Stoopen, G.; Thoen, M.; Wiegers, G.; Jongsma, M.A. Chrysanthemum expressing a linalool synthase gene ‘smells good’ but ‘tastes bad’ to western flower thrips. Plant Biotecnol. J. 2013, 11, 875–882. [Google Scholar] [CrossRef]
- Netam, N. Improving ornamental’s vase life through molecular approaches: A review. Pharm. Phytochem. 2018, 7, 1687–1691. [Google Scholar]
- Chandler, S.F.; Sanchez, C. Genetic modification, the development of transgenic ornamental plant varieties. Plant Biotechnol. J. 2012, 10, 891–903. [Google Scholar] [CrossRef]
- Narumi, T.; Aida, R.; Ohmiya, A.; Satoh, S. Transformation of chrysanthemum with mutated ethylene receptor genes: mDG-ERS1 transgenes conferring reduced ethylene sensitivity and characterization of the transformants. Postharvest Biol. Technol. 2005, 37, 101–110. [Google Scholar] [CrossRef]
- Satoh, S.; Watnabe, M.; Chisaka, K.; Narumi, T. Suppressed leaf senescence in chrysanthemum transformed with a mutated ethylene receptor gene mDG-ERS1 (etr1-4). Plant Biol. 2008, 51, 424–427. [Google Scholar] [CrossRef]
- Bovy, A.G.; Angenent, G.C.; Dons, H.J.; van Altvorst, A.C. Heterologous expression of the Arabidopsis etr1-1 allele inhibits the senescence of carnation flowers. Mol. Breed. 1999, 5, 301–308. [Google Scholar] [CrossRef]
- Sriskandarajah, S.; Mibus, H.; Serek, M. Transgenic Campanula carpatica plants with reduced ethylene sensitivity. Plant Cell Rep. 2007, 26, 805–813. [Google Scholar] [CrossRef]
- Raffeiner, B.; Serek, M.; Winklemann, T. Agrobacterium tumefaciens- mediated transformation of Oncidium and Odontoglossum orchid species with the ethylene receptor mutant gene etr1-1. Plant Cell Tiss. Organ Cult. 2009, 98, 125–134. [Google Scholar] [CrossRef]
- Winkelmann, T.; Warwas, M.; Raffeiner, B.; Serek, M.; Mibus, H. Improved postharvest quality of inflorescences of fbp1::etr1-1 transgenic Burrageara ‘Stefan Isler Lava Flow. ’ J. Plant Growth Regul. 2016, 35, 390–400. [Google Scholar] [CrossRef]
- Gehl, C.; Wamhoff, D.; Schaarschmidt, F.; Serek, M. Improved leaf and flower longevity by expressing the etr1-1 allele in Pelargonium zonale under control of FBP1 and SAG12 promotes. Plant Growth Regul. 2018, 86, 351–363. [Google Scholar] [CrossRef]
- Inokuma, T.; Kinouchi, T.; Satoh, S. Reduced ethylene production in transgenic carnation transformed with ACC oxidase cDNA in sense orientation. J. Appl. Hortic. 2008, 10, 3–7. [Google Scholar] [CrossRef]
- Chang, H.; Jones, M.L.; Banowetz, G.M.; Clark, D.G. Overproduction of cytokinins in petunia flowers transformed with PSAG12-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol. 2003, 132, 2174–2183. [Google Scholar] [CrossRef]
- Zakizadeh, H.; Lutken, H.; Sriskandarajah, S.; Serek, M.; Muller, R. Transformation of miniature potted rose (Rosa hybrida cv. Linda) with PSAG12-ipt gene delays leaf senescence and enhances resistance to exogenous ethylene. Plant Cell Rep. 2013, 32, 195–205. [Google Scholar] [CrossRef]
- Shibuya, K.; Watanabe, K.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. Biochem. 2018, 131, 53–57. [Google Scholar] [CrossRef]
- Xu, J.; Kang, B.C.; Naing, A.H.; Bae, S.J.; Kim, J.S.; Kim, H.; Kim, C.K. CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 2020, 18, 287–297. [Google Scholar] [CrossRef]
- Lin, Y.; Jones, M.L. CRISPR/Cas9-mediated editing of autophagy gene 6 in Petunia decreases flower longevity, seed yield, and phosphorus remobilization by accelerating ethylene production and senescence-related gene expression. Front. Plant Sci. 2022, 13, 840218. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Wang, N.; Yu, Q.; Li, Y.; Gao, J.; Zhou, X.; Ma, N. An efficient CRISPR/Cas9 platform for targeted genome editing in rose (Rosa hybrida). J. Integr. Plant Biol. 2023, 65, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Aida, R.; Komano, M.; Saito, M.; Nakase, K.; Murai, K. Chrysanthemum flower shape modification by suppression of chrysanthemum-AGAMOUS gene. Plant Biotechnol. 2008, 25, 55–59. [Google Scholar] [CrossRef]
- Meng, L.S.; Song, J.P.; Sun, S.B.; Wang, C.Y. The ectopic expression of PttKN1 gene causes pleiotropic alteration of morphology in transgenic carnation (Dianthus caryophyllus L.). Acta Physiol. Plant 2009, 31, 1155–1164. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Yang, C.H. Ectopic expression of two MADS box genes from orchid (Oncidium Gower Ramsey) and lily (Liliumlongiflorum) alters flower transition and formation in Eustoma grandiflorum. Plant Cell Rep. 2009, 28, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Ruokolainene, S.; Ng, Y.P.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Overexpression of the Gerbera hybrida At-SOC1-like1 gene GhSOC1 leads to floral organ identity deterioration. Ann. Bot. 2011, 107, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, X.; Sun, M.; Zhang, T.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 1633. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Zhao, K.; Zhang, X.; Song, A.; Su, J.; Hu, Y.; Zhao, W.; Jiang, J.; Chen, F. Comprehensive characterization of a floral mutant reveals the mechanism of hooked petal morphogenesis in Chrysanthemum morifolium. Plant Biotechnol. J. 2019, 17, 2325–2340. [Google Scholar] [CrossRef]
- Su, S.; Xiao, W.; Guao, W.; Yao, X.; Xiao, J.; Ye, Z.; Wang, N.; Jiao, K.; Lei, M.; Peng, Q.; et al. The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torneria fournieri (Linderniaceae). New Phytol. 2017, 215, 1582–1593. [Google Scholar] [CrossRef]
- Sun, L.; Kao, T.H. CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein-containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflate. Plant Rep. 2018, 31, 129–143. [Google Scholar] [CrossRef]
- Gattolin, S.; Cirilli, M.; Chessa, S.; Stella, A.; Bassi, D.; Rossini, L. Mutations in orthologous PETALOSA TOE-type genes cause dominant double-flower phenotype in phylogenetically distant eudicots. J. Exp. Bot. 2020, 71, 2585–2595. [Google Scholar] [CrossRef]
- Nishihara, M.; Hirabuchi, A.; Goto, F.; Watanabe, A.; Yoshida, C.; Washiashi, R.; Odashima, M.; Nemoto, K. Efficient double-flowered gentian plant production using the CRISPR/Cas9 system. Plant Biotech. 2023, 40, 229–236. [Google Scholar] [CrossRef]
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting and resetting. Plant Cell 2004, 16, S18–S31. [Google Scholar] [CrossRef]
- Shulga, O.A.; Mitiouchkina, T.Y.; Shchennikova, A.V.; Skryabin, K.G.; Dolgov, S.V. Overexpression of AP1-like genes from Asteraceae induces early-flowering in transgenic chrysanthemum plants. In Vitro Cell Dev. Biol. Plant 2011, 47, 553–560. [Google Scholar] [CrossRef]
- Li, X.; Bian, H.; Song, D.; Ma, S.; Ha, N.; Wang, J.; Zhu, M. Flowering time control in ornamental gloxinia (Sinningia speciose) by manipulation of miR159 expression. Ann. Bot. 2013, 111, 791–799. [Google Scholar] [CrossRef]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef]
- Sun, J.; Wang, H.; Ren, L.; Chen, S.; Chen, F.; Jiang, J. CmFTL2 is involved in the photoperiod-and sucrose-mediated control of flowering time in chrysanthemum. Hortic. Res. 2017, 4, 17001. [Google Scholar] [CrossRef]
- Mao, Y.; Sun, J.; Cao, P.; Zhang, R.; Fu, Q.; Chen, S.; Chen, F.; Jiang, J. Functional analysis of alternative splicing of the FLOWERING LOCUS T orthologous gene in Chrysanthemum morifolium. Hortic Res. 2016, 3, 16058. [Google Scholar] [CrossRef]
- Leeggangers, H.A.C.F.; Rosilio-Brami, T.; Bigas-Nadal, J.; Rubin, N.; van Dijk, A.D.J.; Gonzalez, F.F.N.D.C.; Saadon-Shitrit, S.; Nijveen, H.; Hilhorst, F.H.W.M.; Immink, R.G.H.; et al. Tulipa gesneriana and Lilium longiflorum PEBP genes and their putative roles in flowering time control. Plant Cell Physiol. 2018, 59, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Gao, Y.; Gao, Y. Standard genetic transformation protocol for Chrysanthemum cv. ‘Jinba’ with TERMINAL FLOWER1 homolog CmTFL1a. Genes Dev. 2020, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, J.; Ren, L.; Zhou, M.; Han, X.; Ding, L.; Zhang, F.; Guan, Z.; Fang, W.; Chen, S.; et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum. Plant Biotech. J. 2020, 18, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, C.; Xu, Y.; Wei, Q.; Imtiaz, M.; Lan, H.; Gao, S.; Cheng, L.; Wang, M.; Fei, Z. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. J. Plant Cell 2014, 26, 2038–2054. [Google Scholar] [CrossRef]
- Chen, H.; Huang, F.; Liu, Y.; Cheng, P.; Guan, Z.; Fang, W.; Chen, S.; Chen, F.; Jiang, J. Constitutive expression of chrysanthemum CmBBX29 delays flowering time in transgenic Arabidopsis. Can. J. Plant Sci. 2019, 100, 86–94. [Google Scholar] [CrossRef]
- Zhu, L.; Guan, Y.; Liu, Y.; Zhang, Z.; Jaffar, M.A.; Song, A.; Chen, S.; Jiang, J.; Chen, F. Regulation of flowering time in chrysanthemum by the R2R3 MYB transcription factor CmMYB2 is associated with changes in gibberellin metabolism. Hortic. Res. 2020, 7, 96. [Google Scholar] [CrossRef]
- Sankhuan, D.; Ji, M.; Takanashi, S.; Imamura, Y.; Sato, S.; Supaibulwatana, K.; Otani, M.; Nakano, M. Induction of dwarf and early flowering phenotypes in Tricyrtis Sp. by ectopic expression of LEAFY from Arabidopsis thaliana. Plant Biotechnol. 2022, 39, 208. [Google Scholar] [CrossRef]
- Lin, Z.Y.; Zhu, G.F.; Lu, C.Q.; Gao, J.; Li, J.; Xie, Q.; Wei, Y.L.; Jin, J.P.; Wang, F.L.; Yang, F.X. Functional conservation and divergence of SEPTALLATA-like genes in floral development in Cymbidium sinense. Front. Plant Sci. 2023, 14, 1209834. [Google Scholar] [CrossRef]
- Tong, C.G.; Wu, F.H.; Yuan, Y.H.; Chen, Y.R.; Lin, C.S. High efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS genes. Plant Biotech. J. 2020, 18, 889–891. [Google Scholar] [CrossRef]
- Liu, L.; Xue, Y.; Luo, J.; Han, M.; Liu, X.; Jiang, T.; Zhao, Y.; Xu, Y.; Ma, C. Developing a UV-visible reporter-assisted CRISPR-Cas9 gene editing system to alter flowering time in Chrysanthemum indicum. Plant Biotech. J. 2023, 21, 1515–1521. [Google Scholar] [CrossRef]
- Suputri, N.P.A.E.O.; Prasojo, I.S.; Prabowo, L.A.T.; Purwestri, Y.A.; Purnomo; Semiarti, E. Identification of early flowering mutant gene in Phalaenopsis amabilis (L.) Blume for sgRNA construction in CRISPR/Cas9 genome editing system. Braz. J. Biol. 2023, 84, e268133. [Google Scholar] [CrossRef]
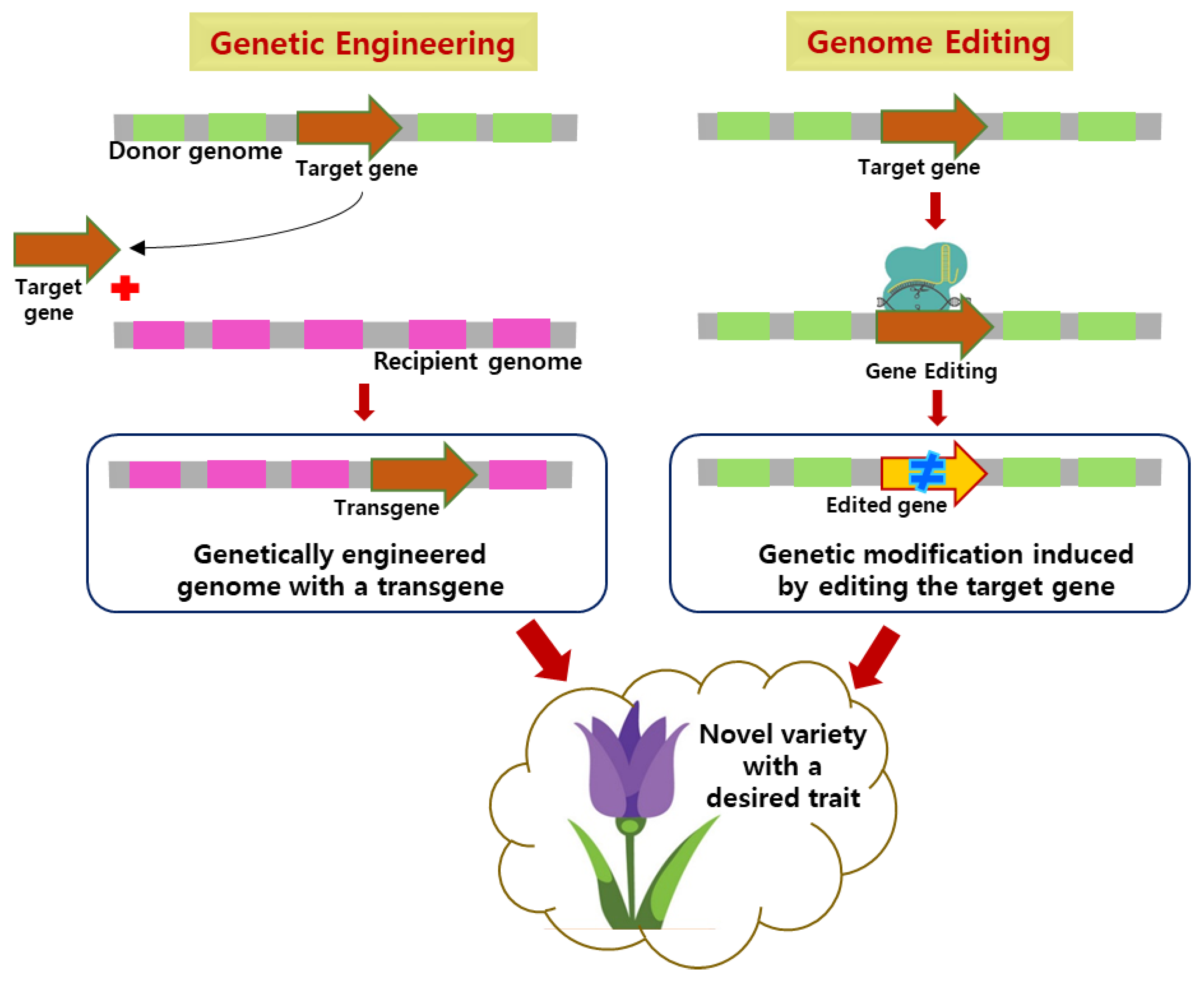
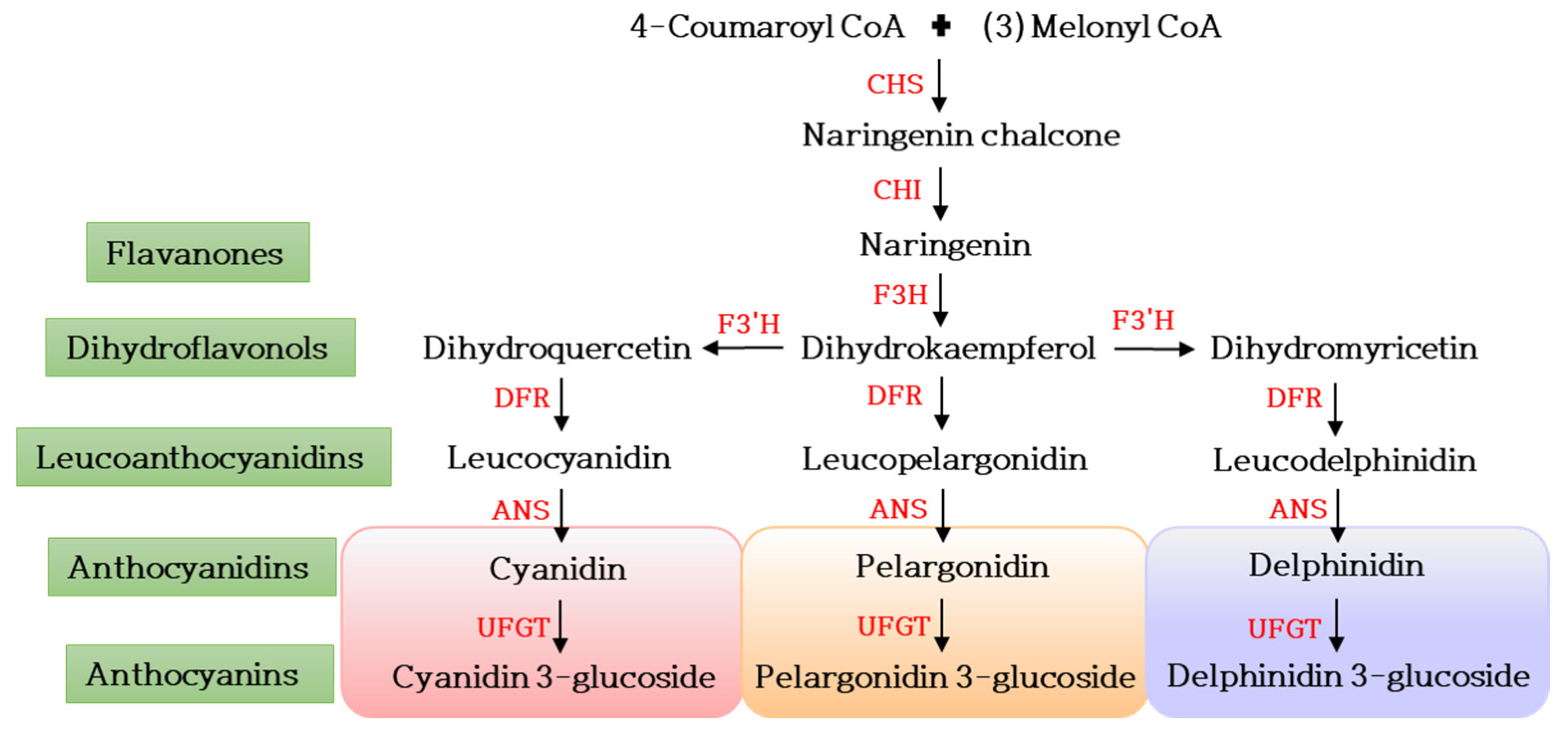
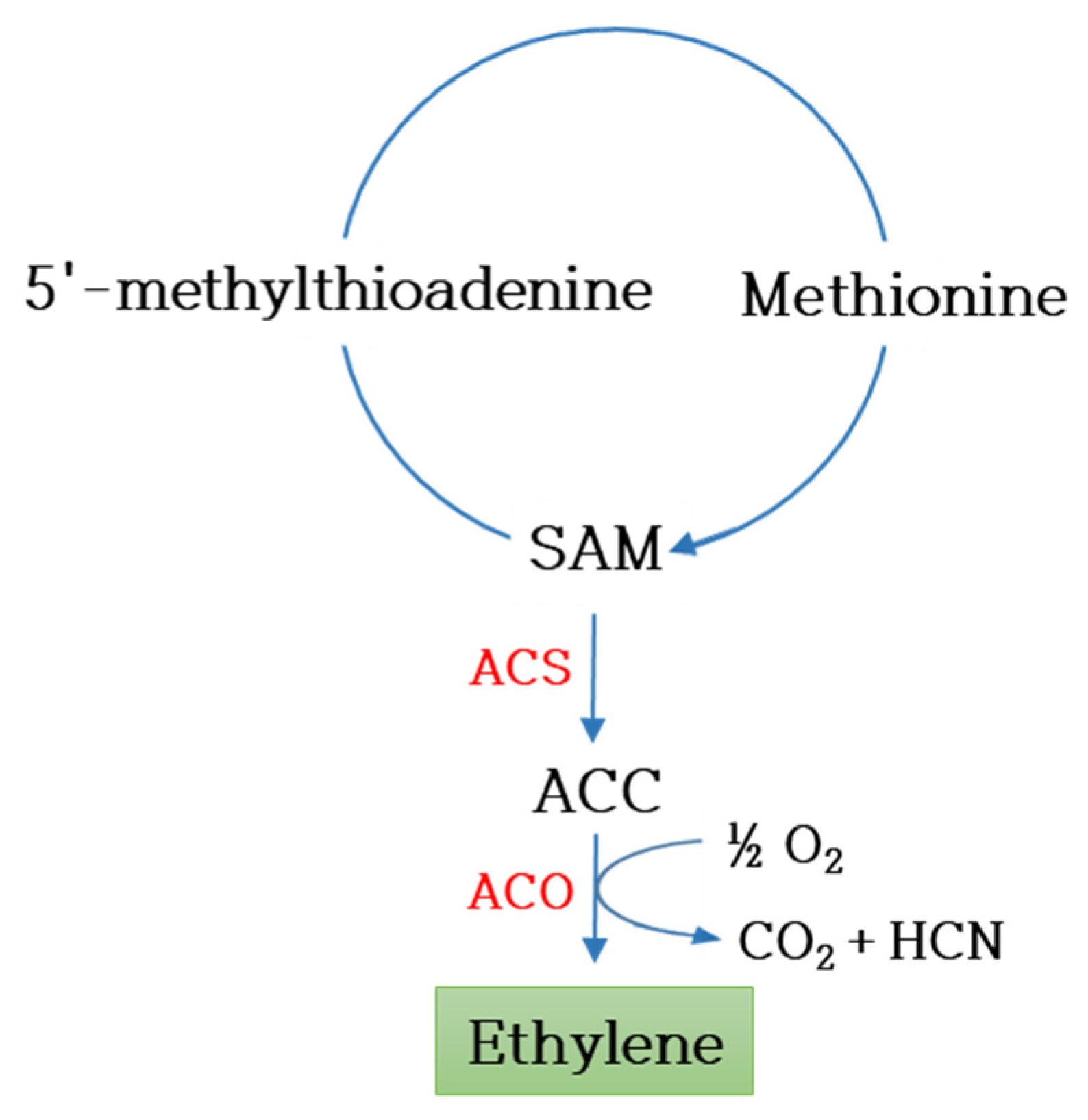
| S. No | Ornamental Plant | Turnover (Million Euros) |
|---|---|---|
| Cut Flowers | ||
| 1 | Rose | 694 |
| 2 | Chrysanthemum | 369 |
| 3 | Tulip | 257 |
| 4 | Lily | 155 |
| 5 | Eustoma russellianum | 149 |
| House Plants | ||
| 6 | Phalaenopsis | 402 |
| 7 | Arrangements | 69 |
| 8 | Anthurium | 64 |
| 9 | Kalanchoe | 64 |
| 10 | Rose | 59 |
| Garden Plants | ||
| 11 | Helleborus | 25 |
| 12 | Hydrangea | 24 |
| 13 | Lavandula | 21 |
| 14 | Carnation | 20 |
| 15 | Bedding plants | 20 |
| Floral Attribute | Genetic Engineering | ||||
|---|---|---|---|---|---|
| Gene | Source | Target Plant | Resulting Trait | Ref. | |
| Floral Color | A1 | Zea mays | Petunia | orange-colored flower | [52] |
| CHS | Petunia | Petunia | star-type pigmentation pattern in corolla | [53] | |
| CHS | Petunia, chrysanthemum | Petunia, chrysanthemum | pigmentation loss and white-colored flower | [54] | |
| CHI | Tobacco | Tobacco | floral color shift to yellow | [55] | |
| F3′5′H | Cyclamen | Cyclamen | floral color shift from purple to red/pink | [56] | |
| DFR | Gerbera | Osteospermum hybrida | shift from delphinidin to pelargonidin | [57] | |
| F3′5′H, DFR | Viola, Iris x hollandica | Rose | blue-colored flower phenotype | [58] | |
| GMYB10 | Gerbera | Gerbera | cyanidin synthesis and increased pigmentation | [59] | |
| F3′5′H | Phalaenopsis | Lilium | floral color shift from pink to purple | [60] | |
| F3′5′H | Senecio cruentus | Chrysanthemum | bright red flower | [61] | |
| F3′5′H | Pansy | Chrysanthemum | violet/blue flower | [62] | |
| ADH+ F3′5′H | Campanula | Chrysanthemum | violet/blue flower | [63] | |
| F3′5′H | Canterbury bells | Chrysanthemum | true-blue-colored flower | [64] | |
| 4′CGT+AS1 | A. majus | African violet | floral color shift from white to yellow | [65] | |
| PhCHS PhF3′5′H | Phalaenopsis | Phalaenopsis, petunia | deeper floral lip color | [66] | |
| RcMYB1 | Rose | Rose, tobacco | increased anthocyanins in white petals | [67] | |
| F3′5′H+NHX | Viola, rose | Rose | floral color shift from white to red-purple | [68] | |
| Floral Scent | BEAT | C. breweri | Lisianthus | floral scent | [77] |
| lis | C. breweri | Carnation | floral scent | [78] | |
| Pap1 | Arabidopsis | Petunia | floral scent | [79] | |
| Pap1 | Arabidopsis | Rose | floral scent | [80] | |
| FaNES1 | Strawberry | Chrysanthemum | floral scent | [81] | |
| Floral Longevity | Mutated etr1-4 | Chrysanthemum | Chrysanthemum | reduced leaf senescence | [85] |
| Mutated etr1-1 | Arabidopsis | Carnation, campanula, orchids | suppressed ethylene susceptibility | [86,87,88,89,90] | |
| ACC oxidase | Carnation | Carnation | lower ethylene production | [91] | |
| PSAG12-IPT | A. tumefaciens | Petunia and rose | delayed senescence | [92,93] | |
| Floral Anatomy | AGAMOUS | Chrysanthemum | Chrysanthemum | floral shape change | [98] |
| PttKN1 | Hybrid aspen | Carnation | modification of phyllotaxis | [99] | |
| MADS1-M | Lily | Lisianthus | altered floral structure | [100] | |
| GhSOC1 | Gerbera | Gerbera | loss of floral distinctiveness | [101] | |
| CmCYC2c | Chrysanthemum | Chrysanthemum | increased ray floret length | [102] | |
| CmYAB1 | Chrysanthemum | Chrysanthemum | reduced petal curvature and pompon flower | [103] | |
| PhCHS5, Ph F3′5′H | Phalaenopsis | Phalaenopsis | increased petals, labial petals | [66] | |
| Flowering Time | AP1 | Asteraceae | Chrysanthemum | early flowering | [109] |
| miR159 | Gloxinia | Gloxinia | flowering time regulation | [110] | |
| CsFTL3 | Chrysanthemum seticuspe | Chrysanthemum | floral bud development | [111] | |
| LlFT | Lilium longiflorum | Lily | early flowering | [114] | |
| CmTFL1a | Chrysanthemum | Chrysanthemum | delayed flowering | [115] | |
| CmBBX8 | Chrysanthemum | Chrysanthemum | early flowering | [116] | |
| CmBBX24 | Chrysanthemum | Chrysanthemum | early flowering | [117] | |
| CmBBX29 | Chrysanthemum | Arabidopsis | delayed flowering | [118] | |
| CmMYB2 | Chrysanthemum | Chrysanthemum | early flowering | [119] | |
| AtLFY | Arabidopsis | Tricyrtis sp. | early flowering | [120] | |
| CsSEP1,2,3,4 | Cymbidium sinense | Arabidopsis | early flowering | [121] | |
| Ornamental Attribute | Genome Editing (CRISPR/Cas9) | |||
|---|---|---|---|---|
| Gene | Target Plant | Resulting Trait | Reference | |
| Floral Color | DFR | Ipomea nil | floral color modification | [69] |
| CCD4 | Ipomea nil | pale yellow-colored petals | [70] | |
| F3H | Torenia fournieri | floral color shift from pale blue to white | [71] | |
| PDS | Lily | albino, albino-green and pale yellow flower pigmentation | [72] | |
| GST1 | Japanese gentian | white and mild blue floral phenotype | [73] | |
| F3HA, F3HB | Petunia | color shift from purple to purplish pink | [74] | |
| DPL | Petunia | vein-associated absence of anthocyanin pattern | [75] | |
| AN4 | Petunia | absence of corolla tube venation | [75] | |
| Floral Longevity | EPH1 | Japanese morning glory | delayed petal senescence | [94] |
| PhACO1 | Petunia | delayed senescence | [95] | |
| PhATG6 | Petunia | accelerated senescence | [96] | |
| RhEIN2 | Rose | ethylene sensitivity | [97] | |
| Floral Anatomy | TfRAD1 | Torenia fournieri | violet color pattern on dorsal petals and ventralized later petals | [104] |
| piSSK1 | Petunia | growth inhibition of pollen tubes | [105] | |
| PET | Tobacco | double-flower phenotype | [106] | |
| AG1 | Gentian | double-flower phenotype | [107] | |
| Flowering Time and Development | MADS | Phalaenopsis | gene editing efficiency in flowering time | [122] |
| CfTFL1a, CiTFL1b | Chrysanthemum indicum | early flowering | [123] | |
| GAI | Phalaenopsis amabilis | early flowering | [124] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mekapogu, M.; Song, H.-Y.; Lim, S.-H.; Jung, J.-A. Genetic Engineering and Genome Editing Advances to Enhance Floral Attributes in Ornamental Plants: An Update. Plants 2023, 12, 3983. https://doi.org/10.3390/plants12233983
Mekapogu M, Song H-Y, Lim S-H, Jung J-A. Genetic Engineering and Genome Editing Advances to Enhance Floral Attributes in Ornamental Plants: An Update. Plants. 2023; 12(23):3983. https://doi.org/10.3390/plants12233983
Chicago/Turabian StyleMekapogu, Manjulatha, Hyun-Young Song, So-Hyeon Lim, and Jae-A Jung. 2023. "Genetic Engineering and Genome Editing Advances to Enhance Floral Attributes in Ornamental Plants: An Update" Plants 12, no. 23: 3983. https://doi.org/10.3390/plants12233983
APA StyleMekapogu, M., Song, H.-Y., Lim, S.-H., & Jung, J.-A. (2023). Genetic Engineering and Genome Editing Advances to Enhance Floral Attributes in Ornamental Plants: An Update. Plants, 12(23), 3983. https://doi.org/10.3390/plants12233983





