Transcriptome Analysis of Maize Ear Leaves Treated with Long-Term Straw Return plus Nitrogen Fertilizer under the Wheat–Maize Rotation System
Abstract
:1. Introduction
2. Results
2.1. Grain Yield and Photosynthetic Parameters under Various Fertilizer Treatments
2.2. Physiological Changes of Ear Leaves Responsive to Various Fertilizer Treatments
2.3. Transcriptome Sequencing
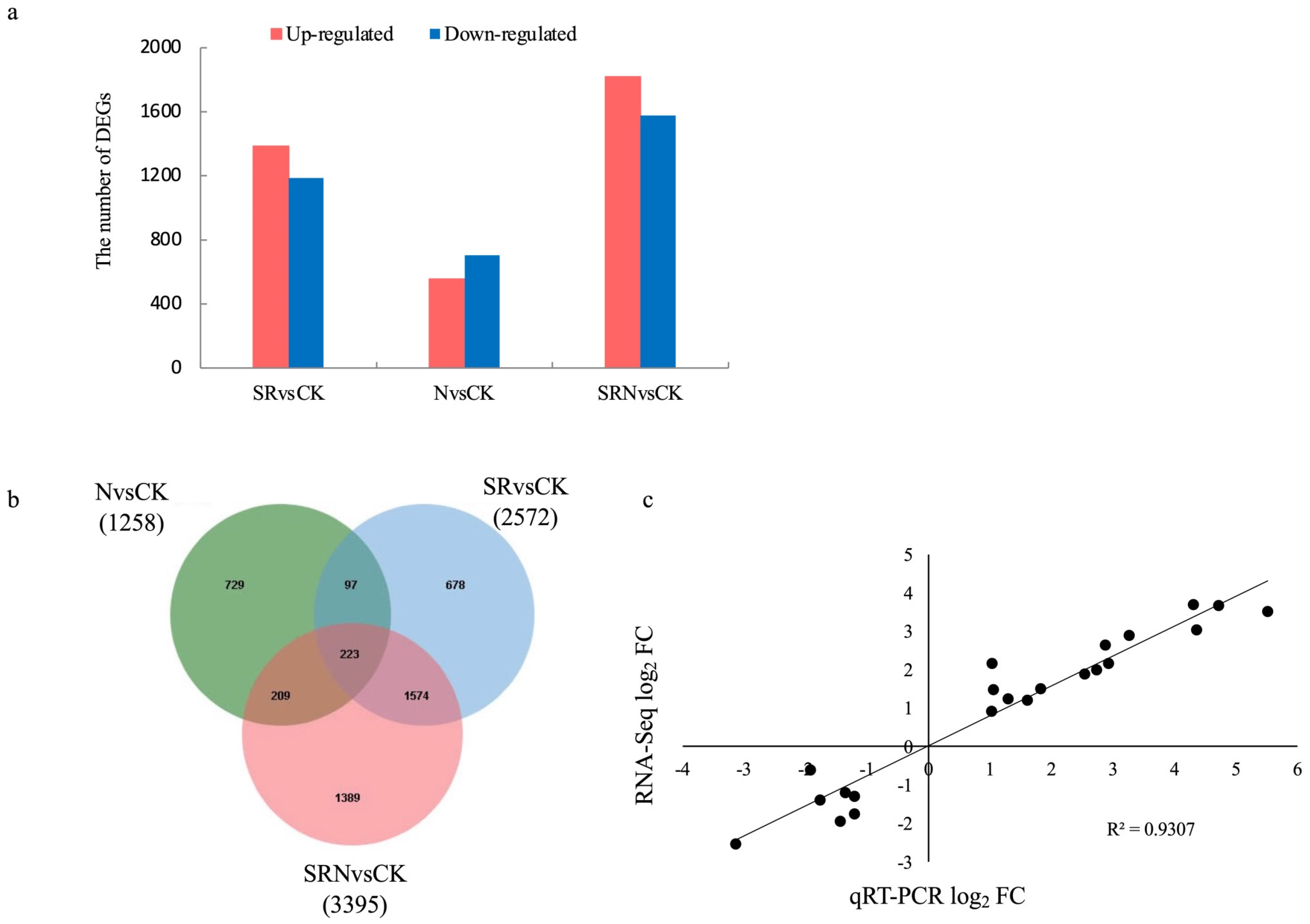
2.4. Identification of DEGs Responsive to Various Fertilizer Treatments
2.5. GO and KEGG Analysis of the DEGs Responsive to Fertilizer Treatments
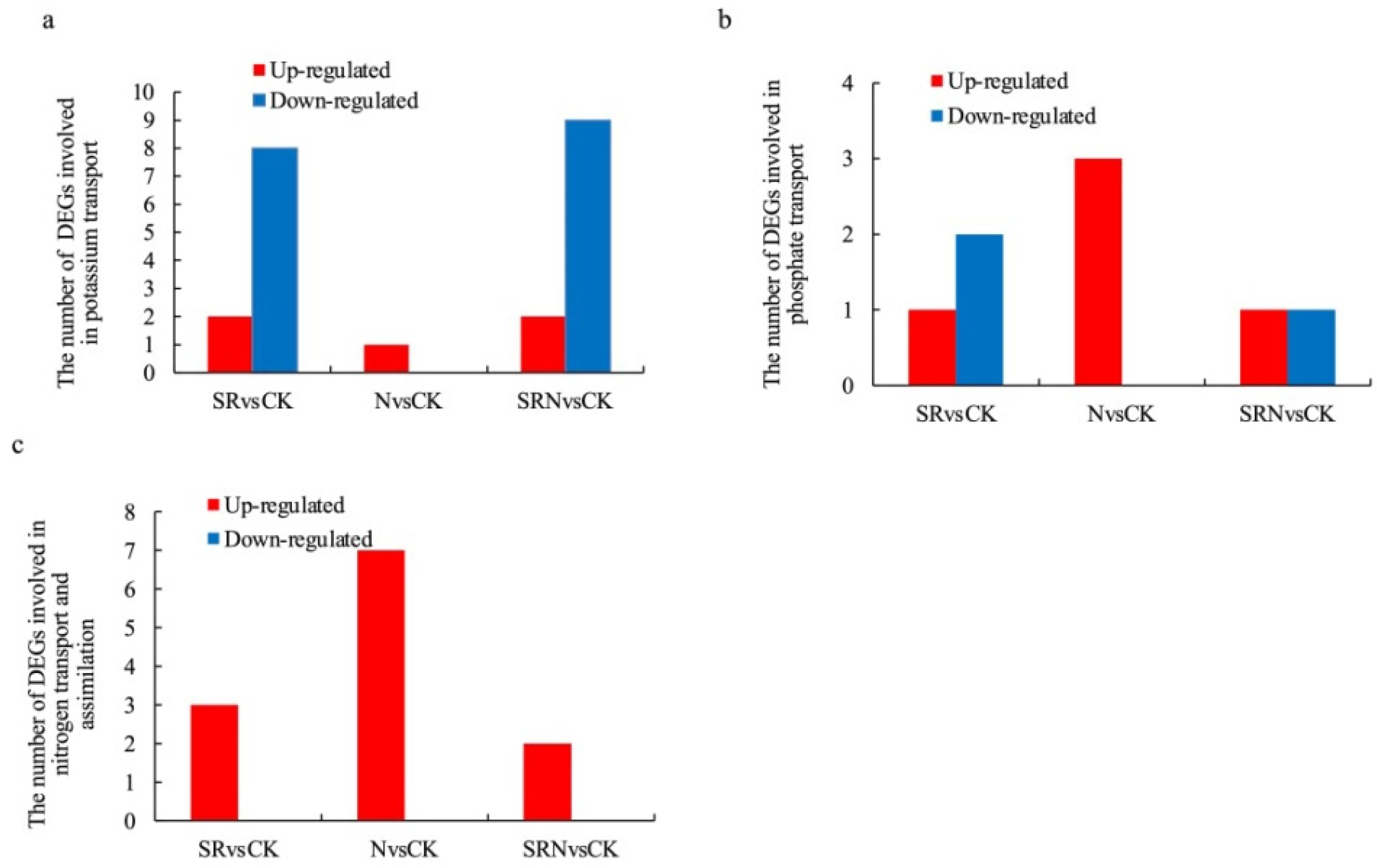
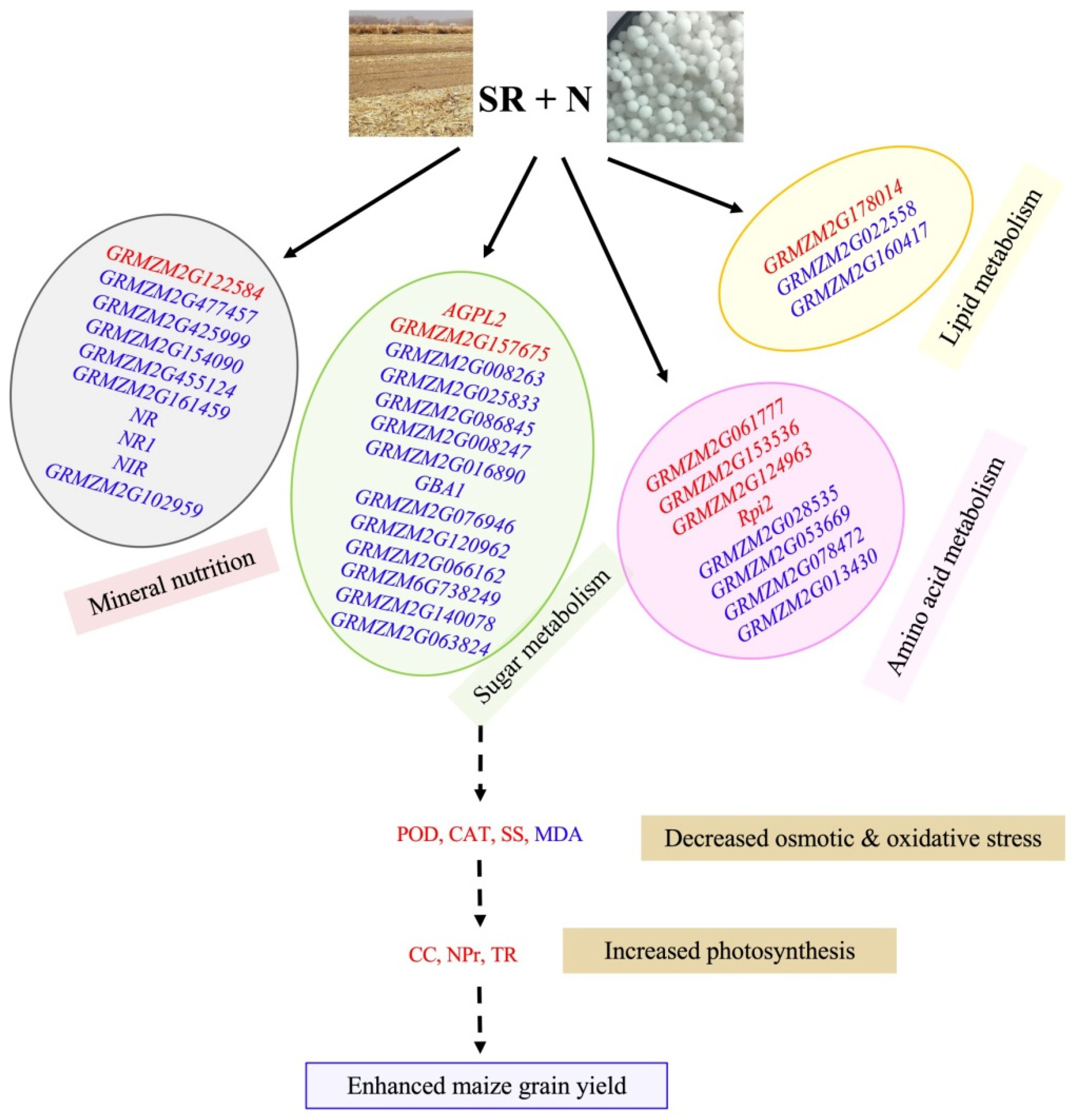
2.6. DEGs Involved in Mineral Nutrient Transport and Assimilation
2.7. DEGs Involved in Sugar Metabolism, Fatty Acid Metabolism and Amino Acids Biosynthesis
3. Discussion
4. Materials and Methods
4.1. Experiment Site, Crop Material and Design
4.2. Measurement of Photosynthetic Parameters and Maize Yield
4.3. Sampling and Assay of Physiological Indexes
4.4. RNA Extraction, cDNA Library Construction, Illumina Sequencing, Read Processing, Assembling and Gene Functional Annotation
4.5. Real-Time Quantitative RT-PCR
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Tilman, D. Global environmental impacts of agricultural expansion: The need for sustainable and efficient practices. Proc. Natl. Acad. Sci. USA 1999, 96, 5995–6000. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.T.; Xing, G.X.; Chen, X.P.; Zhang, S.L.; Zhang, L.J.; Liu, X.J.; Cui, Z.L.; Yin, B.; Christie, P.; Zhu, Z.L.; et al. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc. Natl. Acad. Sci. USA 2009, 106, 3041–3046. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Q.B. Study on potential straw resource nutrient return to field and application 488 technology in China. Soil Fertil. China 2020, 1, 119–126. [Google Scholar]
- Islam, M.U.; Jiang, F.H.; Halder, M.; Liu, S.; Peng, X.H. Impact of straw return combined with different fertilizations on soil organic carbon stock in upland wheat and maize croplands in China: A meta-analysis. Crop Environ. 2023, in press. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, C. Spatial and temporal distribution of air pollutant emissions from open burning of crop residues in China. Sci. Pap. 2008, 3, 329–333. [Google Scholar]
- Li, H.; Dai, M.W.; Dai, S.L.; Dong, X.J. Current status and environment impact of direct straw return in China’s cropland-A review. Ecotoxicol. Environ. Saf. 2018, 189, 293–300. [Google Scholar] [CrossRef]
- Su, Y.; Lv, J.L.; Yu, M.; Ma, Z.H.; Xi, H.; Kou, C.L.; He, Z.C.; Shen, A.L. Long-term decomposed straw return positively affects the soil microbial community. J. Appl. Microbiol. 2020, 28, 138–150. [Google Scholar] [CrossRef]
- Yang, S.Q.; Wang, Y.S.; Liu, R.L.; Xing, L.; Yang, Z.L. Improved crop yield and reduced nitrate nitrogen leaching with straw return in a rice-wheat rotation of Ningxia irrigation district. Sci. Rep. 2018, 8, 9458. [Google Scholar] [CrossRef]
- Cui, H.X.; Luo, Y.L.; Chen, J.; Jin, M.; Li, Y.; Wang, Z.L. Straw return strategies to improve soil properties and crop productivity in a winter wheat-summer maize cropping system. Eur. J. Agron. 2022, 133, 126436. [Google Scholar] [CrossRef]
- Tang, L.; Nie, S.; Li, W.; Fan, C.; Wang, S.; Wu, F.; Pan, K. Wheat straw increases the defense response and resistance of watermelon monoculture to Fusarium wilt. BMC Plant Biol. 2019, 19, 551. [Google Scholar] [CrossRef]
- Chen, Y.; Xin, L.; Liu, J.; Yuan, M.; Liu, S.; Jiang, W.; Chen, J.P. Changes in bacterial community of soil induced by long-term straw returning. Sci. Agric. 2017, 74, 349–356. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Z.; Tan, X.; Jiang, Y.; Gao, J.; Lin, L.; Wang, Z.; Ren, J.; Wang, X.; Qin, L.; et al. Association of the molecular regulation of ear leaf senescence/stress response and photosynthesis/metabolism with heterosis at the reproductive stage in maize. Sci. Rep. 2016, 6, 29843. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.U.; Guo, Z.C.; Jiang, F.H.; Peng, X.H. Does straw return increase crop yield in the Wheat–Maize cropping system in China? A meta-analysis. Field Crops Res. 2022, 279, 108447. [Google Scholar] [CrossRef]
- Liu, J.T.; Zhu, K.; Zhao, H.; Li, Y.; Song, X.; Liu, S.; Li, J. Transcriptome analysis of maize ear leaves under long-term applications of nitrogen fertilizer and its combinations with phosphorus and potassium fertilizers. J. Soil Sci. Plant Nutr. 2022, 22, 112–120. [Google Scholar] [CrossRef]
- Shen, C.; Wang, J.; Shi, X.; Kang, Y.; Xie, C.; Peng, L.; Dong, C.; Shen, Q.; Xu, Y. Transcriptome analysis of differentially expressed genes induced by low and high potassium levels provides insight into fruit sugar metabolism of pear. Front. Plant Sci. 2017, 8, 938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Wei, J. Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 2012, 39, 8465–8473. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lv, S.; Jiang, P.; Li, Y. Roles, regulation and agricultural application of plant phosphate transporters. Front Plant Sci. 2017, 8, 817. [Google Scholar] [CrossRef]
- Chen, Y.F.; Wang, Y.; Wu, W.H. Membrane transporters for nitrogen, phosphate and potassium uptake in plants. J. Integr. Plant Biol. 2008, 50, 835–848. [Google Scholar] [CrossRef]
- Xu, Z.; Zhong, S.; Li, X.; Li, W.; Rothstein, S.J.; Zhang, S.; Bi, Y.; Xie, C. Genome-wide identification of MicroRNAs in response to low nitrate availability in maize leaves and roots. PLoS ONE 2011, 6, e28009. [Google Scholar] [CrossRef]
- Gelli, M.; Duo, Y.; Konda, A.R.; Zhang, C.; Holding, D.; Dweikat, I. Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genom. 2014, 15, 179. [Google Scholar] [CrossRef]
- Kaur, B.; Kaur, G.; Asthir, B. Biochemical aspects of nitrogen use efficiency: An overview. J. Plant Nutr. 2017, 40, 506–523. [Google Scholar] [CrossRef]
- Zheng, L.; Wang, R.; Zhou, P.; Pan, Y.; Shen, R.; Lan, P. Comparative physiological and proteomic response to phosphate deficiency between two wheat genotypes differing in phosphorus utilization efficiency. J. Proteom. 2023, 280, 104894. [Google Scholar] [CrossRef] [PubMed]
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E.; et al. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.C.; Qian, T.F.; Chu, J.P.; Zhang, X.; Liu, Y.J.; Dai, X.L.; He, M.R. Late sowing enhances lodging resistance of wheat plants by improving the biosynthesis and accumulation of lignin and cellulose. J. Integr. Agric. 2023, 22, 1351–1365. [Google Scholar] [CrossRef]
- McMillan, S.D.; Oberlie, N.R.; Hardtke, H.A.; Montes, M.; Brown, D.W.; McQuade, K.L. A secondary function of trehalose-6-phosphate synthase is required for resistance to oxidative and desiccation stress in Fusarium verticillioides. Fungal Biol. 2023, 127, 918–926. [Google Scholar] [CrossRef]
- Zhang, C.K.; Turgeon, R. Mechanisms of phloem loading. Curr. Opin. Plant Biol. 2018, 43, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Liang, K.H.; Sun, Y.J.; Zhang, J.S.; Cao, Y.B.; Zhou, J.; Wang, A.B.; Zhang, L.Y. Evidence of the predominance of passive symplastic phloem loading and sugar transport with leaf ageing in Camellia oleifera. Horti. Plant J. 2023, 9, 811–825. [Google Scholar] [CrossRef]
- Zhou, L.Y.; Li, X.D.; Tang, X.; Lin, Y.J.; Li, Z.F. Effects of different application amount of N, P, K fertilizers on physiological characteristics, yield and kernel quality of peanut. Ying Yong Sheng Tai Xue Bao 2007, 18, 2468–2474. [Google Scholar]
- Shi, J.; Lang, C.; Wu, X.; Liu, R.; Zheng, T.; Zhang, D.; Chen, J.; Wu, G. RNAi knockdown of fatty acid elongase1 alters fatty acid composition in Brassica napus. Biochem. Biophys. Res. Commun. 2015, 466, 518–522. [Google Scholar] [CrossRef]
- Fang, H.; Fu, X.; Ge, H.; Zhang, A.; Shan, T.; Wang, Y.; Li, P.; Wang, B. Genetic basis of maize kernel oil-related traits revealed by high-density SNP markers in a recombinant inbred line population. BMC Plant Biol. 2021, 21, 344. [Google Scholar] [CrossRef]
- McAllister, C.H.; Beatty, P.H.; Good, A.G. Engineering nitrogen use efficient crop plants: The current status. Plant Biotechnol. J. 2012, 10, 1011–1025. [Google Scholar] [CrossRef]
- Forlani, G.; Bertazzini, M.; Cagnano, G. Stress-driven increase in proline levels, and not proline levels themselves, correlates with the ability to withstand excess salt in a group of 17 Italian rice genotypes. Plant Biol. 2019, 21, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Wycisk, K.; Kim, E.J.; Schroeder, J.I.; Krämer, U. Enhancing the first enzymatic step in the histidine biosynthesis pathway increases the free histidine pool and nickel tolerance in Arabidopsis thaliana. FEBS Lett. 2004, 578, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Zhang, G.; Li, H.; Ma, L.; Yang, N. Comparative transcriptomic analysis of Stenotrophomonas sp. MNB17 revealed mechanisms of manganese tolerance at different concentrations and the role of histidine biosynthesis in manganese removal. Ecotoxicol. Environ. Saf. 2022, 244, 114056. [Google Scholar] [CrossRef]
- Yamashita, H.; Sonobe, R.; Hirono, Y.; Morita, A.; Ikka, T. Dissection of hyperspectral reflectance to estimate nitrogen and chlorophyll contents in tea leaves based on machine learning algorithms. Sci. Rep. 2020, 10, 17360. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, M.; Shen, J.L.; Chen, D.; Zheng, Y.; Zhang, W. ZmOST1 mediates abscisic acid regulation of guard cell ion channels and drought stress responses. J. Integr. Plant Biol. 2019, 61, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Gou, T.Y.; Yang, L.; Hu, W.X.; Chen, X.H.; Zhu, Y.X.; Guo, J.; Gong, H.J. Silicon improves the growth of cucumber under excess nitrate stress by enhancing nitrogen assimilation and chlorophyll synthesis. Plant Physiol. Biochem. 2020, 152, 53–61. [Google Scholar] [CrossRef]
- Hui, Z.; Tian, F.X.; Wang, G.K.; Wang, G.P.; Wang, W. The antioxidative defense system is involved in the delayed senescence in a wheat mutant tasg1. Plant Cell Rep. 2012, 31, 1073–1084. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Year | CK | SR | N | SRN |
|---|---|---|---|---|
| 2019 | 3087 ± 24.5 d | 3991 ± 141.1 c | 5276 ± 50.7 b | 7926 ± 118.4 a |
| 2020 | 3542 ± 65.6 d | 3942 ± 169.7 c | 5615 ± 67.9 b | 8180 ± 167.6 a |
| Gene Name | Gene ID | |Log2FC| ≥ 2 | Annotation | ||
|---|---|---|---|---|---|
| SRvsCK | NvsCK | SRNvsCK | |||
| Potassium transport | |||||
| GRMZM2G477457 | 103635197 | −2.29 | −2.23 | HAK3 | |
| GRMZM2G425999 | 103653279 | −2.00 | −2.36 | HAK4 | |
| GRMZM5G838773 | 103636639 | −2.12 | Potassium channel KAT3 | ||
| GRMZM2G122584 | 103651360 | 2.36 | 2.82 | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | |
| Phosphate transport | |||||
| GRMZM2G154090 | 732716 | 1.06 | 2.54 | 1.03 | Pht2 |
| Nitrogen transport and assimilation | |||||
| GRMZM2G455124 | 103636218 | 1.83 | 2.88 | 1.61 | High-affinity nitrate transporter 2.3 |
| GRMZM2G161459 | 103637664 | 2.05 | NRT1/PTR family 6.3 | ||
| NR | 100383210 | 4.42 | 8.13 | 4.31 | Nitrate reductase |
| NR1 | 542278 | 2.96 | Nitrate reductase-like | ||
| NR3 | 109939975 | 2.83 | Nitrate reductase 3 | ||
| NIR | 542264 | 2.7 | Nitrite reductase 2 | ||
| GRMZM2G102959 | 103627781 | 3.89 | Ferredoxin—nitrite reductase | ||
| Gene Name | Gene ID | |Log2FC| ≥ 2 | Annotation | ||
|---|---|---|---|---|---|
| SRvsCK | NvsCK | SRNvsCK | |||
| Sugar metabolism | |||||
| AGPL2 | 103635182 | 2.93 | 2.4 | 2.74 | Glucose-1-phosphate adenylyltransferase large subunit 2 |
| GRMZM2G008263 | 100125638 | −2.08 | −1.47 | Granule-bound starch synthase II, a precursor | |
| GRMZM2G025833 | 100281141 | −3.13 | Beta-amylase | ||
| GRMZM2G086845 | 542107 | −1.68 | −2.29 | Fructokinase 1 | |
| GRMZM2G008247 | 732807 | −2.58 | Beta-D-glucosidase precursor | ||
| GRMZM2G016890 | 542414 | 2.68 | Beta glucosidase 1 | ||
| GBA1 | 103634742 | 2.85 | Beta-glucosidase 1 | ||
| GRMZM2G076946 | 100174972 | −4.84 | Dhurrinase-like B-glucosidase | ||
| GRMZM2G120962 | 103650330 | −2.51 | 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside beta-D-glucosidase 1 | ||
| GRMZM2G066162 | 103641803 | 3.41 | Endoglucanase 12-like | ||
| GRMZM6G738249 | 100273093 | −1.43 | −2.53 | Trehalose-phosphate phosphatase J | |
| GRMZM2G140078 | 107546758 | −2 | Trehalose-6-phosphate phosphatase 2 | ||
| GRMZM2G157675 | 100282631 | −2.08 | SWEET | ||
| GRMZM2G063824 | 100285023 | 4.9 | Carbohydrate transporter/sugar porter | ||
| Fatty acid metabolism | |||||
| GRMZM2G178014 | 100191431 | 1.21 | 2.37 | Alpha/beta-Hydrolases superfamily protein | |
| GRMZM2G022558 | 542018 | −3.7 | −4.54 | Fatty acid elongase 1 | |
| GRMZM2G160417 | 100382638 | −2.07 | Uncharacterized | ||
| Biosynthesis of amino acid | |||||
| GRMZM2G028535 | 100280719 | 1.93 | 3.16 | 2.82 | Delta 1-pyrroline-5-carboxylate synthetase 1 |
| GRMZM2G061777 | 103630430 | 1.89 | 1.5 | 2.08 | Delta-1-pyrroline-5-carboxylate synthase-like |
| GRMZM2G153536 | 100281255 | −2.57 | Branched-chain-amino-acid aminotransferase | ||
| GRMZM2G124963 | 100279149 | 2.14 | 2.34 | alanine aminotransferase 10 | |
| GRMZM2G053669 | 100192350 | 4.99 | Asparagine synthetase 3 | ||
| GRMZM2G078472 | 100192351 | 5.52 | Asparagine synthetase 4 | ||
| Rpi2 | 103626247 | 4.36 | 3.27 | 4.72 | Ribose-5-phosphate isomerase 2 |
| GRMZM2G013430 | 541959 | 2.22 | Serine acetyltransferase 2 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Liu, J.; Zhu, K.; Liu, S. Transcriptome Analysis of Maize Ear Leaves Treated with Long-Term Straw Return plus Nitrogen Fertilizer under the Wheat–Maize Rotation System. Plants 2023, 12, 3868. https://doi.org/10.3390/plants12223868
Li J, Liu J, Zhu K, Liu S. Transcriptome Analysis of Maize Ear Leaves Treated with Long-Term Straw Return plus Nitrogen Fertilizer under the Wheat–Maize Rotation System. Plants. 2023; 12(22):3868. https://doi.org/10.3390/plants12223868
Chicago/Turabian StyleLi, Jun, Jintao Liu, Kaili Zhu, and Shutang Liu. 2023. "Transcriptome Analysis of Maize Ear Leaves Treated with Long-Term Straw Return plus Nitrogen Fertilizer under the Wheat–Maize Rotation System" Plants 12, no. 22: 3868. https://doi.org/10.3390/plants12223868
APA StyleLi, J., Liu, J., Zhu, K., & Liu, S. (2023). Transcriptome Analysis of Maize Ear Leaves Treated with Long-Term Straw Return plus Nitrogen Fertilizer under the Wheat–Maize Rotation System. Plants, 12(22), 3868. https://doi.org/10.3390/plants12223868






