Abstract
Functional defects in key genes for chlorophyll synthesis usually cause abnormal chloroplast development, but the genetic regulatory network for these key genes in regulating chloroplast development is still unclear. Magnesium protoporphyrin IX methyltransferase (ChlM) is a key rate-limiting enzyme in the process of chlorophyll synthesis. Physiological analysis showed that the chlorophyll and carotenoid contents were significantly decreased in the chlm mutant. Transmission electron microscopy demonstrated that the chloroplasts of the chlm mutant were not well developed, with poor, loose, and indistinct thylakoid membranes. Hormone content analysis found that jasmonic acid, salicylic acid, and auxin accumulated in the mutant. A comparative transcriptome profiling identified 1534 differentially expressed genes (DEGs) between chlm and the wild type, including 876 up-regulated genes and 658 down-regulated genes. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that these DEGs were highly involved in chlorophyll metabolism, chloroplast development, and photosynthesis. Protein−protein interaction network analysis found that protein translation played an essential role in the ChlM gene-regulated process. Specifically, 62 and 6 DEGs were annotated to regulate chlorophyll and carotenoid metabolism, respectively; 278 DEGs were predicted to be involved in regulating chloroplast development; 59 DEGs were found to regulate hormone regulatory pathways; 192 DEGs were annotated to regulate signal pathways; and 49 DEGs were putatively identified as transcription factors. Dozens of these genes have been well studied and reported to play essential roles in chlorophyll accumulation or chloroplast development, providing direct evidence for the reliability of the role of the identified DEGs. These findings suggest that chlorophyll synthesis and chloroplast development are actively regulated by the ChlM gene. And it is suggested that hormones, signal pathways, and transcription regulation were all involved in these regulation processes. The accuracy of transcriptome data was validated by quantitative real-time PCR (qRT-PCR) analysis. This study reveals a complex genetic regulatory network of the ChlM gene regulating chlorophyll synthesis and chloroplast development. The ChlM gene’s role in retrograde signaling was discussed. Jasmonic acid, salicylic acid, or their derivatives in a certain unknown state were proposed as retrograde signaling molecules in one of the signaling pathways from the chloroplast to nucleus.
1. Introduction
Chlorophyll is widely found in photosynthetic organisms such as green plants, cyanobacteria, and algae [1]. It plays a central role in photosynthesis, forming complexes with thylakoid membrane proteins such as photosystem I, photosystem II, and cytochrome b6f complexes, which are important for regulating the photosynthetic physiology, yield, and quality in crops [2,3]. The biosynthesis of chlorophyll in higher plants is a complex process involving multiple enzymes, starting with glutamyl-tRNA and ending with the synthesis of chlorophyll b. This process consists of 16 steps, with more than 20 genes encoding 16 enzymes involved in the biosynthesis of chlorophyll [4].
In rice, 11 genes involved in chlorophyll biosynthesis have been cloned. RLIN1 (LOC_Os04g52130, HEMF1) encodes a coproporphyrinogen oxidative decarboxylase, which converts coproporphyrinogen III to protoporphyrinogen IX in the chlorophyll biosynthesis pathway, and the mutation of the RLIN1 gene causes a small distortion in the thylakoid in the chloroplast structure [5,6]. ChlD (LOC_Os03g59640), ChlH (LOC_Os03g20700), and ChlI (LOC_Os03g36540) are predicted to encode the D, H, and I subunits of Mg-chelatase, respectively, which are involved in the synthesis of Mg-protoporphyrin IX from protoporphyrin IX in the chlorophyll biosynthesis pathway. Mutants of these three genes all showed decreased chlorophyll contents and disrupted the thylakoid membranes of the chloroplast [7,8]. CRD1 (LOC_Os01g17170) encodes a monomethyl ester cyclase of magnesium protoporphyrin IX, converting Mg protoporphyrin IX monomethyl ester to form divinyl protochlorophyllide, and the mutation of this gene leads to chlorophyll synthesis deficiency and a lack of stacked grana thylakoids in the chloroplast [9]. PORA (LOC_Os04g58200) and PORB (LOC_Os10g35370) encode NADPH-protochlorophyllide oxidoreductases A and B, catalyzing the synthesis of chlorophyllide a from protochlorophyllide. OsPORB is required for the maintenance of light-dependent chlorophyll synthesis throughout leaf development, especially under high light levels, whereas OsPORA acts primarily in the early stages of leaf development; the OsPORB gene mutant showed severe leaf chlorosis accompanied by barely detectable thylakoid stacking and significantly increased plastoglobuli in the chloroplast [10]. ChlG (LOC_Os05g28200) encodes chlorophyll synthase, catalyzing the synthesis of chlorophyll a or b from chlorophyllide a or chlorophyllide b. The ChlG mutation results in a severe decline in chlorophyll and lacked grana membrane in the chloroplast [11]. Both CAO1/PGL (LOC_Os10g41780) and CAO2 (LOC_Os10g41760) encode chlorophyllide a oxygenase 1, converting chlorophyllide a into chlorophyllide b in the chlorophyll synthesis pathway, and the OsCAO1 mutants show a reduced chlorophyll content and disordered grana thylakoid of chloroplast [12,13]. It is clear from these studies that functional defects of key genes in chlorophyll synthesis also usually cause abnormal chloroplast development, but the downstream genetic networks for these key genes regulating chloroplast development are still unclear.
Magnesium protoporphyrin IX methyltransferase (ChlM) is a key rate-limiting enzyme in the process of chlorophyll synthesis. ChlM is synthesized in the cytoplasm and then transported to the chloroplast, where it is localized to the chloroplast envelope and thylakoid membrane, catalyzing the methyl transfer of S-adenosyl methionine to magnesium protoporphyrin IX (MgP) to form S-adenosyl homocysteine and magnesium protoporphyrin IX methyl ester (MgPME) in the chlorophyll synthesis pathway [14]. Two chlamydomonas reinhardtii mutants defective in the ChlM gene accumulated MgP and showed light-sensitive chlorophyll deficiency, and many photosynthesis-related chloroplast protein-encoding genes were regulated, such as LHCB, ChlH, ChlI, ChlD, CRD1, and HEMA [15]. In tobacco, ChlM has been shown to interact with the ChlH subunit of magnesium-chelatase to function and regulate the expression of chlorophyll-synthesis-related genes, such as HEMA, GSA, and ChlH [16]. Mutation in the ChlM gene in Arabidopsis resulted in the chlm mutant showing yellow–white leaves and accumulated MgP, and it also greatly changed the expression levels of some important chloroplast protein-encoding genes like LHCB, RBCS, and ChlH [17]. In addition, we previously cloned the rice ChlM gene using a yellow–green leaf mutant ygll8, and in vitro enzyme activity assays demonstrated that the YGL18 protein exhibited ChlM enzyme activity, but the mutant ygl18 protein retained only very little ChlM activity [18]. Accordingly, the substrate MgP accumulated in large amounts in ygl18 leaves, while the product MgPME greatly reduced. YGL18 protein is required for light-dependent and photoperiod-regulated chlorophyll synthesis [18]. Taken together, mutants of the ChlM gene in various plant species exhibited severe chlorophyll deficiency and significant expression alteration of many important chloroplast protein-encoding genes. The chlm mutant might be an ideal material to study the genetic regulation network critically linking chlorophyll synthesis and chloroplast development.
In this study, we analyzed the changes in pigment content, chloroplast structure, and hormone content in the rice chlm mutant and then used transcriptomic data to reveal the gene expression network of the ChlM gene regulating chlorophyll synthesis and chloroplast development. Genes related to pigment metabolism, chloroplast development, hormone pathways, signal pathways, and transcriptional regulation were found to play important roles in this regulation process. This study reveals a complex genetic regulatory network linking chlorophyll synthesis and chloroplast development in rice.
2. Results
2.1. Alteration of Pigment Contents and Chloroplast Structure in the Chlm Mutant
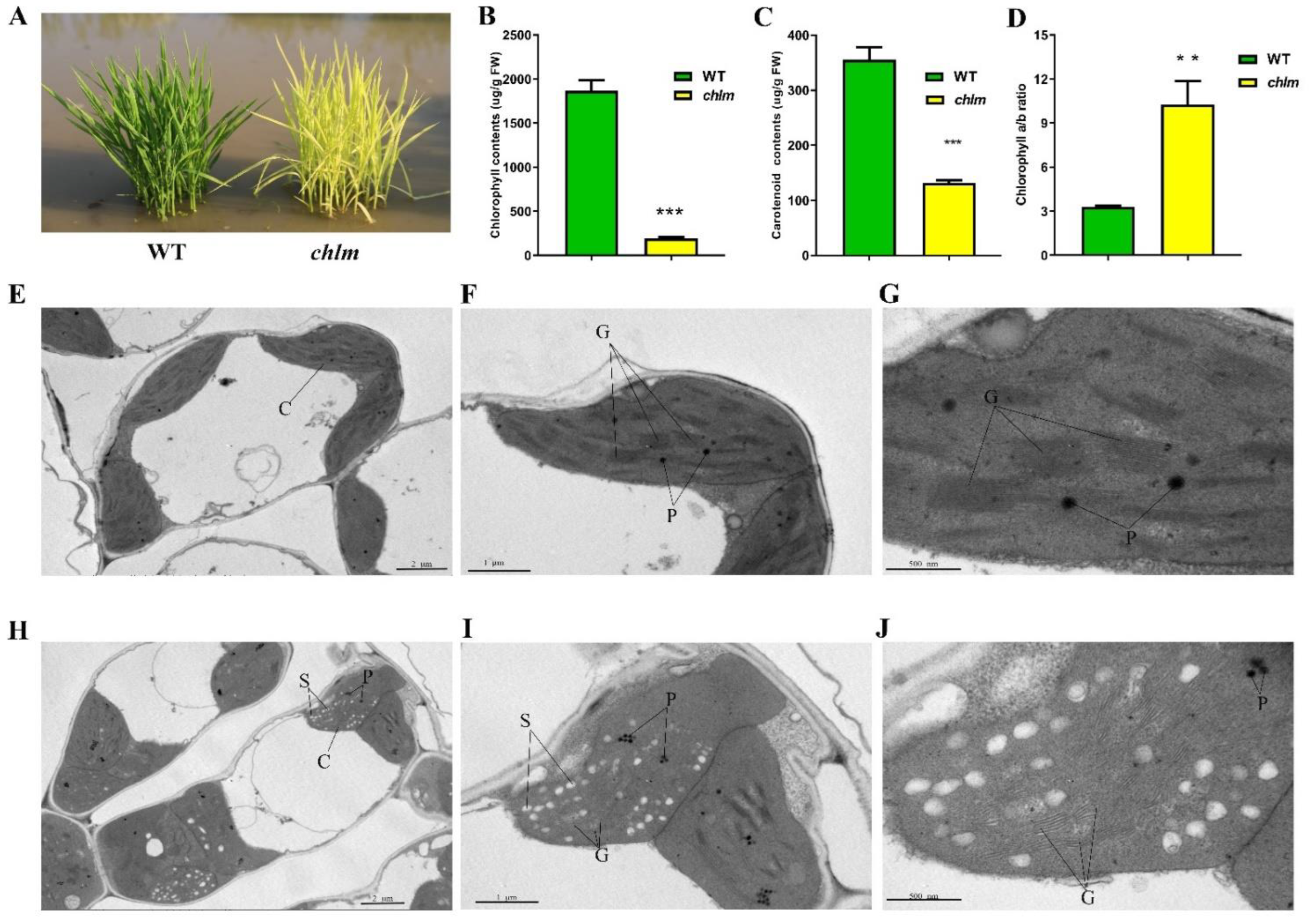
The ChlM gene mutant chlm was isolated from a photo-thermosensitive genic male sterile rice cultivar ‘guangzhan63S’ (Oryza sativa L. subsp. indica) by our previous study [18]. At the seedling stage, the mutant chlm could be easily distinguished by the yellow–green leaf trait (Figure 1A). Thus, the physiological changes of the chlm mutant were investigated. Compared with that of the wild type, the chlorophyll and carotenoid contents in the chlm mutant were significantly reduced (Figure 1B,C), and the ratio of chlorophyll a to chlorophyll b was significantly increased (Figure 1D), which was possibly due to the chlorophyll b synthesis suffering a more severe suppression than chlorophyll a. These results all indicated the impairment of pigment synthesis in the chlm mutant.
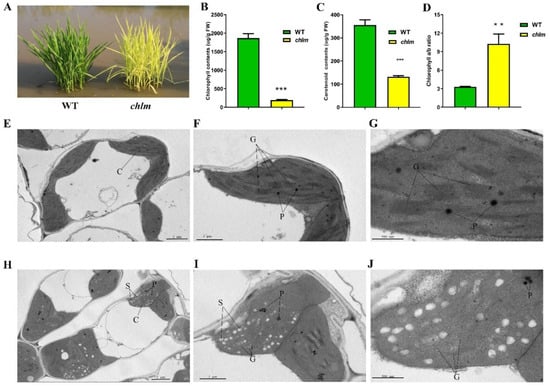
Figure 1.
Phenotypic, physiological and cytological changes between wild-type (WT) and chlm mutant at the seedling stage. (A) Phenotype of wild-type and chlm mutant plants. (B) The chlorophyll contents of wild-type and chlm mutant leaves. (C) The carotenoid contents of wild-type and chlm mutant leaves. (D) The ratio of chlorophyll a to chlorophyll b. In (B–D), all data represent the mean ± SD of three biological replicates, and the asterisk indicates the statistically significant difference between chlm and wild type (** p < 0.005, *** p < 0.0005, Student’s t-test). (E–G) Chloroplast ultrastructure showing typical structures and distinct thylakoid membranes in wild-type leaves. (H–J) Abnormal chloroplast ultrastructure in the chlm mutant leaves. Bars = 2 μm (E,H), 1 μm (F,I), 500 nm (G,J). C, chloroplast. P, plastoglobuli. S, starch granule. G, grana stacks.
To determine whether the ChlM gene mutation affected chloroplast development, chloroplast structure changes of the chlm mutant leaf were additionally observed at the cytological level by using a transmission electron microscope. In the leaves of the wild type, the chloroplast showed the typical structure of well-developed and distinct thylakoid membranes with normal stacked grana (Figure 1E–G). In contrast, ultrastructural analysis of chloroplasts in the chlm mutant leaves revealed poor, loose and indistinct thylakoid membranes and increased osmiophilic plastoglobuli (Figure 1H–J). These results indicated that chloroplast development was significantly affected in the chlm mutant.
2.2. Hormone Changes in the Chlm Mutant
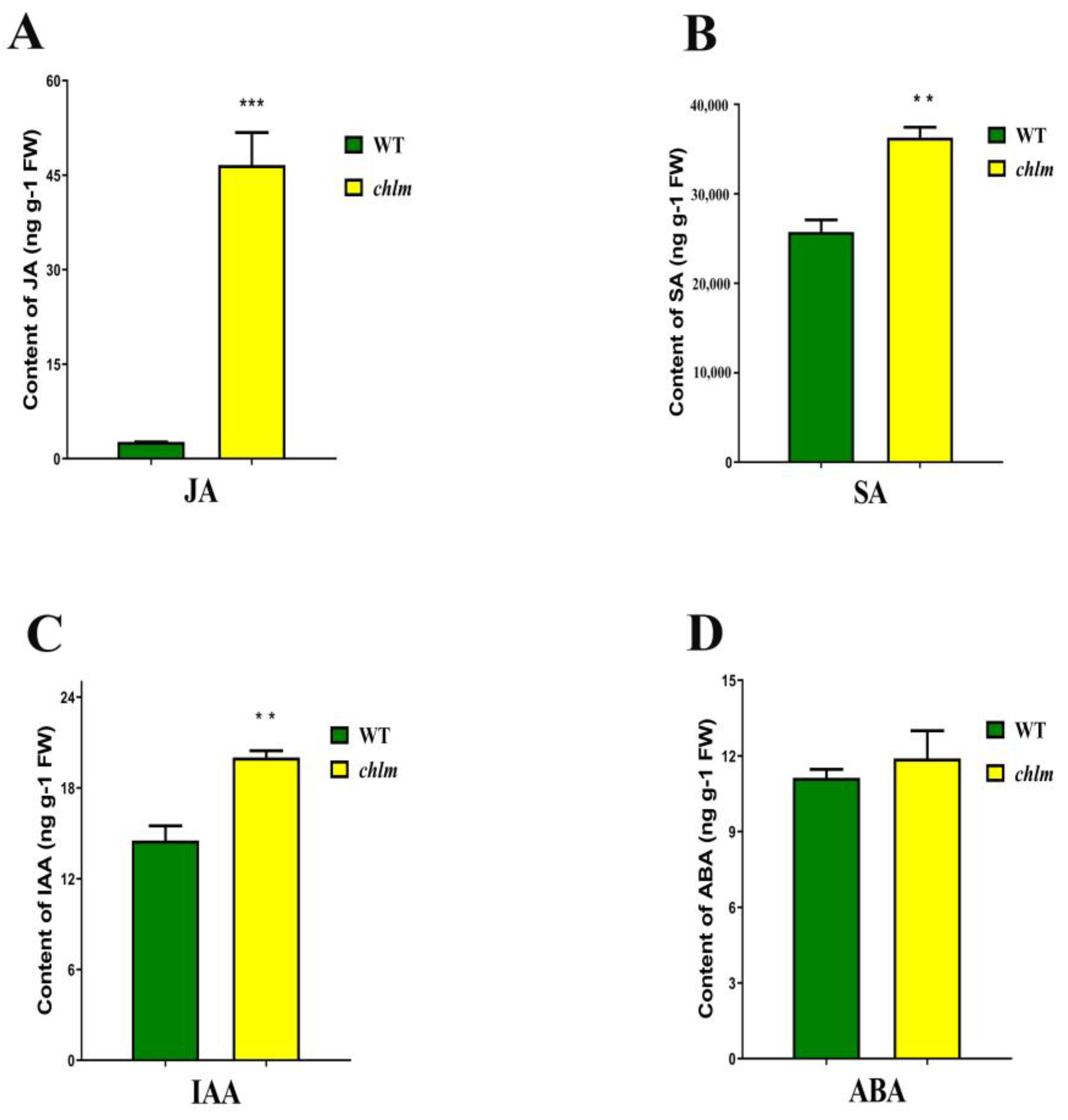
In parallel with the evident chloroplast structure changes in the chlm mutant, the contents of four phytohormones related with chloroplast were measured, including jasmonic acid (JA), salicylic acid (SA), indole acetic acid (IAA) and abscisic acid (ABA). Compared with that of the wild type, the contents of JA, SA and IAA were significantly increased in the chlm mutant, especially the highest increase for JA (Figure 2A–C). Meanwhile, the content of ABA showed no difference between the wild type and the chlm mutant (Figure 2D). The increased JA, SA and IAA may function in the biological process regulated by the ChlM gene.
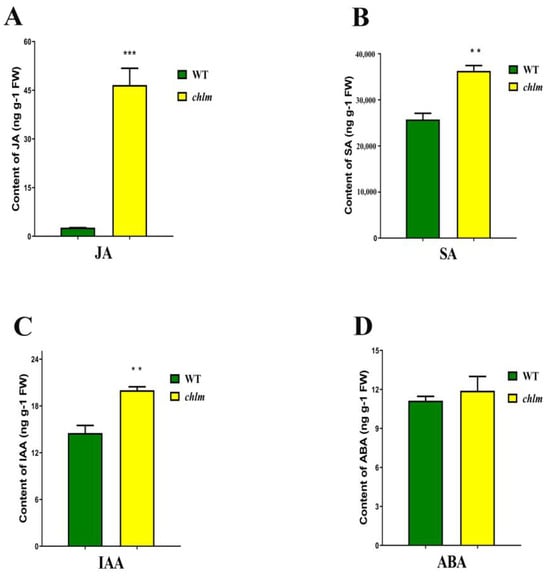
Figure 2.
Changes of hormone contents between wild-type and chlm mutant leaves at the seedling stage. (A) The content of jasmonic acid (JA). (B) The content of salicylic acid (SA). (C) The content of indoleacetic acid (IAA). (D) The content of abscisic acid (ABA). All data represent the mean ± SD of three biological replicates, and the asterisk indicates the statistically significant difference between chlm and the wild type (** p < 0.005, *** p < 0.0005, Student’s t-test).
2.3. Transcriptomic Alterations in Chlm Mutant
Considering the significant changes of phenotypic, physiological and cytological characteristics of the chlm mutant in this study, it was worth investigating the transcriptional regulation network of the ChlM gene. Thus, a transcriptomic comparison between the yellow–green leaves of the chlm mutant with the normal green leaves of the wild type was carried out by RNA sequencing of six libraries with three biological replicates each. A total of 40.10 Gb clean reads (accounting for 92.19% of the raw reads) were obtained by stringent quality check and data cleanup, and the clean reads of each sample were more than 6 Gb. In detail, the total clean reads in each library mapped to the Oryza sativa reference genome ranged 86.72–87.15%, and the unique reads mapped to the rice reference genome ranged 82.61–83.93% (Table 1). In addition, the GC contents of the sequencing outputs were 54.56–55.14%, and the Q30 percentage of clean reads was greater than 85% (Table 1). Accordingly, the throughput and quality of RNA sequencing were high enough to ensure further analysis.

Table 1.
Characteristics of RNA-sequencing data in all six samples.
2.4. Analysis of Differentially Expressed Genes (DEGs)
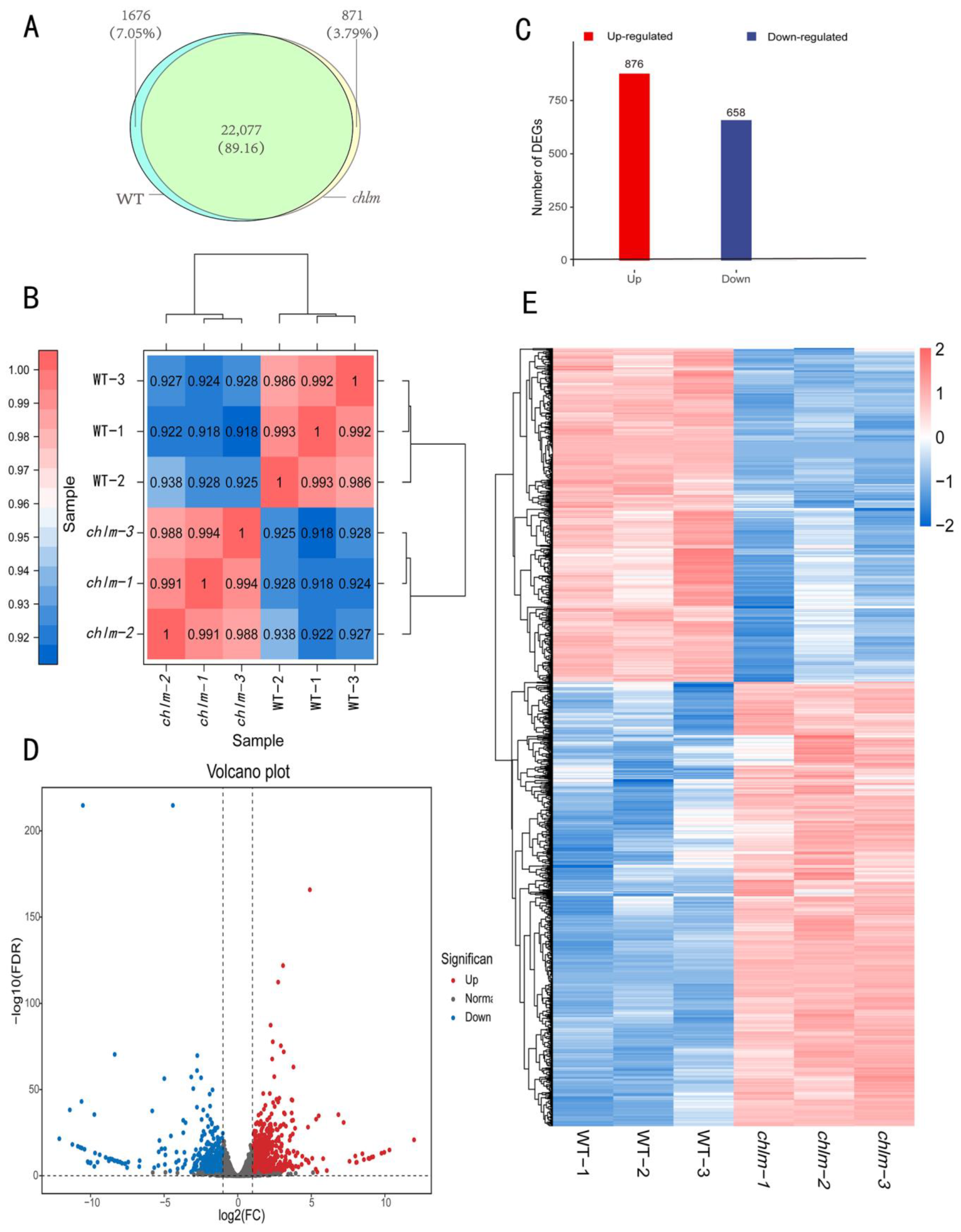
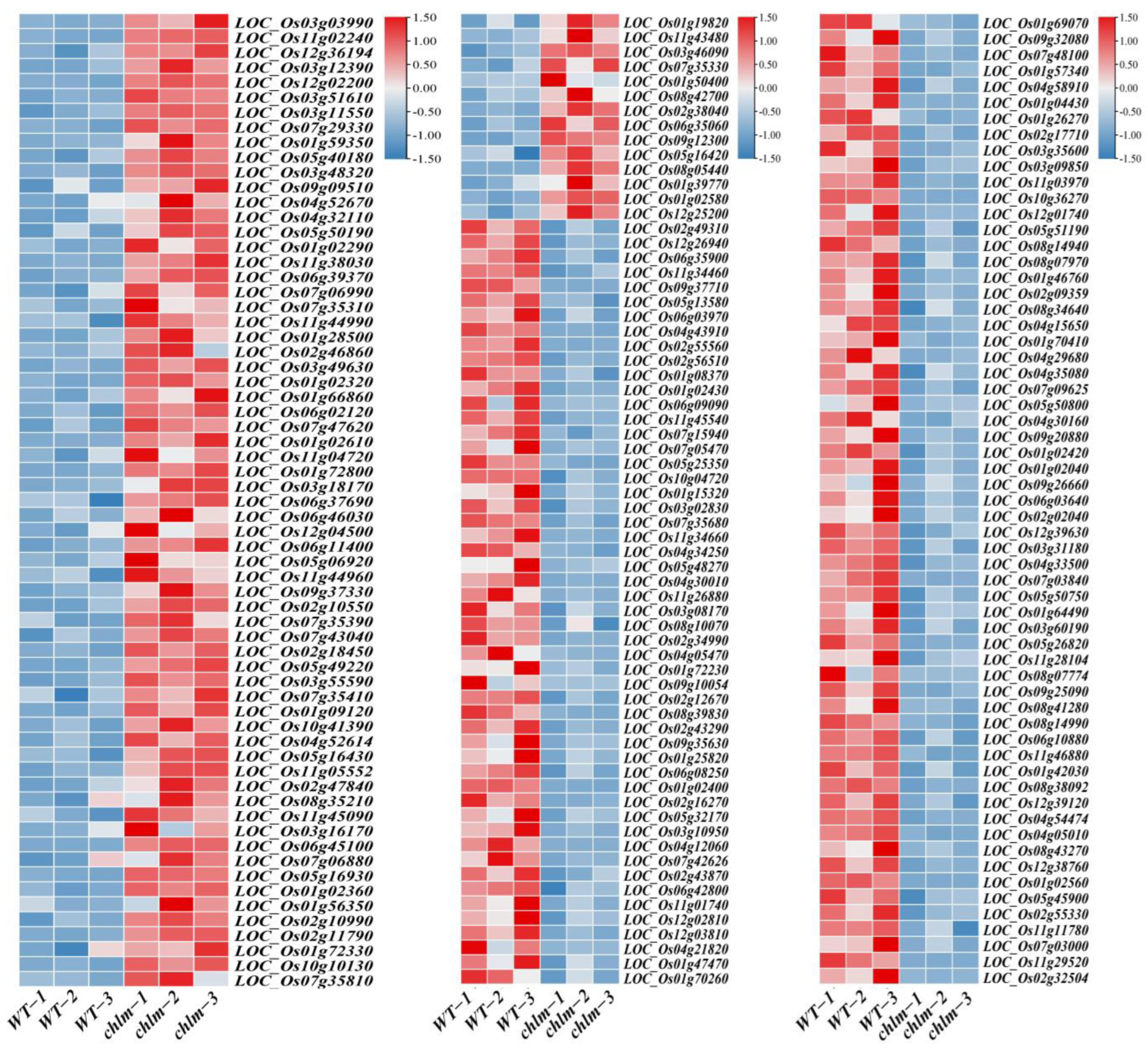
After clean reads were mapped to the Oryza sativa reference genome, the mapped transcripts were assembled and annotated using StringTie software (1.3.4d). Furthermore, FPKM (Fragments Per Kilobase of transcript per Million fragments mapped) methods were used to analyze gene expression patterns in both types of libraries. When comparing the two types of libraries with respect to the FPKM calculation, 23,753 (wild-type) and 22,948 (chlm) genes were identified in the cDNA libraries; meanwhile, 22,077 genes were simultaneously expressed in wild-type and chlm mutant leaves (Figure 3A). Prior to identifying DEGs, the correlation of biological replicates was evaluated. The result displayed a great repeatability (Figure 3B) and thus could be provided for further analysis. According to the screening criteria of false discovery rate (FDR) < 0.01 and fold change ≥ 2, a total of 1534 DEGs were obtained between chlm and the wild type (Table S1). Among them, 876 genes were up-expressed, while 658 genes were down-expressed (Figure 3C). In addition, the volcano plot and hierarchical clustering analysis of DEGs directly showed the differences of gene expression level between the chlm mutant and the wild type (Figure 3D,E).
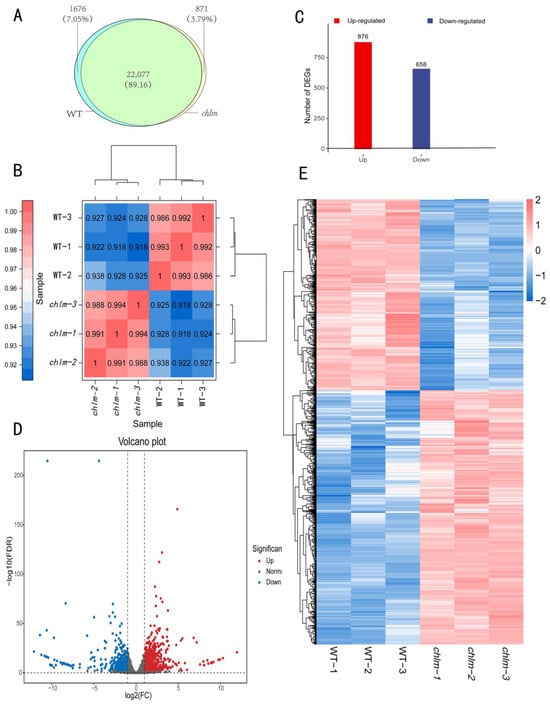
Figure 3.
The identification and analysis of differentially expressed genes (DEGs). (A) Number of genes identified in wild type (WT) and the chlm mutant. (B) The correlation of biological replicates evaluated by Pearson’s correlation coefficient. (C) The number of up- and down-regulated genes. (D) The volcano plot of DEGs. (E) The hierarchical clustering of DEGs.
2.5. Functional Analysis of DEGs
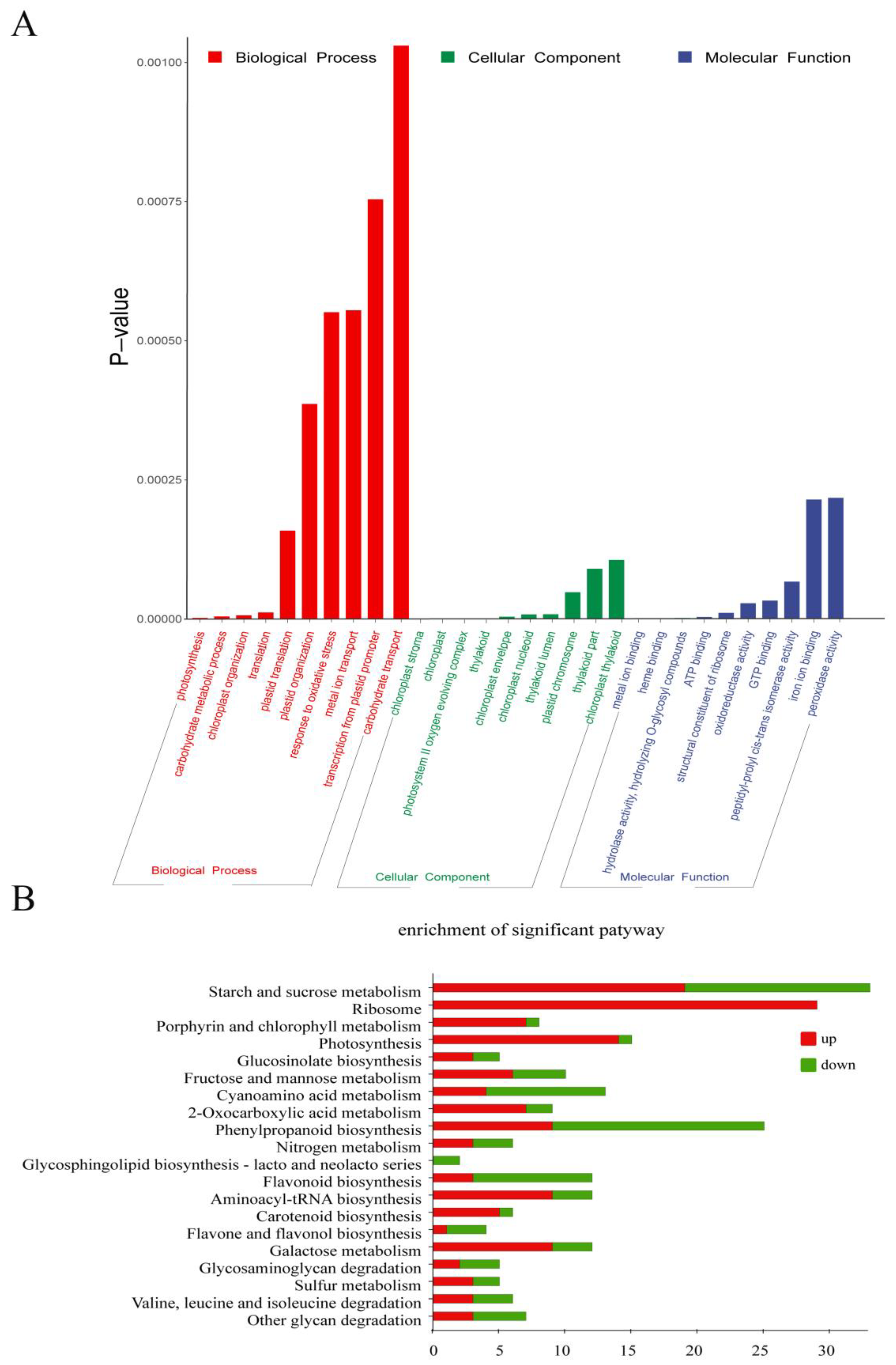
To investigate the potential functions of DEGs involved in the biological process regulated by the ChlM gene, functional enrichment analysis on 1534 DEGs was performed. Gene Ontology (GO) enrichment analysis results showed that a total of 257 GO terms were significantly enriched (p-value < 0.05), among which 112, 39 and 106 were classified under biological process, cellular component, and molecular function, respectively (Table S2). Moreover, a total of 30 significantly top terms were additionally listed (Figure 4A). In biological process terms, ‘photosynthesis (GO:0015979)’, ‘carbohydrate metabolic process (GO:0005975)’, ‘chloroplast organization (GO:0009658)’, ‘translation (GO:0006412)’, ‘plastid translation (GO:0032544)’, ‘plastid organization (GO:0009657)’, ‘response to oxidative stress (GO:0006979)’, ‘metal ion transport (GO:0030001)’, ‘transcription from plastid promoter (GO:0042793)’, and ‘carbohydrate transport (GO:0008643)’ were significantly enriched. In cellular component terms, DEGs were mainly enriched in ‘chloroplast stroma (GO:0009570)’, ‘chloroplast (GO:0009507)’, ‘photosystem II oxygen evolving complex (GO:0009654)’, ‘thylakoid (GO:0009579)’, ‘chloroplast envelope (GO:0009941)’, ‘chloroplast nucleoid (GO:0042644)’, ‘thylakoid lumen (GO:0031977)’, ‘plastid chromosome (GO:0009508)’, ‘thylakoid part (GO:0044436)’, and ‘chloroplast thylakoid (GO:0009534)’, which met the expectation. And in molecular function terms, DEGs were significantly enriched in ‘metal ion binding (GO:0046872)’, ‘heme binding (GO:0020037)’, ‘hydrolase activity, hydrolyzing O-glycosyl compounds (GO:0004553)’, ‘ATP binding (GO:0005524)’, ‘structural constituent of ribosome (GO:0003735)’, ‘oxidoreductase activity (GO:0016491)’, ‘GTP binding (GO:0005525)’, ‘peptidyl-prolyl cis-trans isomerase activity (GO:0003755)’, ‘iron ion binding (GO:0005506)’, and ‘peroxidase activity (GO:0004601)’. Obviously, these GO terms are highly related to chlorophyll metabolism and chloroplast development.
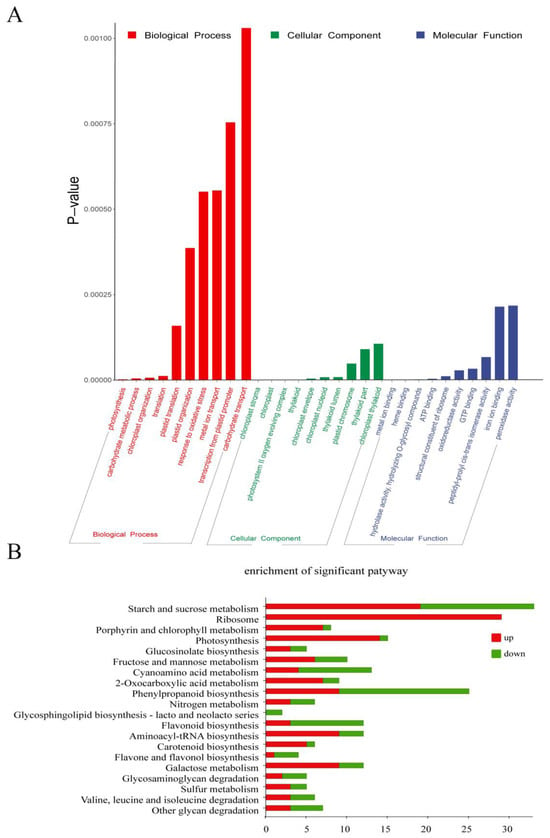
Figure 4.
Functional enrichment analysis of DEGs between the wild type (WT) and the chlm mutant. (A) Gene Ontology (GO) enrichment analysis of DEGs. The 30 most enriched GO terms in three categories are shown. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of DEGs. The top 20 most significant pathways are shown.
Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed to further explore the metabolic pathways of DEGs involved in a ChlM-regulated biological process. At last, a total of 20 pathways were significantly enriched with a p-value < 0.05, in which ‘starch and sucrose metabolism (ko00500)’, ‘ribosome (ko03010)’, ‘porphyrin and chlorophyll metabolism (ko00860)’, ‘photosynthesis (ko00195)’, ‘glucosinolate biosynthesis (ko00966)’, and ‘fructose and mannose metabolism (ko00051)’ were the top six pathways (Figure 4B, Table S2). Apparently, these pathways are also highly related with chlorophyll metabolism and photosynthesis.
2.6. Functional Interaction Network of DEGs
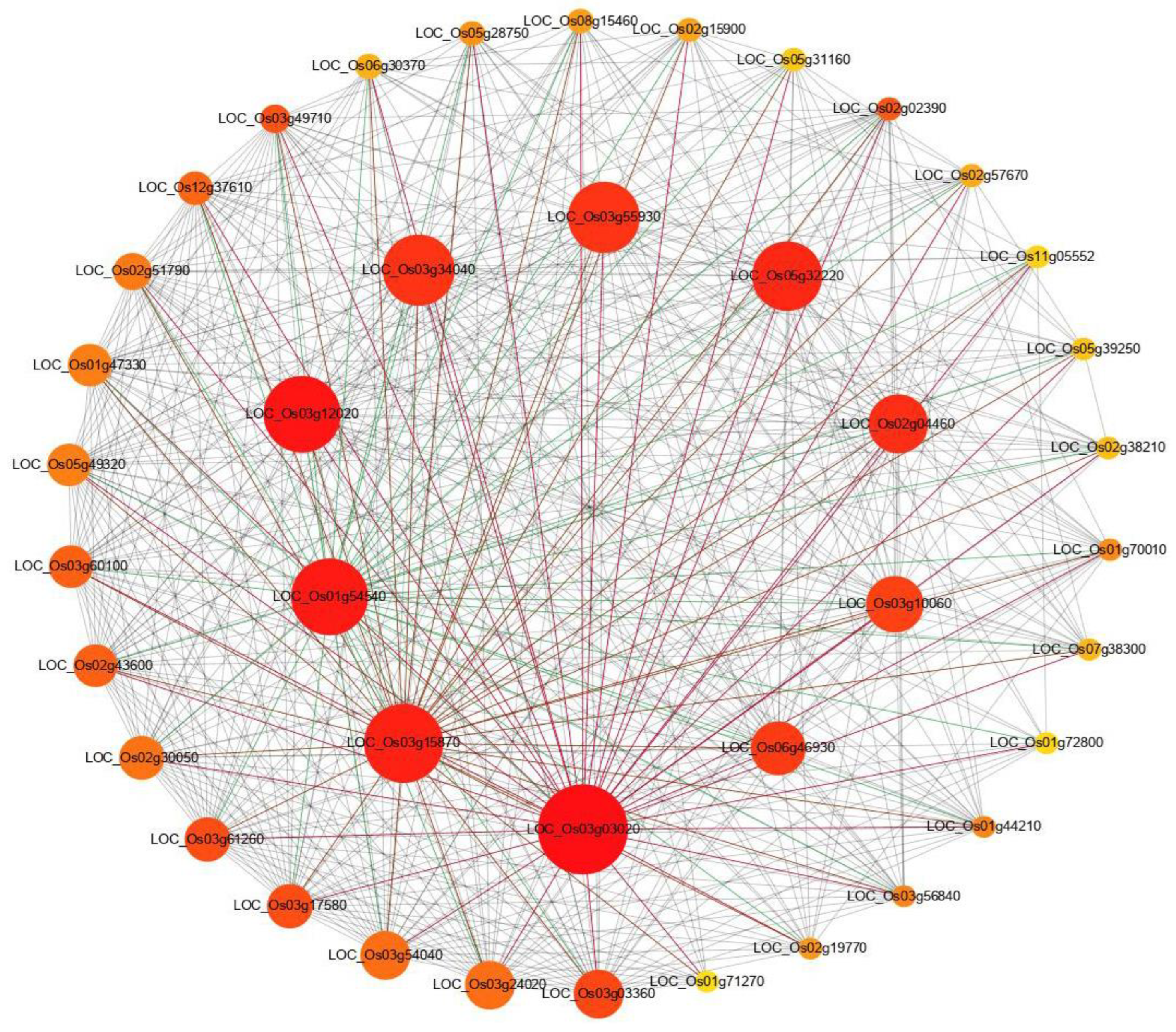
To reveal the functional interaction network of DEGs, the protein–protein interaction (PPI) analysis was conducted using all identified DEGs between the wild type and the chlm mutant. Of the predicted PPI network, highly connected nodes are central to a network’s architecture and function. The top 40 most significant proteins with the highest connectivity were extracted from the predicted network. The result showed that higher interactions were found for proteins encoded by LOC_Os03g03020 (50S ribosomal protein L11), LOC_Os03g15870 (50S ribosomal protein L4), LOC_Os01g54540 (50S ribosomal protein L13), LOC_Os03g12020 (50S ribosomal protein L15), LOC_Os03g34040 (30S ribosomal protein S5), LOC_Os03g55930 (30S ribosomal protein S9), LOC_Os05g32220 (50S ribosomal protein L1), LOC_Os02g04460 (50S ribosomal protein L3), LOC_Os03g10060 (30S ribosomal protein S10), and LOC_Os06g46930 (50S ribosomal protein L24) (Figure 5, Table S3), all being ribosomal proteins for translation. The remaining 30 proteins were also related with ribosome or translation (Table S3). And 39 of these 40 protein-encoding genes were up-regulated in the chlm mutant (Table S3). The above results implied that the translation process played an essential role in chlorophyll synthesis and chloroplast development regulated by the ChlM gene.
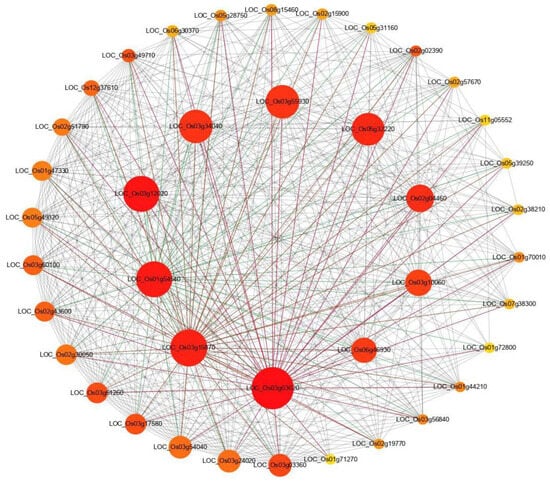
Figure 5.
Protein–protein interaction network analysis of DEGs. STRING V9.1 software was used to predict the protein to protein network of differentially expressed proteins. Differentially accumulated proteins are represented by a node, whereas the different color of lines represents evidence for the predicted functional relationship. The strong interaction is indicated by a redder color. The proteins outside the circle showed weaker interaction.
2.7. Genes Regulating Chlorophyll and Carotenoid Metabolism Were Identified in the Chlm Mutant
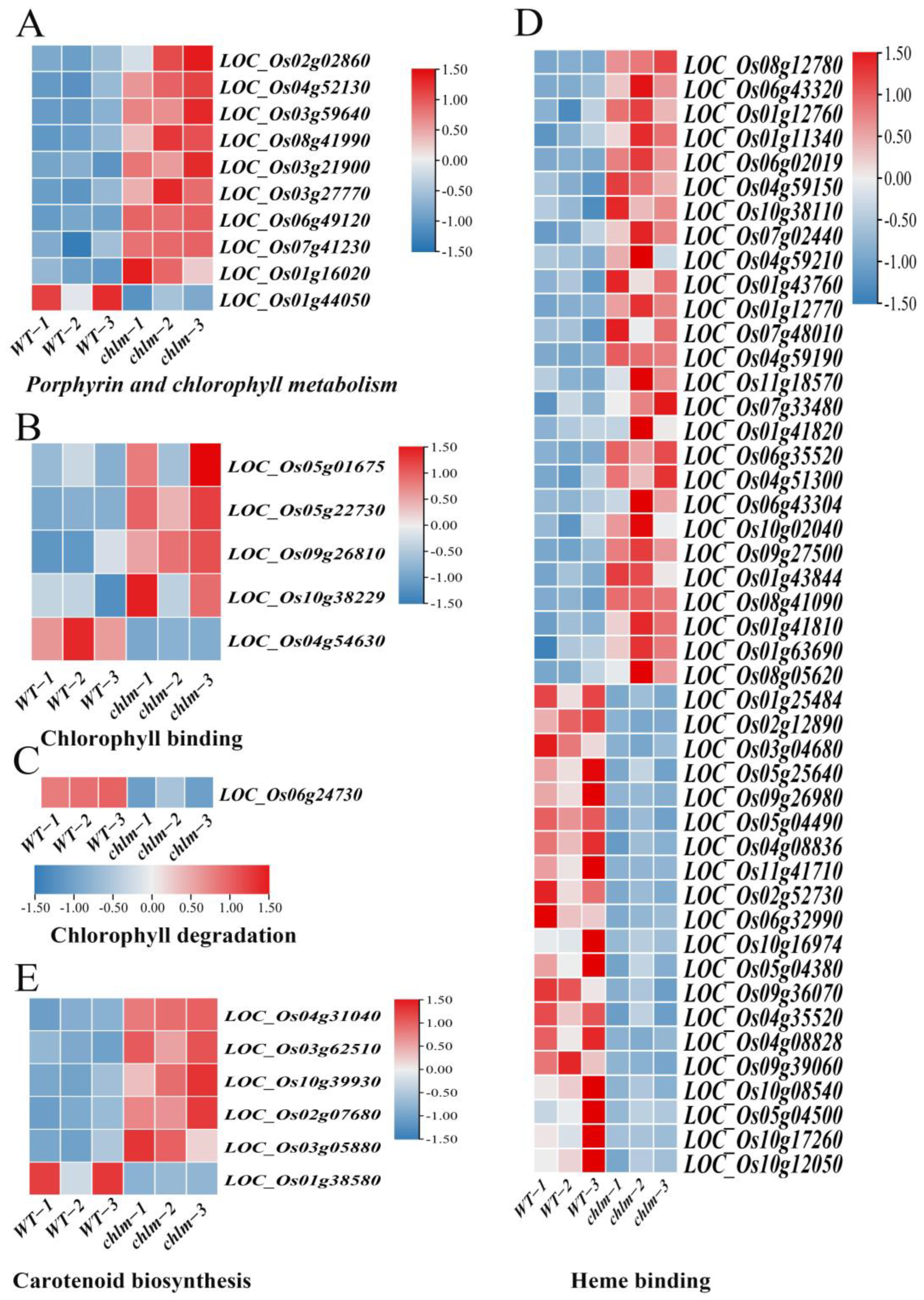
Chlorophyll is the main component of the photosynthetic pigments, which performs important functions in photosynthesis by harvesting light energy and converting it into chemical energy. Chlorophyll synthesis and heme synthesis are two branches in the tetrapyrrole biosynthesis pathway, Mg chelating with protoporphyrin IX to step into the chlorophyll synthesis branch and Fe chelating with protoporphyrin IX to step into the heme synthesis branch [1]. In this study, a total of 62 chlorophyll-related DEGs were identified based on functional annotation, including 39 up-regulated genes and 23 down-regulated genes. These DEGs were assigned to relevant functional terms: specifically, 10 genes involved in porphyrin and chlorophyll metabolism (Figure 6A, Table S4), five genes involved in chlorophyll binding (Figure 6B, Table S4), one gene involved in chlorophyll degradation (Figure 6C, Table S4), and 46 genes involved in heme binding (Figure 6D, Table S4). Among these DEGs, some were cloned and well-studied. The up-expressed genes LOC_Os02g02860 (OsGluRS) encoding a glutamyl tRNA synthetase, LOC_Os04g52130 (RLIN1, LLM1) encoding a putative coproporphyrinogen III oxidase, and LOC_ Os03g59640 (OsChlD) encoding the ChlD subunit of Mg-protoporphyrin IX chelatase were well-studied key enzymes for chlorophyll biosynthesis (Table 2). The down-expressed gene LOC_Os06g24730 (NYC3) encoding an α/β fold hydrolase family protein played an important role in chlorophyll degradation (Table 2). And the up-expressed gene LOC_Os03g27770 (OsHO2) encoding the heme oxygenase was reported to play a key role in heme biosynthesis (Table 2). These results suggested that the chlorophyll metabolism pathway is regulated by the ChlM gene.
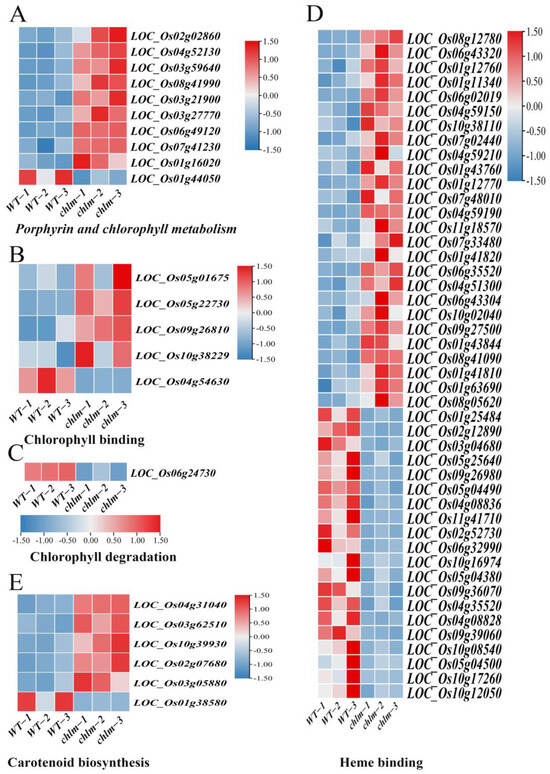
Figure 6.
Expression patterns of DEGs regulating chlorophyll and carotenoid metabolism pathways in the chlm mutant. (A) DEGs involved in porphyrin and chlorophyll metabolism. (B) DEGs involved in chlorophyll binding. (C) DEGs involved in chlorophyll degradation. (D) DEGs involved in heme binding. (E) DEGs involved in carotenoid biosynthesis.

Table 2.
The well-studied functionally validated genes between WT and the chlm mutant.
Furthermore, carotenoids are a diverse group of colorful pigments naturally found in plants, which play essential roles in photosynthesis and plant development [53]. In this study, a total of six carotenoid biosynthesis-related DEGs were identified based on functional annotation, including five up-regulated genes and one down-regulated gene (Figure 6E, Table S4). These results implied that the carotenoid biosynthesis pathway is regulated by the ChlM gene.
These results implied that the information for the decreased content of chlorophyll and carotenoids in the chlm mutant may be perceived by plant cells, which further regulate the expression level of these pigments’ related biosynthetic and metabolic genes, trying to compensate for the reduction in chlorophyll and carotenoids due to the impaired function of the ChlM gene in the chlm mutant.
2.8. Genes Regulating Chloroplast Development Were Identified in the Chlm Mutant
Chloroplasts are the central nodes of the metabolic network in the photosynthetic cells of higher plants, which performs indispensable functions in photosynthesis and other metabolic processes, such as the synthesis of lipid, terpenoids, tetrapyrroles, amino acids and hormones [54]. Many gene mutants in the chlorophyll synthesis pathway have showed defects in chloroplast structure or development. Since the chlm mutant also showed weakened chloroplast development except for the decreased chlorophyll contents, it would be interesting to investigate the expression profile of genes related with chloroplast.
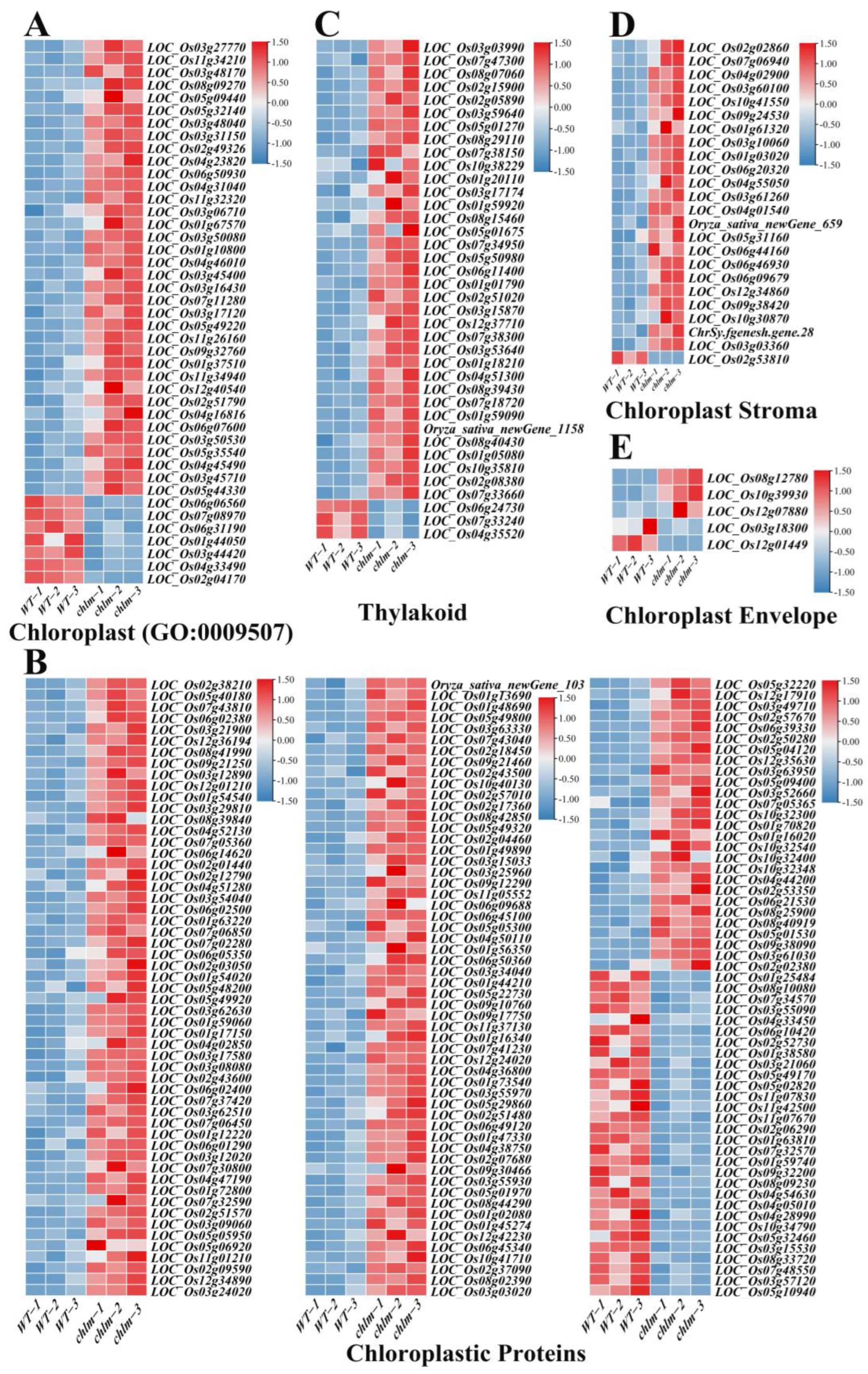
In this study, 43 genes were functionally annotated to be related with cellular component chloroplast (GO:0009507) (Figure 7A, Table S5). And 168 genes were functionally annotated to encode chloroplastic proteins (Figure 7B, Table S5). Specifically, 38, 24 and 5 genes were functionally annotated to be related to the thylakoid, chloroplast stroma, and chloroplast envelope, respectively (Figure 7C–E, Table S5). Obviously, of these 278 chloroplast-related genes, the vast majority (235) were up-regulated in the chlm mutant (Table S5), suggesting the positive response of these genes in chloroplast development. Through searching these 278 chloroplast-related genes on ‘China Rice Data Center’, 26 well-studied genes were reported to be involved in chlorophyll metabolism and chloroplast development (Table 2), including WSP1 (LOC_Os04g51280), OsTrxZ/wp2 (LOC_Os08g29110), WLP2 (LOC_Os01g63220), OsPPR6 (LOC_Os05g49920), CDE4 (LOC_Os08g09270), etl1 (LOC_Os11g01210), etl2 (LOC_Os12g01210), WSL3 (LOC_Os10g32540), OsNUS1 (LOC_Os03g45400), WLP1 (LOC_Os01g54540), ASL2 (LOC_Os02g15900), OsValRS2 (LOC_Os07g06940), ObgC (LOC_Os07g47300), EF-Tu (LOC_Os02g38210), YL1 (LOC_Os02g05890), OscpSRP43 (LOC_Os03g03990), YGL138(t) (LOC_Os11g05552), AL1 (LOC_Os03g31150), OsNOA1 (LOC_Os02g01440), VYL (LOC_Os03g29810), OsFdC1 (LOC_Os03g45710), OsFdC2 (LOC_Os03g48040), OsSTN8 (LOC_Os05g40180), RNRS1 (LOC_Os06g14620), YLC1 (LOC_Os09g21250), and WSL12 (LOC_Os12g36194) (Table 2). These well-studied genes provided direct evidence for the reliability of the role of the identified chloroplast-related genes on chloroplast development. It is speculated that the abnormal chloroplast development of the chlm mutant could be supervised and regulated by plant cells and thus induced the expression changes of chloroplast-related genes, trying to remedy the poor chloroplast state caused by the extreme impaired function of the ChlM gene.
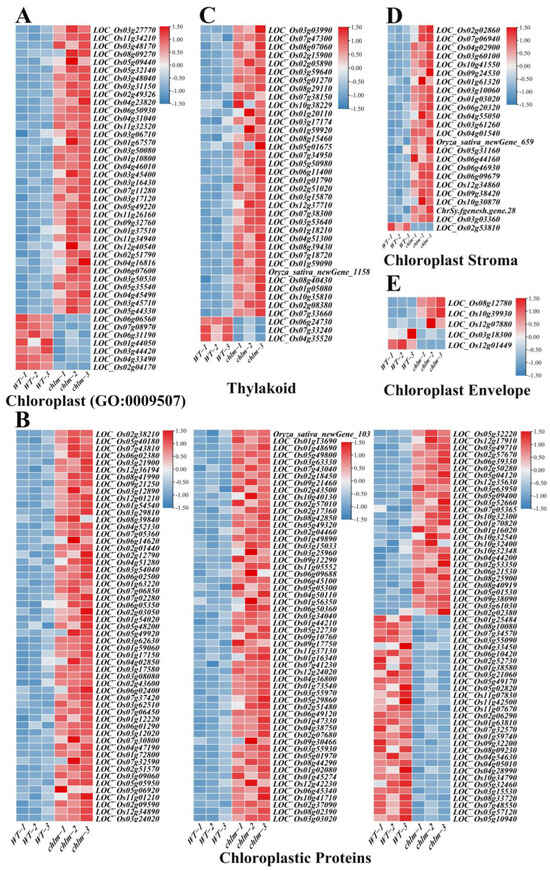
Figure 7.
Expression patterns of DEGs regulating chloroplast development in the chlm mutant. (A) DEGs referring to cellular component chloroplast (GO:0009507). (B) DEGs referring to chloroplastic proteins. (C) DEGs related with thylakoid. (D) DEGs related with chloroplast stroma. (E) DEGs related with chloroplast envelope.
2.9. Genes Regulating Hormone Regulatory Pathways Were Identified in the Chlm Mutant
Phytohormones were reported to perform important functions in regulating chloroplast biogenesis and development [55,56]. Due to the changes of hormone contents in the chlm leaves, expression alterations of hormone-related genes were investigated accordingly.
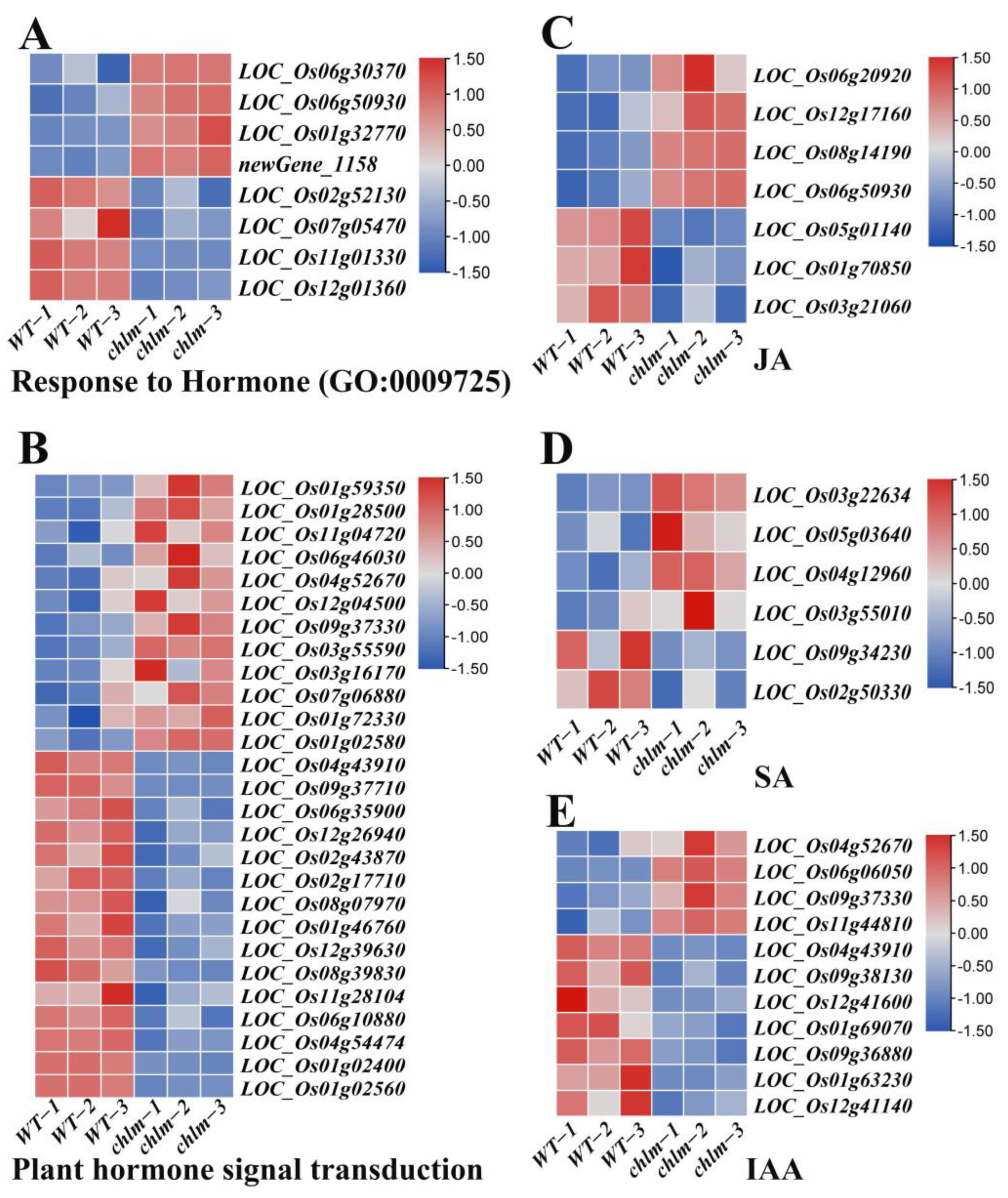
In this study, a total of 59 hormone-related DEGs were identified based on functional annotation, including 28 up-regulated genes and 31 down-regulated genes (Table S6). Of these 59 DEGs, eight genes were annotated to respond to hormone (GO:0009725) (Figure 8A, Table S6), and 27 genes were found to be involved in plant hormone signal transduction (ko04075) (Figure 8B, Table S6). Importantly, seven genes were annotated to be associated with JA metabolism or response, in which LOC_Os06g20920, LOC_Os12g17160, LOC_Os08g14190, LOC_Os05g01140, and LOC_Os01g70850 were involved in JA metabolism, while LOC_Os06g50930 and LOC_Os03g21060 were involved in JA response (Figure 8C, Table S6). Six genes were annotated to be associated with SA metabolism or response, in which LOC_Os03g22634, LOC_Os05g03640, LOC_Os04g12960, LOC_Os03g55010, and LOC_Os09g34230 were involved in SA metabolism, while LOC_Os02g50330 was involved in SA response (Figure 8D, Table S6). And 11 genes were annotated to be associated with IAA response or transport, in which LOC_Os04g52670, LOC_Os09g37330, LOC_Os11g44810, LOC_Os04g43910, LOC_Os12g41600, LOC_Os01g69070, and LOC_Os01g63230 were involved in IAA response, while LOC_Os06g06050, LOC_Os09g38130, LOC_Os09g36880, and LOC_Os12g41140 were involved in IAA transport (Figure 8E, Table S6). Among these genes, two genes LOC_Os06g20920 and LOC_Os05g01140 were identified and cloned as the OsJMT gene involving in JA metabolism (Table 2).
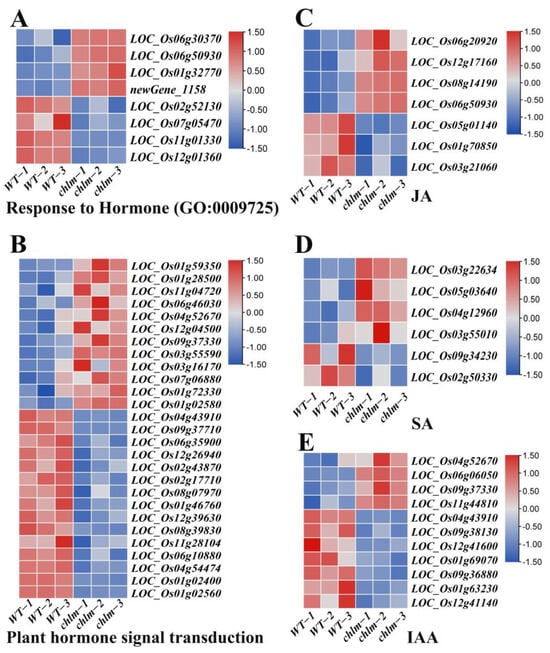
Figure 8.
Expression patterns of DEGs involved in hormone pathways in the chlm mutant. (A) DEGs involved in response to hormone. (B) DEGs involved in plant hormone signal transduction pathways. (C) DEGs involved in JA metabolism or response. (D) DEGs involved in SA metabolism or response. (E) DEGs involved in IAA response or transport.
These results showed that the expression of hormone-related genes was regulated in the chlm mutant, implying that hormones might play important roles in pigment metabolism and chloroplast development regulated by the ChlM gene.
2.10. Genes Regulating Signal Pathways Were Identified in the Chlm Mutant
Signal transmission and transduction play an essential role in plant development [57]. It would be interesting to investigate the signal-related genes participating in the biological process regulated by the ChlM gene.
In this study, a total of 192 genes were annotated to be involved in signal recognition or transduction pathways, in which 79 genes displayed up-expression while 113 genes showed down-expression (Figure 9, Table S7). According to the search results from the China Rice Data Center, four signal-related genes including LOC_Os03g03990 (OscpSRP43), LOC_Os11g05552 (YGL138(t)), LOC_Os05g40180 (OsSTN8), and LOC_Os12g36194 (WSL12) were reported to play an essential role in chloroplast development and pigment accumulation (Table 2). These results showed that the expression of signal-related genes was regulated in the chlm mutant, implying that the signal pathway might play important roles in pigment metabolism and chloroplast development.
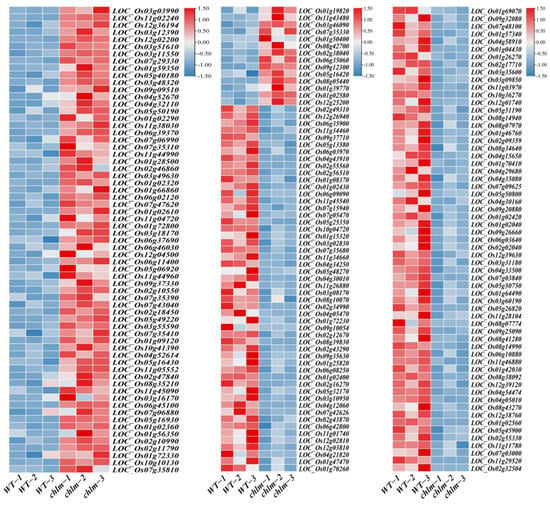
Figure 9.
Expression patterns of DEGs involved in signal pathways in the chlm mutant. Genes referring to signal recognition or transduction are shown.
2.11. Transcription Factors Were Identified in the Chlm Mutant
Transcription factors are important regulators that activate or repress gene expression in a sequence-specific manner, playing an important role in gene expression regulation to various biological processes, including plant growth, development and stress responses [58,59]. It is speculated that transcription factors may be involved in regulating the gene expression of the chlm mutant.
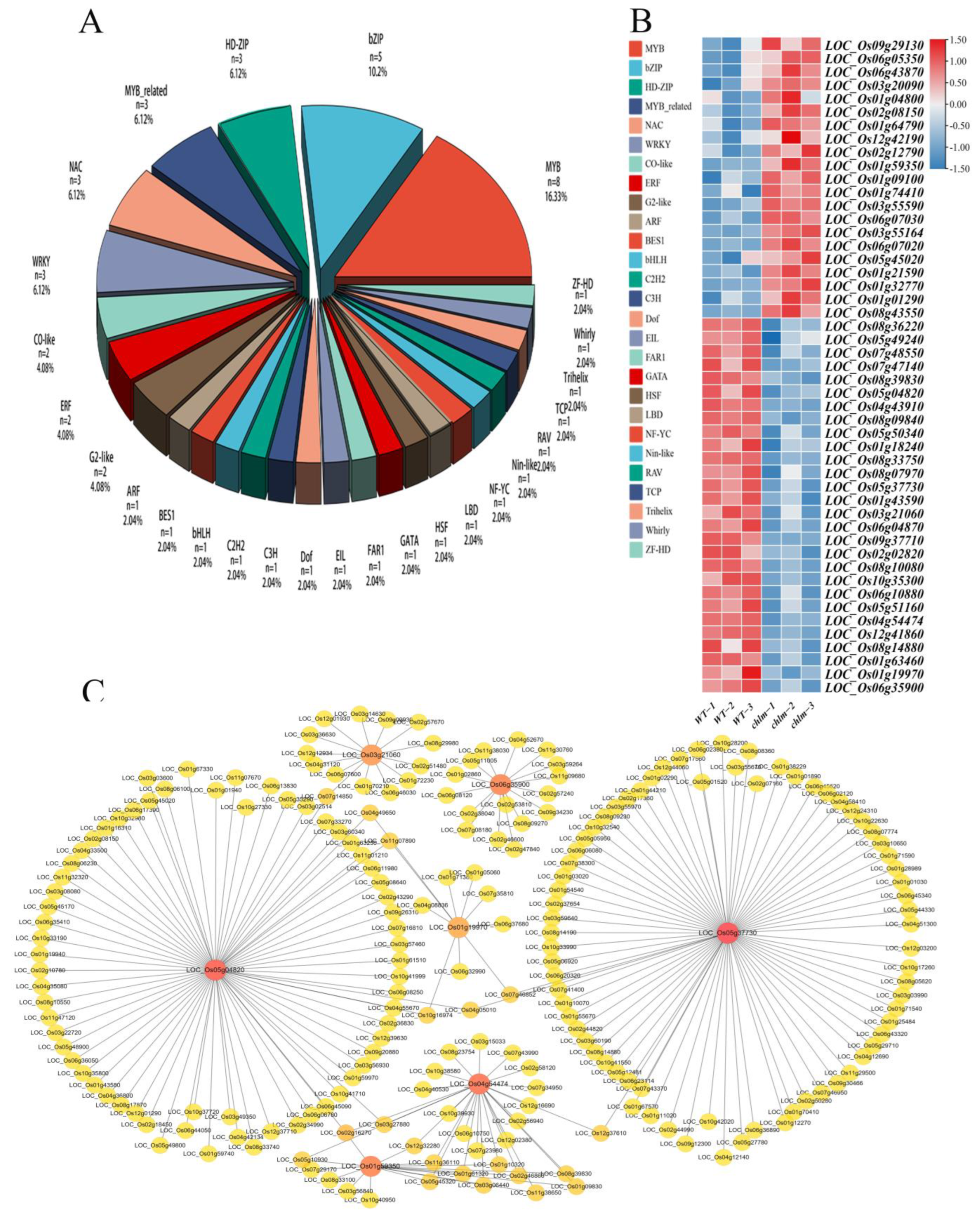
Among the 1507 DEGs in this study, 49 DEGs were putatively identified as transcription factors associated with 27 families by searching the Plant Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn/). The most abundant transcription factor family was the MYB superfamily (16.3%), which was followed by the bZIP family (10.2%), HD-ZIP family (6.1%), MYB-related family (6.1%), WRKY family (6.1%), and NAC family (6.1%) (Figure 10A). Furthermore, a total of 21 DEGs encoding transcription factors exhibited up-expression, while 28 transcription factors showed down-expression in the chlm mutant (Figure 10B, Table S8).
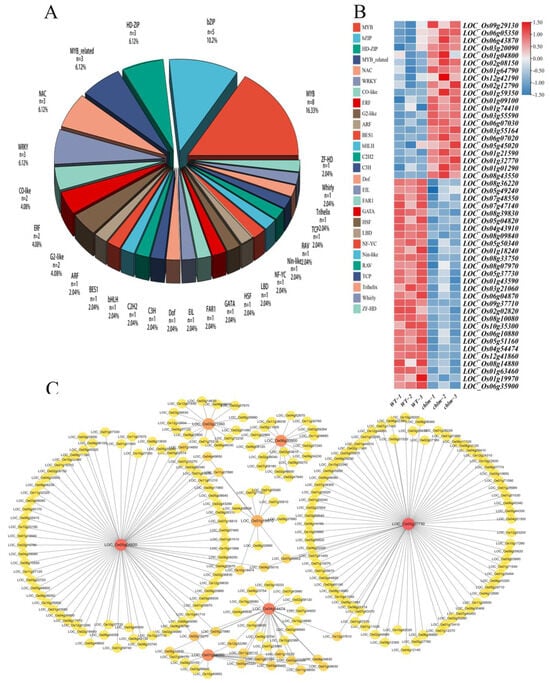
Figure 10.
Identification of differentially expressed transcription factor genes in the chlm mutant. (A) Proportion of transcription factors. (B) Expression patterns of transcription factors in the wild type and chlm mutant. (C) The potential regulatory roles between seven transcription factors with identified DEGs.
To further identify the potential regulatory roles between transcription factors with identified DEGs, all 1507 DEGs were investigated by searching the Plant Transcription Factor Database Regulatory Prediction Tool (http://planttfdb.cbi.pku.edu.cn/, accessed on 26 April 2023). Seven transcription factors (LOC_Os05g37730, LOC_Os05g04820, LOC_Os04g54474, LOC_Os06g35900, LOC_Os01g59350, LOC_Os01g19970, and LOC_Os03g21060) were found to be significantly associated with the tested DEGs (Figure 10C, Table S9). The MYB-type transcription factor LOC_Os05g37730 was predicted to regulate 76 DEGs, including six pigment metabolism-related genes, 19 chloroplast development-related genes, one hormone pathway-related gene, and six signal pathway-related genes (Figure 10C, Table S9). The MYB-type transcription factor LOC_Os05g04820 was predicted to regulate 73 DEGs, including one pigment metabolism-related gene, 10 chloroplast development-related genes, two hormone pathway-related genes, and seven signal pathway-related genes (Figure 10C, Table S9). The bZIP-type transcription factor LOC_Os04g54474 was predicted to regulate 26 DEGs, including one pigment metabolism-related gene, three chloroplast development-related genes, one hormone pathway-related gene, and two signal pathway-related genes (Figure 10C, Table S9). The bES1-type TF LOC_Os06g35900 was predicted to regulate 16 DEGs, including two chloroplast development-related genes, two hormone pathway-related genes, and three signal pathway-related genes (Figure 10C, Table S9). The bZIP-type TF LOC_Os01g59350 was predicted to regulate 16 DEGs, including one chloroplast development-related gene, one hormone pathway-related genes, and two signal pathway-related genes (Figure 10C, Table S9). The MYB-type TF LOC_Os01g19970 was predicted to regulate 10 DEGs, including three pigment metabolism-related genes and one signal pathway-related gene (Figure 10C, Table S9). The NAC-type TF LOC_Os03g21060 was predicted to regulate 14 DEGs, including three chloroplast development-related genes, one hormone pathway-related gene, and one signal pathway-related gene (Figure 10C, Table S9). These transcription factors might play important roles in chlorophyll synthesis and chloroplast development by regulating their target genes.
In addition, it is worth demonstrating that two transcription factors, LOC_Os06g05350 (OsWHY1, WHIRLY family) and LOC_Os02g12790 (OsCGA1, GATA family), were functionally validated and reported to be involved in chlorophyll synthesis and chloroplast development (Table 2). These results all imply that transcription regulation plays an important role in chlorophyll synthesis and chloroplast development regulated by the ChlM gene.
2.12. Verification by qRT-PCR of Some DEGs
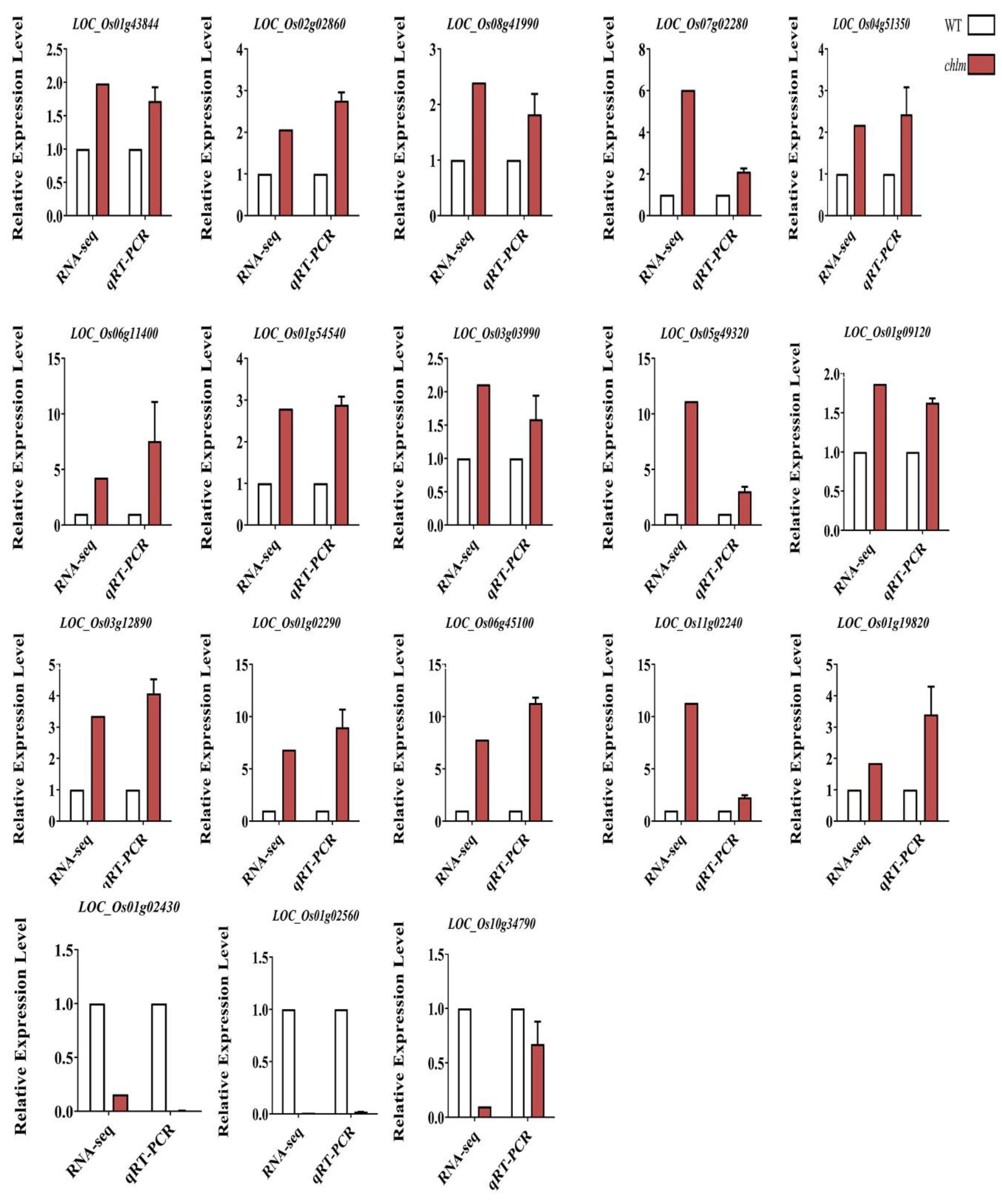
To identify the reliability of the transcriptomic data, 18 DEGs were randomly selected to detect their expression in the chlm leaves by qRT-PCR. The results showed that the expression patterns of all 18 genes detected by qRT-PCR were highly consistent with those from the Illumina sequencing data except for one gene LOC_Os10g34790 (Figure 11). These results demonstrated that the transcriptomic data are reliable.
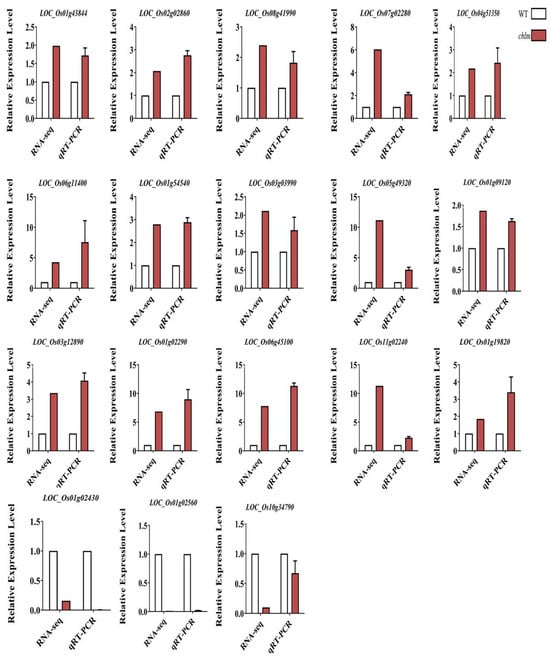
Figure 11.
Validation of the RNA-Seq results by qRT-PCR. Eighteen DEGs were randomly selected for detection. Error bars indicate means ± SD of three biological replicates.
3. Discussion
Chlorophyll content and chloroplast development are two critical factors for normal green leaf phenotype and photosynthesis. It is reported that gene mutation in the chlorophyll synthesis pathway usually leads to abnormal chloroplast development and eventually affects photosynthesis. It seems that these genes can also regulate the development of the chloroplast besides their roles in chlorophyll synthesis. However, the genetic regulatory network linking chlorophyll synthesis and chloroplast development is not very clear. In our previous study, map-based cloning of the chlm mutant identified the gene ChlM (LOC_Os06g04150) encoding the magnesium protoporphyrin IX methyltransferase (ChlM), which catalyzes the formation of MgPME from MgP and is a key speed-limiting enzyme in the chlorophyll synthesis pathway [18]. In this study, a physiological, cytological, and transcriptomic analysis of the chlm mutant was conducted to reveal the complex genetic regulatory network of the ChlM gene linking chlorophyll synthesis and chloroplast development.
3.1. Chlorophyll Metabolism Were Regulated by the ChlM Gene
Chlorophylls are the most abundant pigments used for harvesting energy from visible light in plants and then utilized for photosynthesis to support life forms on earth [60,61]. Due the lack of ChlM gene function, the chlm mutant leaves owned reduced chlorophyll content and thus showed a yellow leaf color (Figure 1A,B). This phenomenon is similar to other gene mutations in the chlorophyll synthesis pathway, such as ChlD [8], ChlI [8], CRD1 [9], ChlG [11] and OsCAO1 [13]. Chlorophyll metabolism is determined by complex biological processes, and genes in this process can be regulated in the chlm mutant. This study identified 62 chlorophyll-related DEGs in the chlm mutant leaves (Figure 6A–D). Glutamyl-tRNA is the initial substrate for the chlorophyll synthesis. OsGluRS was cloned to encode glutamyl-tRNA synthetase, and the mutation of this gene led to a chlorophyll deficiency phenotype [19]. LOC_Os02g02860 (OsGluRS) was found to be up-regulated in the chlm mutant (Table 2). Coproporphyrinogen III oxidase encoded by gene RLIN1 could transform coproporphyrinogen III into protoporphyrinogen IX, which played an important role in the tetrapyrrole biosynthetic pathway [5]. In our study, LOC_Os04g52130 was identified to be gene RLIN1, which was up-expressed in the chlm mutant (Table 2). OsChlD encoded one of the subunits of the magnesium chelatase, which could convert protoporphyrin IX into Mg-protoporphyrin IX during chlorophyll synthesis [8]. Interestingly, LOC_Os03g59640 was identified to be OsChlD in this study (Table 2), and its expression level was up-regulated in the chlm mutant. Examining the motivation of the well-studied chlorophyll synthesis genes OsGluRS, RLIN1, and OsChlD in the chlm mutant implied that the functional decline of the ChlM protein could regulate the chlorophyll synthesis pathway itself to promote chlorophyll accumulation. Stroma-localized rice heme oxygenase 2 (OsHO2) is required for the oxidative cleavage of heme to biliverdin, which is the other branch of tetrapyrrole biosynthesis besides the chlorophyll biosynthesis branch [20]. And OsHO2 (LOC_Os03g27770) was identified to be up-regulated in the chlm mutant (Table 2), which may be a feedback regulation pattern to reduce the accumulation of intermediate metabolites in chlorophyll biosynthesis branch due to the greatly weakened function of the chlm protein in the chlm mutant. OsNYC3 encodes an alpha/beta fold hydrolase family protein, playing important roles in chlorophyll degradation [21]. OsNYC3 (LOC_Os06g24730) was found to be down-regulated in the chlm leaves (Table 2), which might prevent the degradation of chlorophyll in this mutant.
In summary, the functional decline of the ChlM protein and decrease in chlorophyll content in the chlm mutant could cause the regulation of chlorophyll metabolism-related genes to compensate for the defect of chlorophyll synthesis in this mutant.
3.2. Chloroplast Development Was Regulated by the ChlM Gene
Chloroplasts are the place for photosynthesis, which usually consists of the chloroplast membrane, thylakoid and stroma in plants. The normal development of chloroplasts in higher plants requires the coordination of chloroplast genes and nuclear genes. Impaired function of the ChlM gene has caused the abnormal chloroplast development in the chlm mutant (Figure 1H–J). Whether the chlorophyll synthesis gene ChlM regulates chloroplast development-related genes is an interesting question.
Based on the transcriptomic data, a total of 278 chloroplast-related genes were identified to be regulated in the chlm mutant leaves (Figure 7). And interestingly, many of these genes were well studied and reported to be involved in chloroplast development. WSP1 encoding a multiple organellar RNA editing factor protein is essential for chloroplast development by regulating plastid RNA editing and chloroplast ribosome biogenesis [22]. OsTrxZ/wp2 encoding thioredoxin z regulates plastid RNA editing by interacting with multiple organellar RNA editing factors and contributes to chloroplast biogenesis in rice with an albino seedling lethality phenotype for the mutant [23,24]. WLP2 encoding a plastid-encoded RNA polymerase-associated protein is required for chloroplast biogenesis under heat stress through interacting with the OsTrxZ protein in rice [25]. OsPPR6 encoding a pentatricopeptide repeat protein for editing and splicing chloroplast RNA is required for chloroplast biogenesis in rice [26]. CDE4 encodes a pentatricopeptide repeat protein involved in chloroplast RNA splicing and affects chloroplast development under low-temperature conditions in rice; and the gene mutant shows white leaf and defective chloroplast development at low temperature [27]. Two complementary recessive genes etl1 and etl2 encoding the homologous protein of HCF152 in Arabidopsis required for chloroplast RNA processing are found to control etiolation and chloroplast thylakoid development [28]. WSL3 encoding a non-core subunit of plastid-encoded RNA polymerase is essential for early chloroplast development by interacting with subunits of the plastid-encoded RNA polymerase complex in rice [29]. OsNUS1 encoding a plastid protein is involved in the regulation of chloroplast RNA metabolism and is essential for the build-up of the plastid genetic system during early chloroplast development under cold stress conditions [30]. The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low-temperature conditions, and mutation of this gene leads to white leaf and abnormal chloroplast development [31]. Mutation of the rice ASL2 gene encoding plastid ribosomal protein L21 caused chloroplast developmental defects and albino seedling death, suggesting the important role of this gene in chloroplast development [32]. OsValRS2, encoding a Val-tRNA synthetase for regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development [33]. ObgC encoding a spo0B-related guanosine triphosphate binding protein that is crucial for chloroplast development and leaf greening plays a major role in the biosynthesis of plastid ribosomes during rice chloroplast development [34]. Nuclear encoded elongation factor EF-Tu is required for chloroplast development in rice grown under low-temperature conditions, and the gene mutant shows temperature-sensitive albino and disrupted chloroplast without thylakoid [35]. The nucleus-encoded chloroplast protein gene YL1 is involved in chloroplast development and the efficient biogenesis of chloroplast ATP synthase in rice, and the mutation of this gene causes yellow leaf and abnormal chloroplast morphology [36]. OscpSRP43 encoding a chloroplast signal recognition particle 43 protein is required for chloroplast development and photosynthesis, and the mutation of OscpSRP43 induces a yellow–green leaf phenotype and impaired chloroplast development [37]. YGL138(t) encoding a putative signal recognition particle 54 kDa protein is involved in chloroplast development in rice [38]. AL1 that encodes the sole octotricopeptide repeat protein plays an essential role in chloroplast development in rice, and the mutation of this gene results in albino leaves and a disrupted thylakoid structure [39]. OsNOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis, rubisco formation and chloroplast development in rice, and the gene mutant has yellow leaves and vague thylakoid membranes [40]. The VYL gene encoding a plastid protein homologous to the Arabidopsis ClpP6 subunit plays an important role in the assembly of plastid caseinolytic protease and chloroplast development, and the gene mutant shows yellow leaves and a reduced thylakoid membrane [41]. OsFd1 encoding the ferredoxin participates in photosynthetic electron transport, and the lost function of OsFd1 leads to chloroplast degradation and leaf chlorosis [42]. The OsFdC2 gene encoding a ferredoxin-like protein causes yellow leaves and an impaired chloroplast structure [43]. OsSTN8 participates in the photosystem II repair mechanism and thus affects the chloroplast structure in rice [44]. RNRS1 encoding the ribonucleotide reductase subunit for plastid DNA synthesis is necessary for chloroplast biogenesis during early leaf development [45]. YLC1 encoding a DUF3353 superfamily protein is suggested to be involved in chlorophyll and lutein accumulation and chloroplast development at early leaf development in rice [46]. WSL12 encoding a nucleoside diphosphate kinase 2 plays an important role in chloroplast development and chlorophyll synthesis in rice [47]. In this study, WSP1 (LOC_Os04g51280), OsTrxZ/wp2 (LOC_Os08g29110), WLP2 (LOC_Os01g63220), OsPPR6 (LOC_Os05g49920), CDE4 (LOC_Os08g09270), etl1 (LOC_Os11g01210), etl2 (LOC_Os12g01210), WSL3 (LOC_Os10g32540), OsNUS1 (LOC_Os03g45400), WLP1 (LOC_Os01g54540), ASL2 (LOC_Os02g15900), OsValRS2 (LOC_Os07g06940), ObgC (LOC_Os07g47300), EF-Tu (LOC_Os02g38210), YL1 (LOC_Os02g05890), OscpSRP43 (LOC_Os03g03990), YGL138(t) (LOC_Os11g05552), AL1 (LOC_Os03g31150), OsNOA1 (LOC_Os02g01440), VYL (LOC_Os03g29810), OsFdC1 (LOC_Os03g45710), OsFdC2 (LOC_Os03g48040), OsSTN8 (LOC_Os05g40180), RNRS1 (LOC_Os06g14620), YLC1 (LOC_Os09g21250), and WSL12 (LOC_Os12g36194) were all identified to be up-regulated in the chlm mutant (Table 2). So many well-studied genes were found in this study, which provided effective evidence for the roles of identified chloroplast-related DEGs on chloroplast development. It is speculated that the chlm mutant regulates the genes probably trying to compensate for chloroplast structure defects due to the impaired function of the ChlM protein in the mutant. It is proposed that chloroplast development was regulated by the ChlM gene in rice.
3.3. JA, SA and IAA May Play an Important Role in the Regulation Process of ChlM Gene to Chlorophyll Synthesis and Chloroplast Development
The biosynthesis of some plant hormones is highly related with chloroplasts. JA biosynthesis from linolenic acid begins in chloroplasts [62]. Plants synthesize SA through two pathways: the isochorismate acid synthase pathway and the phenylalanine ammonia lyase pathway, both of which originate from chloroplasts, use chorismic acid as a precursor, and refer to multiple enzyme catalysis [63]. Tryptophan dependent auxin synthesis is related to chloroplasts because tryptophan biosynthesis occurs in chloroplasts [64]. Plants synthesize ABA using the carotenoid pathway initiated from the cleavage of a C40 precursor known as β-carotene, and this process is mainly carried out in chloroplasts [65]. Accompanied by the changes of chloroplast structure, the increase in contents for JA, SA and IAA were observed in the chlm leaves, especially the significant increase for JA (Figure 2). And accordingly, 59 hormone-related DEGs were identified to be regulated in the chlm leaves (Figure 8, Table S6). DEGs related with JA, SA and IAA were mainly referring to the metabolism, response and transport of these hormones. However, the DEGs for the synthesis of JA, SA and IAA were not identified in the chlm mutant. The impaired function of the ChlM gene might not regulate these synthesis-related genes. The accumulation of these three hormones was possibly attributed to the increase in their substrate contents, which might be affected due to the impeded chlorophyll synthesis and chloroplast development of the chlm mutant. This speculation needs further measured experiments for related metabolites. The function of the accumulated JA, SA and IAA in the chlm mutant was an interesting question. OsJMT encoding the jasmonic acid carboxyl methyltransferase is involved in methylating JA to methyl jasmonate [48,49]. LOC_Os06g20920 and LOC_Os05g01140 were both identified as OsJMT genes in this study (Table 2). Methyl jasmonate and auxin were found to regulate the expression of chloroplast genes in barley [66]. Exogenous methyl jasmonate also played an important role in regulating the expression of chlorophyll synthesis genes to accumulate chlorophyll and improve the photosynthesis capacity in citrus [67]. And exogenous SA effectively improved the growth, photosynthesis, antioxidant enzyme activity, and stoma and chloroplast development in Dianthus superbus [68]. SA played protective roles in maintaining the integrity and function of photosynthetic photosystems in Medicago sativa [69]. It is reported that impaired chloroplast proteostasis (specifically for PSII proteins) may activate the chloroplast-established isochorismate pathway to produce SA; thus, SA is proposed to serve as a retrograde signaling molecule [70]. Considering the increases in JA, SA and IAA contents, and the expression changes of hormone-related genes, it is suggested that these three hormones may play an important role in the genetic network of the ChlM gene, regulating chlorophyll synthesis and chloroplast development.
3.4. Signal Pathways May Play an Important Role in the Regulation Process of ChlM Gene to Chlorophyll Synthesis and Chloroplast Development
The development and function of chloroplasts needs the involvement of enormous proteins encoded by the nucleus. And the status of chloroplasts is usually monitored by the nucleus. The processes that regulate the communication between nucleus and chloroplast, and the subsequent function execution of chloroplast, must need the involvement of massive genes related with signal pathways [71]. As expected, a large number of genes related with signal recognition and transduction were identified in the chlm mutant (Figure 9, Table S7). Some genes were cloned and verified to participate in chloroplast function. For example, rice chloroplast signal recognition particle 43 encoding gene OscpSRP43 is required for chlorophyll synthesis, chloroplast development and photosynthesis [37]. In this study, LOC_Os03g03990 was identified to be OscpSRP43 (Table 2). Similarly, a putative signal recognition particle 54 kDa protein-encoding gene YGL138(t) is involved in chlorophyll accumulation and chloroplast development [38]. And LOC_Os11g05552 was identified to be YGL138(t) in this study (Table 2). OsSTN8 affects the phosphorylation of the core protein of photosystem II and is crucial for the repair of photosystem II in rice [44]. In the present study, an up-expressed gene LOC_Os05g40180 was identified as OsSTN8 and was annotated to be involved in signal transduction mechanisms (Table 2, Table S7). The WSL12 locus encoding the nucleoside diphosphate kinase OsNDPK2, with its mutation resulting in a white stripe leaf phenotype, plays an important role in chlorophyll biosynthesis and chloroplast development by regulating the expression of related genes [47]. In this study, an up-expressed gene LOC_Os12g36194 was identified to be gene WSL12 and was annotated to be involved in the MAPK signaling pathway (Table 2, Table S7). It is suggested that signal pathway-related genes may play an important role in the regulation process of the ChlM gene to chlorophyll synthesis and chloroplast development.
3.5. Transcription Regulation May Play an Important Role in the Regulation Process of ChlM Gene to Chlorophyll Synthesis and Chloroplast Development
Transcription factors have been found to function on chlorophyll biosynthesis and chloroplast development in plants in a positive or negative manner [72,73]. In the chlm mutant, there were 49 transcription factors identified to show up- or down-expression (Figure 10, Table S8). Through predicting their target genes, seven transcription factors were found to target abundant DEGs, including those identified to be involved in pigment metabolism, chloroplast development, hormone regulation, and the signal pathways (Table S9). It is suggested that transcription factors can play an important role in the regulation process of the ChlM gene to chlorophyll synthesis and chloroplast development by regulating their target genes. In rice, some transcription factors were also cloned and found to be involved in chlorophyll synthesis and chloroplast development. OsWHY1 encodes a WHIRLY family transcription factor protein, which interacts with OsTRXz to function on chloroplast RNA editing and splicing and eventually regulates chloroplast development by affecting the expression of chloroplast and ribosome development-related genes and chlorophyll synthesis-related genes [50]. In this study, LOC_Os06g05350 was identified as the OsWHY1 gene, which was up-regulated in the chlm mutant (Table 2). OsCGA1 encodes a GATA family transcription factor protein and is co-expressed with important nucleus-encoded chloroplast-localized genes, regulating chlorophyll accumulation and chloroplast biogenesis [51]. The ectopic expression of the OsCGA1 gene in the rice bundle sheath could enhance its chloroplast biogenesis and increase the expression of photosynthesis-associated nuclear genes; and further activation of the endogenous OsCGA1 gene by engineering its promoter could directly enhance chloroplast development [52]. In this study, LOC_Os02g12790 was identified as the OsCGA1 gene, which was up-regulated in the chlm mutant (Table 2). These functionally validated transcription factor genes provided reliable evidence that transcription regulation played an important role in the regulation process of the ChlM gene, affecting chlorophyll synthesis and chloroplast development.
3.6. Possible Patterns for the Involvement of the ChlM Gene in the Retrograde Signaling Pathway
Plant cells coordinate their regulation of the expression of nuclear and plastid genes that encode components of the photosynthetic apparatus. The retrograde signaling from chloroplasts to the nucleus plays an important role in this coordinate control. It is proposed that MgP, the substrate of the ChlM gene, can act as a signaling molecule in the retrograde signaling pathway, and the accumulation of MgP is necessary to regulate the expression of many nuclear genes encoding photosynthesis-related chloroplastic proteins, including LHCB1 [74]. However, a study re-evaluated this hypothesis by quantifying the correlation between the steady-state levels of MgP and MgPME with altered plastid signaling responses as monitored by the expression of LHCB1, RBCS, and HEMA1 genes in a range of Arabidopsis mutants and conditions in which the steady-state levels of MgP and MgPME have been modified, and they found that there was no correlation between the steady-state levels of MgP (MgPME) with LHCB1 expression or with any of the other genes tested [75]. Another study developed a sensitive liquid chromatography–mass spectrometry method to measure tetrapyrrole intermediates, and they also showed that there is no correlation between nuclear gene LHCB1 expression with any of the chlorophyll biosynthetic intermediates including MgP over a range of growth conditions and treatments [76]. Consequently, these two studies negated the hypothesis that MgP acts as the retrograde signaling molecule to repress the expression of photosynthesis-related nuclear genes. Actually, in the rice chlm mutant, the expression level of a large number of pigment metabolism-related genes (Figure 6) and chloroplast protein-encoding genes (Figure 7) were regulated due to the impaired function of the ChlM gene, implying the fact that the retrograde signaling regulating this process absolutely exists. In this study, JA accumulated greatly in the chlm leaves (Figure 2A), and interestingly, the chlm mutant plants show an active growth state but not leaf senescence or death. Moreover, two OsJMT genes involved in methylating JA to form methyl jasmonate were also regulated in the chlm mutant (Table 2). Methyl jasmonate was found to function by regulating the expression of chloroplast genes in barley [66] and chlorophyll synthesis genes in citrus [67]. It is seemed that the accumulated JA or its downstream methyl jasmonate would play an important role in the regulatory process of chlorophyll synthesis and chloroplast development of the chlm mutant. Accordingly, we proposed that JA or its derivatives (such as methyl jasmonate) in a certain unknown state might act as a retrograde signaling molecule in one of the signaling pathways from the chloroplast to the nucleus. A previous report proposed that SA might serve as a retrograde signaling molecule in plants [70]. SA was found to accumulated in the chlm leaves (Figure 2B). And we also proposed that SA or its derivatives in a certain unknown state might act as a retrograde signaling molecule in one of the signaling pathways from chloroplasts to the nucleus. However, these speculations need further experiments for validation.
4. Materials and Methods
4.1. Plant Material and Sample Collection
The spontaneous mutant chlm in this study was derived from a photo-thermosensitive genic male sterile rice cultivar “guangzhan63S” (Oryza sativa L. subsp. indica) [18]. This mutant showed a yellow–green leaf phenotype and could be inherited stably. In our experiments, the seeds of wild-type and chlm mutant plants were soaked in a growth incubator at 37 °C until germinated. Then, the germinated seeds were grown in the experimental field with natural conditions. After 20 days of growth, the chlm mutant plants were easily distinguished by the yellow–green phenotype. For physiological, cytological, and RNA-seq experiments, the leaves of the wild-type and chlm mutant plants at the seedling stage were separately sampled. All the samples were collected from the uniform growth plants, flash frozen in liquid nitrogen and stored at −80 °C until use. Three biological replicates were performed in all experiments.
4.2. Determination of Chlorophyll and Hormone Contents
The chlorophyll content was determined according to the method as previously described [77]. In detail, the leaves sampled from the wild type and the chlm mutant were powdered in a pre-chilled mortar with liquid nitrogen. Then, approximately 0.1 g powder samples were mixed with 80% acetone to extract chlorophyll until the powder became white. During this process, the samples were kept in dark and periodic oscillation conditions. Next, the absorbance values of the extract were examined by a spectrophotometer at 663, 647 and 470 nm. Finally, the chlorophyll content was calculated by their corresponding formula.
Phytohormones including jasmonic acid (JA), salicylic acid (SA), indoleacetic acid (IAA), and abscisic acid (ABA) were extracted and measured following a previously described method [78]. In brief, the powder samples were mixed with Bieleski solvent (methanol/water) to extract hormones, after which we added the internal standards of these four hormones, and next, the extract solution was detected using a nano-LC-ESI-Q-TOF-MS system to determine hormones.
4.3. Transmission Electron Microscopic Observation
The leaves dissected from the wild-type and the chlm mutant were cut into smaller segments. Then, the leaf segments were fixed with 2.5% glutaraldehyde in sodium phosphate buffer (pH 7.2) for 4 h at 4 °C and postfixed with 1% (v/v) osmium tetroxide in PBS for an additional 2 h. Then, the tissues were dehydrated through an acetone series and embedded in Spurr’s resin according to the previously described method [46]. Subsequently, for ultrastructural observations of leaf chloroplasts, 70 nm thick sections were cut with a Leica EM UC6 ultramicrotome and stained with 1% (w/v) uranyl acetate and 1% (w/v) lead citrate. At last, leaf cells were observed and photographed using a transmission electron microscope.
4.4. RNA Extraction and Preparation of cDNA Library
The total RNA was extracted from leaves of the wild type and the chlm mutant using TRIZOL reagent following the manufacturer’s instructions. Three biological replicates were analyzed for each RNA sample. RNA degradation and contamination were monitored on 1% agarose gel electrophoresis. RNA purity was tested by a NanoPhotometer spectrophotometer (IMPLEN, Munich, Germany). RNA concentration and integrity were measured using a Qubit RNA Assay Kit in a Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). RNA concentration and purity were measured using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
After qualification of the RNA quality test, the libraries were constructed following the procedures. A 1 µg RNA sample was used as the input material for library preparation. Sequencing libraries were generated using an NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. In detail, mRNA was purified using poly (dT) oligo-attached magnetic beads. Afterwards, mRNA was randomly broken into fragments. Then, the first-strand cDNA was synthesized using a random hexamer primer and M-MLV reverse transcriptase. Second-strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Next, the double-strand cDNA was subjected to end-repair, dA tailing and adaptor ligation. Finally, suitable fragments were isolated and then enriched using polymerase chain reaction amplification. Furthermore, the quality of obtained libraries was assessed on the Agilent Bioanalyzer 2100 system.
4.5. Illumina Deep Sequencing and Data Analysis
After the libraries were qualified, the clustering of the index-coded samples was performed on a cBot cluster generation system using a TruSeq PE Cluster Kit v4-cBot-HS (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. After cluster generation, the constructed libraries were sequenced on an Illumina Hiseq 2500 platform, and paired-end reads were generated. Library construction and sequencing were performed by Beijing Biomarker (BMKCloud) Technology Co., Ltd. (Beijing, China). Finally, the raw reads were generated.
To obtain the clean reads, the adaptor sequences, reads containing ploy-N and low-quality reads were removed from raw reads using Perl script. Simultaneously, the GC content, Q20 and Q30 of the clean reads were calculated. All the clean reads with high quality were used for the identification of mRNAs. Subsequently, the clean reads were mapped to the rice reference genome (http://rice.plantbiology.msu.edu/, accessed on 1 August 2022) using TopHat2 V 2.1.0 software [79]. The read counts for each gene were calculated with Cufflinks V2.0 software, and gene expression levels were estimated as Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) values.
4.6. Identification and Functional Enrichment Analysis of Differentially Expressed Genes
Prior to identifying the interested and reliable differentially expressed genes (DEGs), three biological replicates were performed. The correlations of samples were evaluated by Pearson’s correlation coefficient [80]. Furthermore, DESeq R package [81] was performed to identify DEGs between the wild type and the chlm mutant. DESeq provides statistical routines for identifying differential expression in gene expression data using a model based on the negative binomial distribution. To control for the false discovery rate (FDR), p-values obtained from DESeq were adjusted using the edgeR approach. Genes with the fold change ≥ 2 and FDR < 0.01 were considered to be differentially expressed.
To explore the potential functions of DEGs, the functional enrichment analysis was performed. Gene Ontology (GO) enrichment analysis was performed using the GOseq R package [82]. And GO terms with p-values < 0.05 were considered significantly enriched. To further investigate the involvement in pathways of DEGs, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed using KOBAS V3.0 software. And the pathways with the p-value < 0.05 were considered to be statistically significantly enriched. In addition, the heatmap analysis of DEGs was carried out using TBtools V2.012 software. The FPKM values of DEGs were imported into TBtools, and the data were calculated by Log2 and then clustered to obtain the resulting figures. Furthermore, protein–protein interaction analysis of DEGs was performed, and the result was visualized through Cytoscape V3.9.1 software.
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
To validate the expression patterns of DEGs, quantitative real-time PCR (qRT-PCR) was performed. The total RNA of rice leaf samples was extracted using a TRIZOL reagent (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. After extraction, the total RNA was subjected to reverse transcription using a PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Beijing, China) according to manufacturer’s instructions. Then, qRT-PCR was performed using a SYBR Green PCR kit (Takara, Beijing, China) on a CFX96 Real-Time PCR system (Bio-Rad, Hercules, CA, USA) to validate the expression levels of DEGs. Three genes, UBC (LOC_Os02g42314), ARF (LOC_Os05g41060) and Profilin-2 (LOC_Os06g05880) were used as internal reference genes to normalize the qRT-PCR data [83]. Furthermore, qRT-PCR conditions were as follows: 10 min at 95 °C for pre-denaturation followed by 40 cycles of 10 s at 95 °C, 10 s at 60 °C, and 15 s at 72 °C and then a melt curve from 65 to 95 °C. The relative RNA expression levels were calculated by using the CFX Manager 3.1 (Bio-Rad) with the 2−ΔΔCT method. The reaction was carried out using three biological replicates with three technical replicates. All primers are shown in Table S10.
5. Conclusions
In this study, we analyzed changes in pigment contents, chloroplast structure and hormone contents in the chlm mutant and then used transcriptomic data to reveal the ChlM gene involved regulatory network linking chlorophyll synthesis with chloroplast development. Genes related to pigment metabolism, chloroplast development, hormone pathways, signal pathways and transcriptional regulation were found to function in this regulation process, revealing a complex genetic regulatory network linking chlorophyll synthesis and chloroplast development. The ChlM gene involved in retrograde signaling was discussed. Jasmonic acid, salicylic acid or their derivatives in a certain unknown state were proposed as retrograde signaling molecules in one of the signaling pathways from the chloroplast to the nucleus.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants12213785/s1, Table S1. The detailed information of 1534 DEGs between wild type and chlm mutant; Table S2. GO and KEGG enrichment analysis of 1534 DEGs; Table S3. The detailed information of the top 40 PPI protein-encoding genes; Table S4. The detailed information of genes related to pigment metabolism; Table S5. The detailed information of genes related to chloroplast development; Table S6. The detailed information of genes related to hormone pathways; Table S7. The detailed information of genes related to signal pathways; Table S8. The detailed information of genes related to transcription factors; Table S9. The potential regulatory roles between transcription factors with DEGs; Table S10. All primers used for qRT-PCR analysis.
Author Contributions
Y.Y. and H.Z. contributed to bioinformatics analysis and original draft preparation. R.G. performed qRT-PCR experiments and participated in original draft preparation. J.F. and S.L. participated in bioinformatics analysis. J.L. helped in bioinformatic analysis. Y.H. helped in data consolidation and manuscript revision. Z.W. conceptualized and supervised this study and performed manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 32260495, 32060071, 31960062), the Science Fund for Distinguished Young Scholars of Jiangxi Province (20224ACB215003), Postgraduate Innovation Special Fund Project of Jiangxi Province (YC2022-s429), the Natural Science Foundation of Jiangxi Province (20212BAB205008) and National College Students Innovation and Entrepreneurship Training Program (202210410006).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data described in the manuscript are available from the corresponding author on reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ChlM | Magnesium protoporphyrin IX methyltransferase |
| MgP | Magnesium protoporphyrin IX |
| MgPME | Magnesium protoporphyrin IX methyl ester |
| WT | Wild type |
| JA | Jasmonic acid |
| SA | Salicylic acid |
| IAA | Indole acetic acid |
| ABA | Abscisic acid |
| DEGs | Differentially expressed genes |
| FPKM | Fragments Per Kilobase of transcript per Million fragments mapped |
| FDR | False discovery rate |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | Protein–protein interaction |
| qRT-PCR | Quantitative real-time PCR |
References
- Czarnecki, O.; Grimm, B. Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 2012, 63, 1675–1687. [Google Scholar] [CrossRef]
- Fromme, P.; Melkozernov, A.; Jordan, P.; Krauss, N. Structure and function of photosystem I: Interaction with its soluble electron carriers and external antenna systems. FEBS Lett. 2003, 555, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Rye, J.J.E.; Sobotka, R.; Sobotka, R. Editorial: Assembly of the Photosystem II Membrane-Protein Complex of Oxygenic Photosynthesis %J Frontiers in Plant Science. Front. Plant Sci. 2017, 8, 884. [Google Scholar]
- Beale, S.I.J. Green genes gleaned. Trends Plant Sci. 2005, 10, 309–312. [Google Scholar] [CrossRef]
- Sun, C.; Liu, L.; Tang, J.; Lin, A.; Zhang, F.; Fang, J.; Zhang, G.; Chu, C. RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J. Genet. Genom. 2011, 38, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ye, B.; Yin, J.; Yuan, C.; Zhou, X.; Li, W.; He, M.; Wang, J.; Chen, W.; Qin, P. Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol. Biochem. 2015, 97, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Hur, J.; Ryu, C.H.; Choi, Y.; Chung, Y.Y.; Miyao, A.; Hirochika, H.; An, G. Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, J.; Yoo, J.-H.; Yoo, S.-C.; Cho, S.-H.; Koh, H.-J.; Seo, H.S.; Paek, N.-C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef]
- Kong, W.; Yu, X.; Chen, H.; Liu, L.; Xiao, Y.; Wang, Y.; Wang, C.; Lin, Y.; Yu, Y.; Wang, C.; et al. The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Mol. Biol. 2016, 92, 177–191. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.H.; Kim, Y.S.; Koh, H.J.; Yoo, S.C.; Paek, N.C. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.H.; Yoo, E.S.; Lee, C.H.; Hirochika, H.; An, G. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol. Biol. 2005, 57, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, J.; Huang, L.; Leng, Y.; Dai, L.; Rao, Y.; Chen, L.; Wang, Y.; Tu, Z.; Hu, J. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J. Exp. Bot. 2016, 67, 1297–1310. [Google Scholar] [CrossRef]
- SHEPHERD, M.; REID, J.D.; HUNTER, C.N. Purification and kinetic characterization of the magnesium protoporphyrin IX methyltransferase from Synechocystis PCC6803. Biochem. J. 2003, 371, 351–360. [Google Scholar] [CrossRef]
- Meinecke, L.; Alawady, A.; Schroda, M.; Willows, R.; Kobayashi, M.C.; Niyogi, K.K.; Grimm, B.; Beck, C.F. Chlorophyll-deficient mutants of Chlamydomonas reinhardtii that accumulate magnesium protoporphyrin IX. Plant Mol. Biol. 2010, 72, 643–658. [Google Scholar] [CrossRef]
- Alawady, A.E.; Grimm, B. Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 2005, 41, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.; Albrieux, C.; Joyard, J.; Lagrange, T.; Block, M.A. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis: Effects on chloroplast development and on chloroplast-to-nucleus signaling. J. Biol. Chem. 2007, 282, 2297–2304. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hong, X.; Hu, K.; Wang, Y.; Wang, X.; Du, S.; Li, Y.; Hu, D.; Cheng, K.; An, B.; et al. Impaired Magnesium Protoporphyrin IX Methyltransferase (ChlM) Impedes Chlorophyll Synthesis and Plant Growth in Rice. Front. Plant Sci. 2017, 8, 1694. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Fu, Y.; Hu, G.; Si, H.; Zhu, L.; Wu, C.; Sun, Z. Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.). Planta 2007, 226, 785–795. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, F.-Y.; Gao, X.; Sun, Y.; Li, S.; Tao, Y.; Lo, C.; Liu, H. Young Leaf Chlorosis 2 encodes the stroma-localized heme oxygenase 2 which is required for normal tetrapyrrole biosynthesis in rice. Planta 2014, 240, 701–712. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, X.; Wang, Y.; Wu, J.; Gu, X.; Lu, T. The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol. Plant 2017, 10, 86–98. [Google Scholar] [CrossRef]
- He, L.; Zhang, S.; Qiu, Z.; Zhao, J.; Nie, W.; Lin, H.; Zhu, Z.; Zeng, D.; Qian, Q.; Zhu, L. Fructokinase-Like Protein 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 2018, 60, 94–111. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Ren, Y.; Duan, E.; Zhu, X.; Hao, Y.; Zhu, J.; Chen, R.; Lei, J.; Teng, X. white panicle2 encoding thioredoxin z, regulates plastid RNA editing by interacting with multiple organellar RNA editing factors in rice. New Phytol. 2021, 229, 2693–2706. [Google Scholar] [CrossRef]
- Lv, Y.; Shao, G.; Qiu, J.; Jiao, G.; Sheng, Z.; Xie, L.; Wu, Y.; Tang, S.; Wei, X.; Hu, P. White Leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J. Exp. Bot. 2017, 68, 5147–5160. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhang, W.; Wen, K.; Chen, G.; Sun, J.; Tian, Y.; Tang, W.; Yu, J.; An, H.; Wu, T. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 2017, 95, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, X.; Cao, R.; Jiao, G.; Hu, S.; Shao, G.; Sheng, Z.; Xie, L.; Tang, S.; Wei, X.; et al. CDE4 encodes a pentatricopeptide repeat protein involved in chloroplast RNA splicing and affects chloroplast development under low-temperature conditions in rice. J. Integr. Plant Biol. 2021, 63, 1724–1739. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, H.; Liu, T.; Yang, G.; Xing, Y. Two complementary recessive genes in duplicated segments control etiolation in rice. Theor. Appl. Genet. 2011, 122, 373–383. [Google Scholar] [CrossRef]
- Wang, L.; Wang, C.; Wang, Y.; Niu, M.; Ren, Y.; Zhou, K.; Zhang, H.; Lin, Q.; Wu, F.; Cheng, Z. WSL3, a component of the plastid-encoded plastid RNA polymerase, is essential for early chloroplast development in rice. Plant Mol. Biol. 2016, 92, 581–595. [Google Scholar] [CrossRef]
- Kusumi, K.; Sakata, C.; Nakamura, T.; Kawasaki, S.; Yoshimura, A.; Iba, K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 2011, 68, 1039–1050. [Google Scholar] [CrossRef]
- Song, J.; Wei, X.; Shao, G.; Sheng, Z.; Chen, D.; Liu, C.; Jiao, G.; Xie, L.; Tang, S.; Hu, P. The rice nuclear gene WLP1 encoding a chloroplast ribosome L13 protein is needed for chloroplast development in rice grown under low temperature conditions. Plant Mol. Biol. 2014, 84, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Jiang, Q.; Zheng, K.; Chen, S.; Zhou, H.; Gong, X.; Xu, J.; Teng, S.; Dong, Y. Mutation of the rice ASL 2 gene encoding plastid ribosomal protein L21 causes chloroplast developmental defects and seedling death. Plant Biol. 2015, 17, 599–607. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Zheng, M.; Lyu, J.; Xu, Y.; Li, X.; Niu, M.; Long, W.; Wang, D.; Wang, H. WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol. 2016, 170, 2110–2123. [Google Scholar] [CrossRef]
- Bang, W.Y.; Chen, J.; Jeong, I.S.; Kim, S.W.; Kim, C.W.; Jung, H.S.; Lee, K.H.; Kweon, H.-S.; Yoko, I.; Shiina, T.; et al. Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J. 2012, 71, 122–134. [Google Scholar] [CrossRef]
- Cai, L.; Liu, Z.; Cai, L.; Yan, X.; Hu, Y.; Hao, B.; Xu, Z.; Tian, Y.; Liu, X.; Liu, L.; et al. Nuclear encoded elongation factor EF-Tu is required for chloroplast development in rice grown under low-temperature conditions. J. Genet. Genom. 2022, 49, 502–505. [Google Scholar] [CrossRef]
- Chen, F.; Dong, G.; Wu, L.; Wang, F.; Yang, X.; Ma, X.; Wang, H.; Wu, J.; Zhang, Y.; Wang, H. A nucleus-encoded chloroplast protein YL1 is involved in chloroplast development and efficient biogenesis of chloroplast ATP synthase in rice. Sci. Rep. 2016, 6, 32295. [Google Scholar] [CrossRef]
- Lv, X.; Shi, Y.; Xu, X.; Wei, Y.; Wang, H.; Zhang, X.; Wu, J. Oryza sativa chloroplast signal recognition particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PLoS ONE 2015, 10, e0143249. [Google Scholar] [CrossRef]
- Zhang, F.; Luo, X.; Hu, B.; Wan, Y.; Xie, J. YGL138 (t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 2013, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tan, J.; Shi, Z.; Xie, Q.; Xing, Y.; Liu, C.; Chen, Q.; Zhu, H.; Wang, J.; Zhang, J. Albino Leaf1 that encodes the sole octotricopeptide repeat protein is responsible for chloroplast development. Plant Physiol. 2016, 171, 1182–1191. [Google Scholar] [PubMed]
- Yang, Q.; He, H.; Li, H.; Tian, H.; Zhang, J.; Zhai, L.; Chen, J.; Wu, H.; Yi, G.; He, Z.-H. NOA1 functions in a temperature-dependent manner to regulate chlorophyll biosynthesis and Rubisco formation in rice. PLoS ONE 2011, 6, e20015. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Fei, G.; Wu, C.; Wu, F.; Sun, Y.; Chen, M.; Ren, Y.; Zhou, K.; Cheng, Z.; Wang, J. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 2013, 162, 1867–1880. [Google Scholar] [CrossRef]
- He, L.; Li, M.; Qiu, Z.; Chen, D.; Zhang, G.; Wang, X.; Chen, G.; Hu, J.; Gao, Z.; Dong, G. Primary leaf-type ferredoxin 1 participates in photosynthetic electron transport and carbon a7ssimilation in rice. Plant J. 2020, 104, 44–58. [Google Scholar] [CrossRef]
- Li, C.; Hu, Y.; Huang, R.; Ma, X.; Wang, Y.; Liao, T.; Zhong, P.; Xiao, F.; Sun, C.; Xu, Z. Mutation of FdC2 gene encoding a ferredoxin-like protein with C-terminal extension causes yellow-green leaf phenotype in rice. Plant Sci. 2015, 238, 127–134. [Google Scholar] [CrossRef]
- Nath, K.; Poudyal, R.S.; Eom, J.S.; Park, Y.S.; Zulfugarov, I.S.; Mishra, S.R.; Tovuu, A.; Ryoo, N.; Yoon, H.S.; Nam, H.G. Loss-of-function of OsSTN 8 suppresses the photosystem II core protein phosphorylation and interferes with the photosystem II repair mechanism in rice (Oryza sativa). Plant J. 2013, 76, 675–686. [Google Scholar] [CrossRef]
- Yoo, S.-C.; Cho, S.-H.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.-J.; Iba, K.; Paek, N.-C. Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Ren, Y.; Lv, J.; Wang, Y.; Liu, F.; Zhou, F.; Zhao, S.; Chen, S.; Peng, C.; Zhang, X. Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 2013, 237, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Hu, S.; Wu, L.; Ge, C.; Cui, Y.; Chen, P.; Wang, X.; Xu, J.; Ren, D.; Dong, G. White stripe leaf 12 (WSL12), encoding a nucleoside diphosphate kinase 2 (OsNDPK2), regulates chloroplast development and abiotic stress response in rice (Oryza sativa L.). Mol. Breed. 2016, 36, 57. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Kim, Y.S.; Park, S.; Koo, Y.J.; Choi, Y.D.; Chung, Y.; Lee, I.; Kim, J. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef]
- Qi, J.; Li, J.; Han, X.; Li, R.; Wu, J.; Yu, H.; Hu, L.; Xiao, Y.; Lu, J.; Lou, Y. Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 2016, 58, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Chen, D.; Teng, L.; Guan, P.; Yu, G.; Zhang, P.; Song, J.; Zeng, Q.; Zhu, L. OsWHY1 Interacts with OsTRX z and is Essential for Early Chloroplast Development in Rice. Rice 2022, 15, 50. [Google Scholar] [CrossRef] [PubMed]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.-M.; Rothstein, S.J. Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef]
- Lee, D.Y.; Hua, L.; Khoshravesh, R.; Giuliani, R.; Kumar, I.; Cousins, A.; Sage, T.L.; Hibberd, J.M.; Brutnell, T.P. Engineering chloroplast development in rice through cell-specific control of endogenous genetic circuits. Plant Biotechnol. J. 2021, 19, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Plant J. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 1017–1024. [Google Scholar] [CrossRef] [PubMed]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. Found. 2022, 233, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lin, N.; Liu, X.; Yang, S.; Wang, W.; Wan, X. From chloroplast biogenesis to chlorophyll accumulation: The interplay of light and hormones on gene expression in Camellia sinensis cv. Shuchazao Leaves. Front. Plant Sci. 2020, 11, 256. [Google Scholar] [CrossRef]
- Sparks, E.; Wachsman, G.; Benfey, P.N. Spatiotemporal signalling in plant development. Nat. Rev. Genet. 2013, 14, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The Multifaceted Roles of HY5 in Plant Growth and Development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Murchie, E.H.; Niyogi, K.K. Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 2011, 155, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.E.; Sullivan, S.; Hermanowicz, P.; Petersen, J.; Diaz-Ramos, L.A.; Hoey, D.J.; Łabuz, J.; Christie, J.M. Engineering the phototropin photocycle improves photoreceptor performance and plant biomass production. Proc. Natl. Acad. Sci. USA 2019, 116, 12550–12557. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, G.; Cao, C.; Liu, P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021, 2, 100231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Zubo, Y.O.; Yamburenko, M.V.; Kusnetsov, V.V.; Börner, T. Methyl jasmonate, gibberellic acid, and auxin affect transcription and transcript accumulation of chloroplast genes in barley. J. Plant Physiol. 2011, 168, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Xu, Y.; Xiong, B.; Dai, L.; Huang, S.; Dong, T.; Sun, G.; Liao, L.; Deng, Q.; Wang, X. Effects of exogenous methyl jasmonate on the synthesis of endogenous jasmonates and the regulation of photosynthesis in citrus. Physiol. Plant. 2020, 170, 398–414. [Google Scholar] [CrossRef]
- Ma, X.; Zheng, J.; Zhang, X.; Hu, Q.; Qian, R. Salicylic acid alleviates the adverse effects of salt stress on Dianthus superbus (Caryophyllaceae) by activating photosynthesis, protecting morphological structure, and enhancing the antioxidant system. Front. Plant Sci. 2017, 8, 600. [Google Scholar] [CrossRef]
- Cheng, X.; Fang, T.; Zhao, E.; Zheng, B.; Huang, B.; An, Y.; Zhou, P. Protective roles of salicylic acid in maintaining integrity and functions of photosynthetic photosystems for alfalfa (Medicago sativa L.) tolerance to aluminum toxicity. Plant Physiol. Biochem. 2020, 155, 570–578. [Google Scholar] [CrossRef]
- Dogra, V.; Kim, C. Chloroplast protein homeostasis is coupled with retrograde signaling. Plant Signal. Behav. 2019, 14, 1656037. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Verdeja, T.; Strand, Å. Retrograde signals navigate the path to chloroplast development. Plant Physiol. 2018, 176, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, X.; Yan, W.; Zhang, Y.; Song, P.; Guo, Y.; Xie, K.; Hu, J.; Hou, J.; Wu, Y. Melon yellow-green plant (Cmygp) encodes a Golden2-like transcription factor regulating chlorophyll synthesis and chloroplast development. Theor. Appl. Genet. 2023, 136, 66. [Google Scholar] [CrossRef] [PubMed]
- Gang, H.; Li, R.; Zhao, Y.; Liu, G.; Chen, S.; Jiang, J. Loss of GLK1 transcription factor function reveals new insights in chlorophyll biosynthesis and chloroplast development. J. Exp. Bot. 2019, 70, 3125–3138. [Google Scholar] [CrossRef] [PubMed]
- Strand, Å.; Asami, T.; Alonso, J.; Ecker, J.R.; Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 2003, 421, 79–83. [Google Scholar] [CrossRef]
- Mochizuki, N.; Tanaka, R.; Tanaka, A.; Masuda, T.; Nagatani, A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 15184–15189. [Google Scholar] [CrossRef] [PubMed]
- Moulin, M.; McCormac, A.C.; Terry, M.J.; Smith, A.G. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. USA 2008, 105, 15178–15183. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Chen, M.; Fu, X.; Liu, J.; Ye, T.; Hou, S.; Huang, Y.; Yuan, B.; Wu, Y.; Feng, Y. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC–ESI-Q-TOF-MS analysis. J. Chromatogr. B 2012, 905, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Schulze, S.K.; Kanwar, R.; Gölzenleuchter, M.; Therneau, T.M.; Beutler, A.S. SERE: Single-parameter quality control and sample comparison for RNA-Seq. BMC Genom. 2012, 13, 524. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Yang, J.; Hu, K.; An, B.; Deng, X.; Li, Y. Reliable selection and holistic stability evaluation of reference genes for rice under 22 different experimental conditions. Appl. Biochem. Biotechnol. 2016, 179, 753–775. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).