Genome Analysis of Thinopyrum intermedium and Its Potential Progenitor Species Using Oligo-FISH
Abstract
:1. Introduction
2. Results
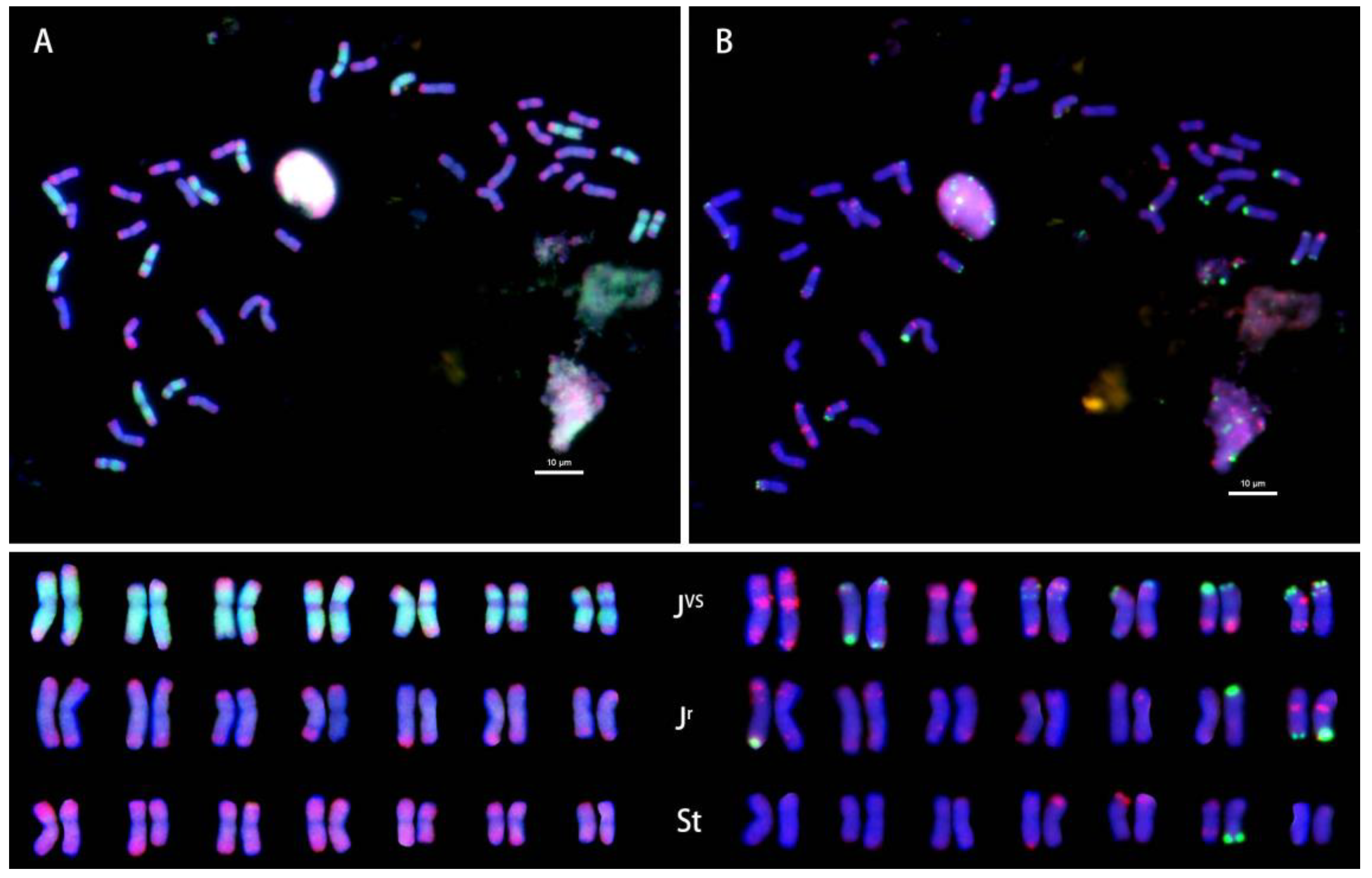
2.1. OligoFISH of Ten Accessions of IWG
2.2. OligoFISH of Three Tetraploid Species of Thinopyrum Genus
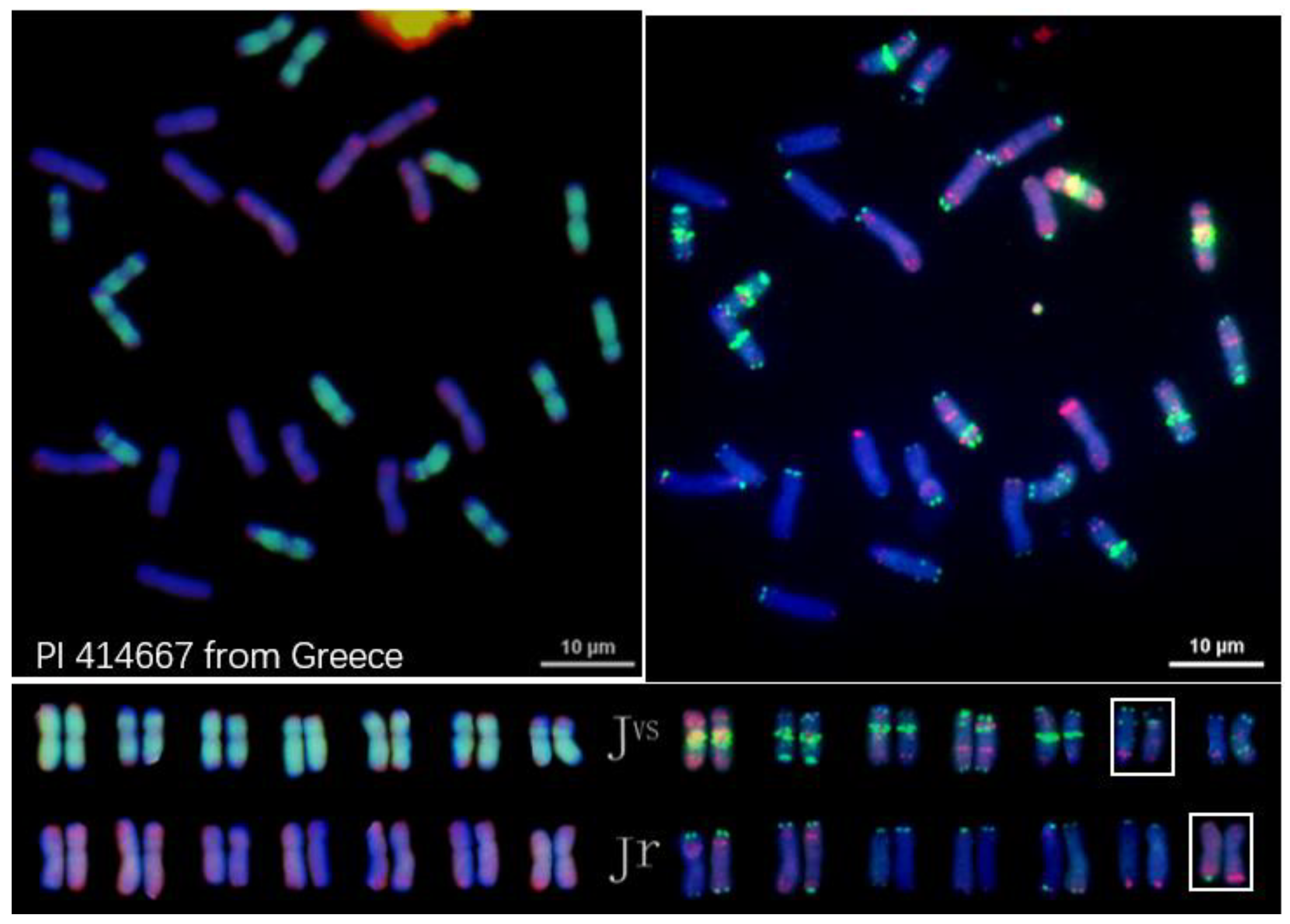
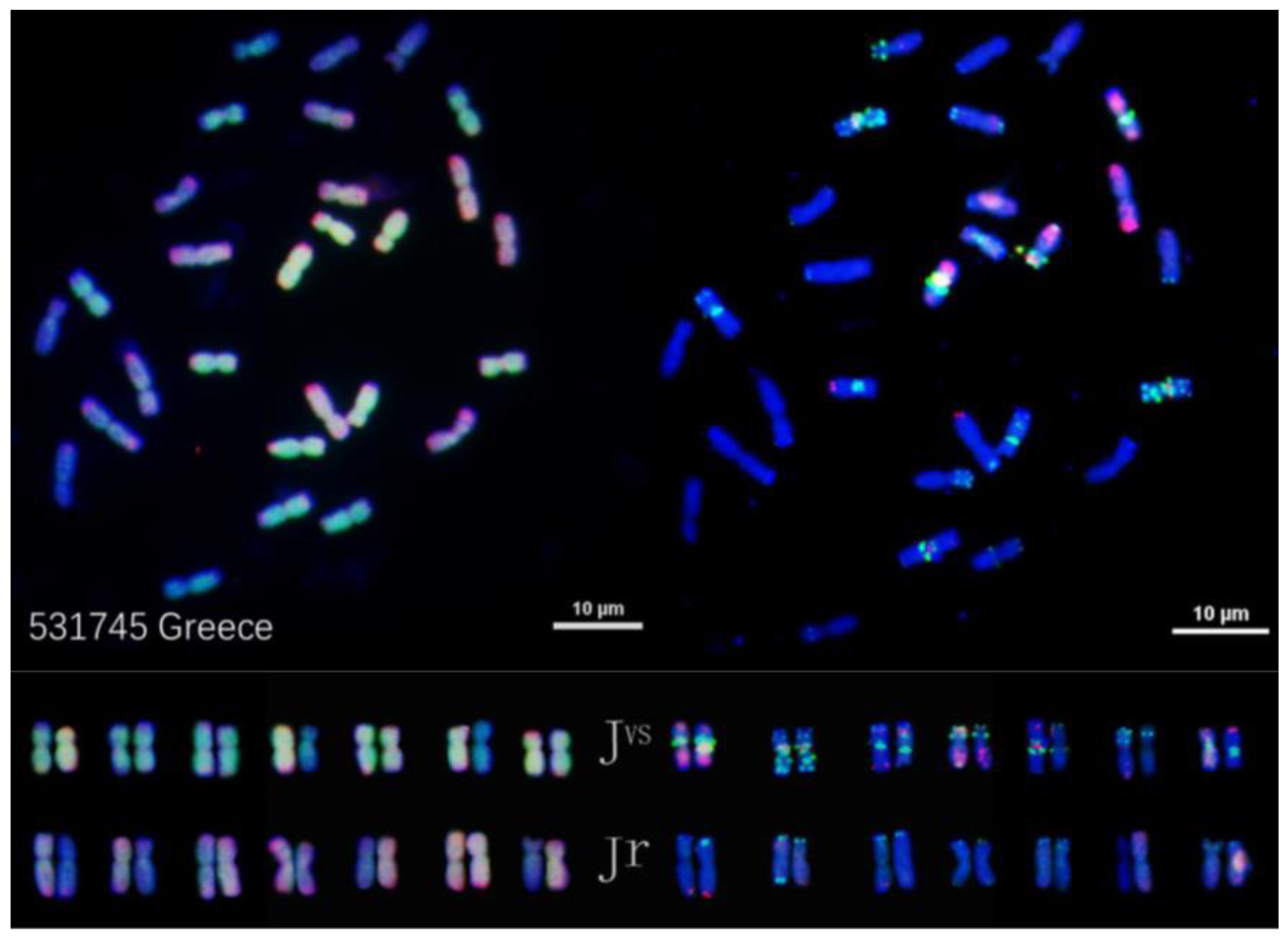
2.3. OligoFISH of Four Diploid Species That Were Implicated as Progenitors of IWG
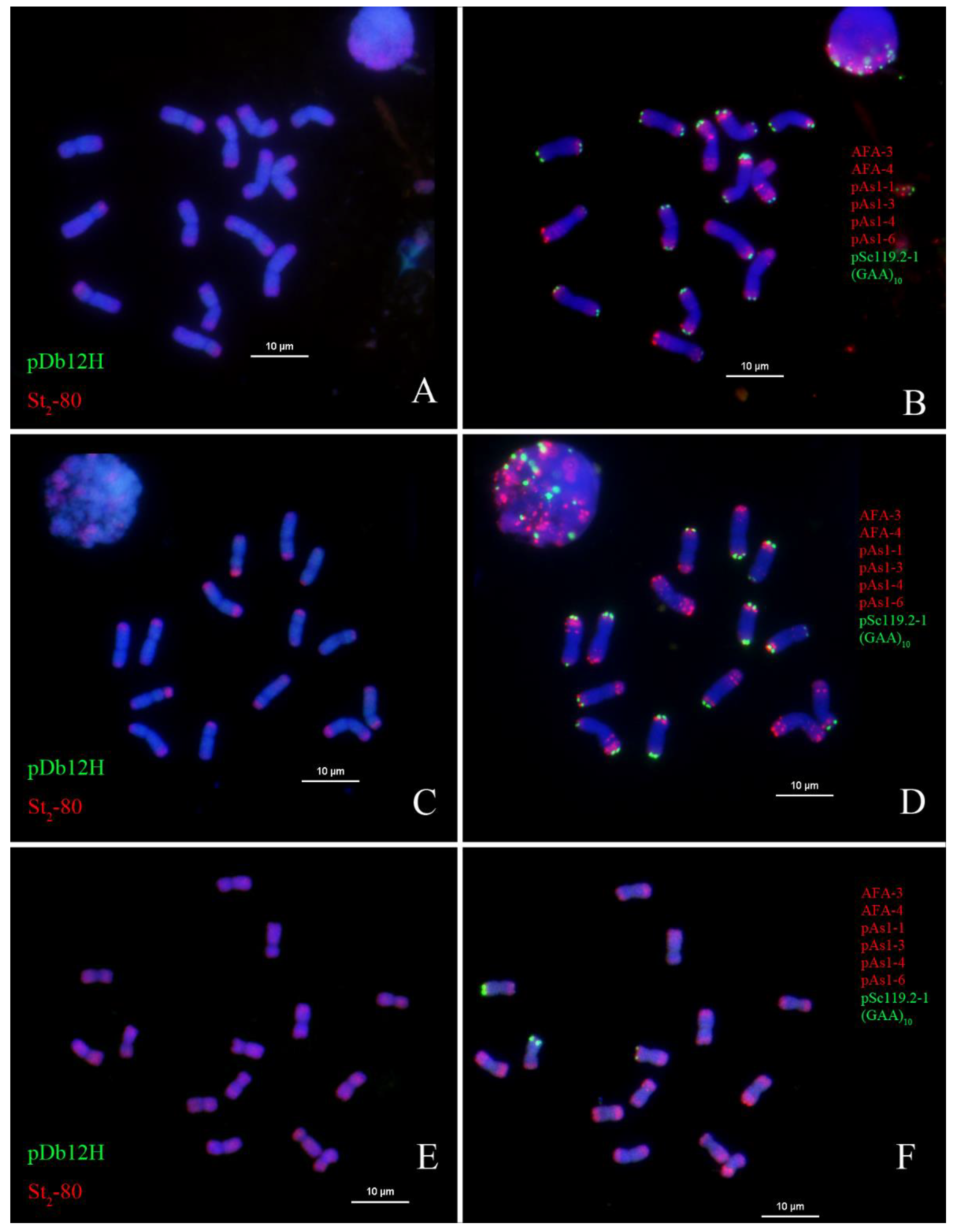
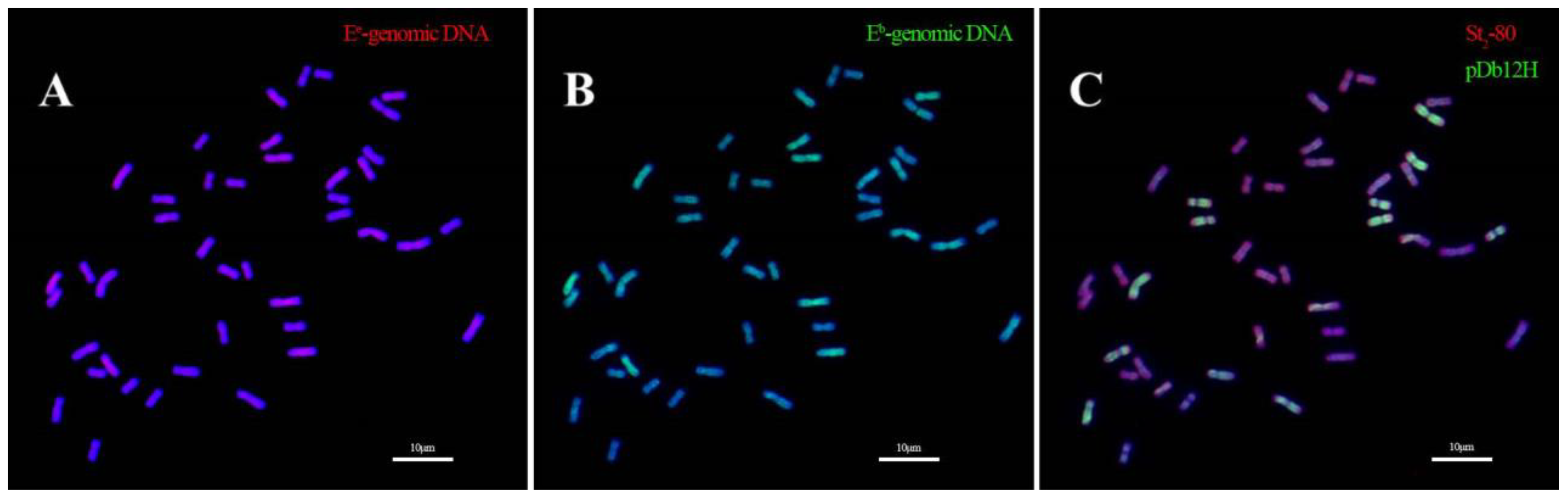
2.4. In Situ Hybridization of Th. intermedium Using Genomic DNA of Th. elongatum and Th. bessarabicum, and Oligo Probes pDb12H and St2-80
2.5. In Situ Hybridization of Th. intermedium Using Genomic DNA of Dasypyrum villosum and Oligo Probes pDb12H and St2-80
3. Discussion
3.1. Prior Studies on Thinopyrum intermedium and Related Species
3.2. Current Studies on Thinopyrum intermedium and Related Species
3.3. Future Studies on Thinopyrum intermedium and Related Species Needed
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extraction and Probe Preparation
4.3. Chromosome Preparation and GISH, FISH Protocol
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kumar, A.; Choudhary, A.; Kaur, H.; Mehta, S. A walk towards Wild grasses to unlock the clandestine of gene pools for wheat improvement: A review. Plant Stress 2022, 3, 100048. [Google Scholar] [CrossRef]
- Altendorf, K.R.; DeHaan, L.R.; Heineck, G.C.; Zhang, X.; Anderson, J.A. Floret site utilization and reproductive tiller number are primary components of grain yield in intermediate wheatgrass spaced plants. Crop Sci. 2021, 61, 1073–1088. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Kazi, A.G.; Dundas, I.; Rasheed, A.; Ogbonnaya, F.; Kishii, M.; Bonnett, D.; Wang, R.R.C.; Xu, S.; Chen, P.; et al. Genetic diversity for wheat improvement as a conduit to food security. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 122, pp. 179–258. [Google Scholar]
- Li, W.; Danilova, T.; Rouse, M.N.; Bowden, R.L.; Friebe, B.; Gill, B.S.; Pumphrey, M.O. Development and characterization of a compensating wheat-Thinopyrum intermedium Robertsonian translocation with Sr44 resistance to stem rust (Ug99). Theor. Appl. Genet. 2013, 126, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Ren, Y.K.; Bao, Y.G.; Nan, H.; Tang, Z.H.; Guo, Q.; Niu, Y.Q.; Yan, W.Z.; Sun, Y.; Li, H.J. Assessment of Resistance to Cereal Cyst Nematode, Stripe Rust, and Powdery Mildew in Wheat-Thinopyrum intermedium Derivatives and Their Chromosome Composition. Plant Disease 2021, 105, 2898–2906. [Google Scholar] [CrossRef]
- Ivanova, Y.N.; Rosenfread, K.K.; Stasyuk, A.I.; Skolotneva, E.; Silkova, O.G. Raise and characterization of a bread wheat hybrid line (Tulaykovskaya 10 × Saratovskaya 29) with chromosome 6Agi2 introgressed from Thinopyrum intermedium. Vavilov J. Genet. Breed. 2021, 25, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; DeHaan, L.R.; Higgins, L.A.; Markowski, T.W.; Wyse, D.L.; Anderson, J.A. New insights into high-molecular-weight glutenin subunits and subgenomes of the perennial crop Thinopyrum intermedium (Triticeae). J. Cereal Sci. 2014, 59, 203–210. [Google Scholar] [CrossRef]
- Marti, A.; Bock, J.; Pagani, M.; Ismail, B.; Seetharaman, K. Structural characterization of proteins in wheat flour doughs enriched with intermediate wheatgrass (Thinopyrum intermedium) flour. Food Chem. 2016, 194, 994–1002. [Google Scholar] [CrossRef]
- Crain, J.; Larson, S.R.; Dorn, K.M.; Dehaan, L.; Poland, J. Genetic architecture and QTL selection response for Kernza perennial grain domestication traits. Theor. Appl. Genet. 2022, 135, 2769–2784. [Google Scholar] [CrossRef]
- Locatelli, A.; Gutierrez, L.; Picasso Risso, V.D. Vernalization requirements of Kernza intermediate wheatgrass. Crop Sci. 2022, 62, 524–535. [Google Scholar] [CrossRef]
- de Oliveira, G.; Brunsell, N.A.; Sutherlin, C.E.; Crews, T.E.; DeHaan, L.R. Energy, water and carbon exchange over a perennial Kernza wheatgrass crop. Agric. For. Meteorol. 2018, 249, 120–137. [Google Scholar] [CrossRef]
- Dewey, D.R. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In Gene Manipulation in Plant Improvement; Gustafson, J.P., Ed.; Plenum Publishing Corporation: New York, NY, USA, 1984; pp. 209–279. [Google Scholar]
- Wang, R.R.-C.; von Bothmer, R.; Dvorák, J.; Fedak, G.; Linde-Laursen, I.; Muramatsu, M. Genome symbols in the Triticeae (Poaceae). In Proceedings of the 2nd International Triticeae Symposium, Logan, UT, USA, 20–24 June 1994; Wang, R.R.-C., Jensen, K.B., Jaussi, C., Eds.; Utah State University Publication Design and Production: Logan, UT, USA, 1995; pp. 29–34. [Google Scholar]
- Wang, R.R.-C.; Zhang, X.-Y. Characterization of the translocated chromosome using fluorescence in situ hybridization and random amplified polymorphic DNA on two Triticum aestivum-Thinopyrum intermedium translocation lines resistant to wheat streak mosaic or barley yellow dwarf virus. Chromosome Res. 1996, 4, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Conner, R.L.; Laroche, A.; Thomas, J.B. Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome 1998, 41, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Kishii, M.; Wang, R.R.-C.; Tsujimoto, H. GISH analysis revealed new aspect of genomic constitution of Thinopyrum intermedium. Czechoslov. J. Genet. Plant Breed. 2005, 41, 92–95. [Google Scholar] [CrossRef]
- Mahelka, V.; Kopecký, D.; Paštová, L. On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol. Biol. 2011, 11, 127. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.X.; Yang, Z.J.; Fu, S.L.; Yang, M.Y.; Li, G.R.; Zhang, H.Q.; Tan, F.Q.; Ren, Z. A new long terminal repeat (LTR) sequence allows the identify J genome from Js and St genomes of Thinopyrum intermedium. J. Appl. Genet. 2011, 52, 31–33. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.L.; Bai, L.L.; Fu, S.L.; Yin, W.B.; Zhang, Y.X.; Chen, Y.H.; Wang, R.R.-C.; Zhang, X.Q.; Han, F.P.; Hu, Z.M. Microdissection and chromosome painting of the alien chromosome in an addtition line of wheat-Thinopyrum intermedium. PLoS ONE 2013, 8, e72564. [Google Scholar] [CrossRef]
- Wang, L.; Shi, Q.; Su, H.; Wang, Y.; Sha, L.; Fan, X.; Kang, H.; Zhang, H.; Zhou, Y. St2-80: A new FISH marker for St genome and genome analysis in Triticeae. Genome 2017, 60, 553–563. [Google Scholar] [CrossRef]
- Yang, Z.-J.; Liu, C.; Feng, J.; Li, G.-R.; Zhou, J.-P.; Deng, K.-J.; Ren, Z.-L. Studies on genome relationship and species-specific PCR marker for Dasypyrum breviaristatumin Triticeae. Hereditas 2006, 143, 47–54. [Google Scholar] [CrossRef]
- Liu, C.; Yang, Z.J.; Jia, J.Q.; Li, G.R.; Zhou, J.P.; Ren, Z.L. Genomic distribution of a long end repeat (LTR) Sabrina-like retrotransposon in Triticeae species. Cereal Res. Commun. 2009, 37, 363–372. [Google Scholar] [CrossRef]
- Wang, R.R.-C.; Larson, S.R.; Jensen, K.B.; Bushman, B.S.; DeHaan, L.R.; Wang, S.; Yan, X. Genome evolution of intermediate wheatgrass as revealed by EST-SSR markers developed from its three progenitor diploid species. Genome 2015, 58, 63–70. [Google Scholar] [CrossRef]
- Cseh, A.; Yang, C.Y.; Hubbart-Edwards, S.; Scholefield, D.; Ashling, S.S.; Burridgem, A.J.; Wilkinson, P.A.; King, I.P.; King, J.; Grewal, S. Development and validation of an exome-based SNP marker set for identification of the St, Jr and Jvs genomes of Thinopyrym intermedium in a wheat background. Theor. Appl. Genet. 2019, 132, 1555–1570. [Google Scholar] [CrossRef] [PubMed]
- Divashuk, M.G.; Karlov, G.I.; Kroupin, P.Y. Copy Number Variation of Transposable Elements in Thinopyrum intermedium and Its Diploid Relative Species. Plants 2019, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-M.; Lee, B.S.; Mammadov, A.C.; Koo, B.-C.; Mott, I.W.; Wang, R.R.-C. CAPS markers specific to Eb, Ee and R genomes in the tribe Triticeae. Genome 2007, 50, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Kantarski, T.; Larson, S.; Zhang, X.; DeHaan, L.; Borevitz, J.; Anderson, J.; Poland, J. Development of the first consensus genetic map of intermediate wheatgrass (Thinopyrum intermedium) using genotyping-by-sequencing. Theor. Appl. Genet. 2017, 130, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.; DeHaan, L.; Poland, J.; Zhang, X.; Dorn, K.; Kantarski, T.; Anderson, J.; Schmutz, J.; Grimwood, J.; Jenkins, J.; et al. Genome mapping of quantitative trait loci (QTL) controlling domestication traits of intermediate wheatgrass (Thinopyrum intermedium). Theor. Appl. Genet. 2019, 132, 2325–2351. [Google Scholar] [CrossRef] [PubMed]
- DeHaan, L.; Christians, M.; Crain, J.; Poland, J. Development and evolution of an intermediate wheatgrass domestication program. Sustainability 2018, 10, 1499. [Google Scholar] [CrossRef]
- Qiao, L.; Liu, S.; Li, J.; Li, S.; Yu, Z.; Liu, C.; Li, X.; Liu, J.; Ren, Y.; Zhang, P.; et al. Development of Sequence-Tagged Site Marker Set for Identification of J, JS, and St Sub-genomes of Thinopyrum intermedium in Wheat Background. Front. Plant Sci. 2021, 12, 685216. [Google Scholar] [CrossRef]
- Gao, L.; Ma, Q.; Liu, Y.; Xin, Z.; Zhang, Z. Molecular characterization of the genomic region harboring the BYDV-resistance gene Bdv2 in wheat. J. Appl. Genet. 2009, 50, 89–98. [Google Scholar] [CrossRef]
- Ceoloni, C.; Kuzmanovi’c, L.; Gennaro, A.; Forte, P.; Giorgi, D.; Grossi, M.R.; Bitti, A. Genomes, Chromosomes and Genes of the Wheatgrass Genus Thinopyrum: The Value of their Transfer into Wheat for Gains in Cytogenomic Knowledge and Sustainable Breeding. In Genomics of Plant Genetic Resources; Tuberosa, R., Graner, A., Frison, E., Eds.; Springer: Dordrecht, The Netherlands, 2014; Chapter 14; pp. 333–358. [Google Scholar]
- Guo, X.; Huang, Y.; Wang, J.; Fu, S.; Wang, C.; Wang, M.; Zhou, C.; Hu, X.; Wang, T.; Yang, W.; et al. Development and cytological characterization of wheat– Thinopyrum intermedium translocation lines with novel stripe rust resistance gene. Front. Plant Sci. 2023, 14, 1135321. [Google Scholar] [CrossRef]
- Wang, R.R.-C.; Lu, B.R. Biosystematics and evolutionary relationships of perennial Triticeae species revealed by genomic analyses. J. Syst. Evol. 2014, 52, 697–705. [Google Scholar] [CrossRef]
- Liu, Z.W.; Wang, R.R.-C. Genome analysis of Elytrigia caespitosa, Lophopyrum nodosum, Pseudoroegneria geniculate ssp. scythica, and Thinopyrum intermedium. Genome 1993, 36, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-W.; Wang, R.R.-C. Genome analysis of Thinopyrum junceiforme and T. sartorii. Genome 1992, 35, 758–764. [Google Scholar] [CrossRef]
- Pienaar, R.d.V.; Littlejohn, G.M.; Sears, E.R. Genomic relationships in Thinopyrum. S. Afr. J. Bot. 1988, 54, 541–550. [Google Scholar] [CrossRef]
- Jensen, K.B.; Zhang, Y.F.; Dewey, D.R. Mode of pollination of perennial species of the Triticeae in relation to genomically defined genera. Can. J. Plant Sci. 1990, 70, 215–225. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Dong, Y.C.; Wang, R.R.-C. Characterization of genomes and chromosomes in partial amphiploids of the hybrid Triticum aestivum ×Thinopyrum ponticum by in situ hybridization, isozyme analysis, and RAPD. Genome 1996, 39, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Koul, A.; Petroski, R.; Ouellet, T.; Fedak, G.; Dong, Y.S.; Wang, R.R.-C. Molecular verification and characterization of BYDV-resistant germplasms derived from hybrids of wheat with Thinopyrum ponticum and Th. intermedium. Theor. Appl. Genet. 1996, 93, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q. Detection of alien chromatin introgression from Thinopyrum into wheat using S genomic DNA as a probe—A landmark approach for Thinopyrum genome research. Cytogenet. Genome Res. 2005, 109, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S.; Morishita, M. Genome relationship in the genus of Dasypyrum (Gramineae). Hereditas 2001, 135, 101–110. [Google Scholar] [CrossRef]
- Li, G.; Gao, D.; Zhang, H.; Li, J.; Wang, H.; La, S.; Ma, J.; Yang, Z. Molecular cytogenetic characterization of Dasypyrum breviaristatum chromosomes in wheat background revealing the genomic divergence between Dasypyrum species. Mol. Cytogenet. 2016, 9, 6. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Q.; Wang, S.; Langham, M.A.; Singh, D.; Bowden, R.L.; Xu, S.S. Development and characterization of wheat-sea wheatgrass (Thinopyrum junceiforme) amphiploids for biotic stress resistance and abiotic stress tolerance. Theor. Appl. Genet. 2019, 132, 163–175. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, H.; Yang, E.; Li, G.; Yang, Z. Precise Identification of Chromosome Constitution and Rearrangements in Wheat–Thinopyrum intermedium Derivatives by ND-FISH and Oligo-FISH Painting. Plants 2022, 11, 2109. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Han, Y.S.; Xiao, H.; Zou, B.C.; Wu, D.D.; Sha, L.A.; Yang, C.R.; Liu, S.Q.; Cheng, Y.R.; Wang, Y.; et al. Chromosome-specific painting in Thinopyrum species using bulked oligonucleotides. Theor. Appl. Genet. 2023, 136, 8. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhuang, L.; Wang, Y.; Yuan, L.; Wang, Q.; Wang, D.; Dawadondup, X.; Tan, L.; Shen, J.; Xu, H.; et al. Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 2017, 60, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Kato, A. Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem. 1999, 74, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Han, F.P.; Liu, B.; Fedak, G.; Liu, Z.H. Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 2004, 109, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Xing, P.; Bao, Y.; Ren, M.; Liu, S.; Wang, Y.; Li, X.; Wang, H. Chromosome pairing in hybrid progeny between Triticum aestivum and Elytrigia elongata. Front. Plant Sci. 2017, 8, 2161. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Xing, P.; Qi, X.; Bao, Y.; Wang, H.; Wang, R.R.-C.; Li, X.F. Characterization of chromosome constitution in three wheat—Thinopyrum intermedium amphiploids revealed frequent rearrangement of alien and wheat chromosomes. BMC Plant Biol. 2021, 21, 129. [Google Scholar] [CrossRef]



| Species | ID | Chr Number | Origin | Note |
|---|---|---|---|---|
| Thinopyrum intermedium (Host) Barkworth & D. R. Dewey | PI 109219 | 42 | District of Columbia, United States | |
| PI 206259 | 42 | Turkey | ||
| PI 210990 | 42 | Afghanistan | ||
| PI 228386 | 42 | Iran | ||
| PI 249146 | 42 | Portugal | ||
| PI 297876 | 42 | Former, Soviet Union | ||
| PI 325190 | 42 | Stavropol, Russian Federation | ||
| PI 401204 | 42 | Iran | ||
| PI 440036 | 42 | Kazakhstan | ||
| PI 634290 | 42 | Krym, Ukraine | ||
| Th. junceiforme (A. & D. Löve) A. Löve | PI 414667 | 28 | Greece | listed as Thinopyrum junceum (L.) Á. Löve |
| Th. sartorii (Bioss. & Heldr.) Á. Löve | PI 531745 | 28 | Greece | |
| Th. scirpeum (C. Presl) D. R. Dewey | PI 531750 | 28 | Greece | |
| Th. bessarabicum (Savul. & Rayss) A. Löve | PI 531712 | 14 | Estonia | |
| Th. elongatum (Host) D. R. Dewey | PI 340063 | 14 | Turkey | |
| Pseudoroegneria spicata Pursh) Á. Löve | PI 563869 | 14 | Oregon, USA | |
| Dasypyrum villosum (L.) Candargy | 14 | From X-F Li’s collection | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, F.; Liang, S.; Xing, P.; Bao, Y.; Wang, R.R.-C.; Li, X. Genome Analysis of Thinopyrum intermedium and Its Potential Progenitor Species Using Oligo-FISH. Plants 2023, 12, 3705. https://doi.org/10.3390/plants12213705
Qi F, Liang S, Xing P, Bao Y, Wang RR-C, Li X. Genome Analysis of Thinopyrum intermedium and Its Potential Progenitor Species Using Oligo-FISH. Plants. 2023; 12(21):3705. https://doi.org/10.3390/plants12213705
Chicago/Turabian StyleQi, Fei, Shuang Liang, Piyi Xing, Yinguang Bao, Richard R.-C. Wang, and Xingfeng Li. 2023. "Genome Analysis of Thinopyrum intermedium and Its Potential Progenitor Species Using Oligo-FISH" Plants 12, no. 21: 3705. https://doi.org/10.3390/plants12213705
APA StyleQi, F., Liang, S., Xing, P., Bao, Y., Wang, R. R.-C., & Li, X. (2023). Genome Analysis of Thinopyrum intermedium and Its Potential Progenitor Species Using Oligo-FISH. Plants, 12(21), 3705. https://doi.org/10.3390/plants12213705







