The Past, Present, and Future of Wheat Dwarf Virus Management—A Review
Abstract
1. Introduction
2. Wheat Dwarf Virus (WDV)
2.1. Classification and Genomic Organization of WDV
2.2. Life Cycle of the Virus
2.3. Phylogenetics
3. Wheat Dwarf Disease (WDD)
3.1. History
3.2. Host Range
3.3. Symptoms of WDD
4. WDV and Its Vector
4.1. Taxonomy and Virus Transmission of P. alienus
4.2. Morphology of P. alienus
4.3. Life Cycle of P. alienus
4.4. Process of Virus Transmission
4.5. Host Range and Wild Reservoirs
4.6. Studies of Insect-Plant Interactions
5. Management of WDD, Its Vector and Virus
6. Resistance Research and Status Quo in Wheat
| Time | Event | Reference |
|---|---|---|
| 1982 | Report: WDV shows tendencies to prefer different wheat varieties. | [288] |
| 2000 | Screening: Description of five Russian varieties as well as ten Slovakian and Czech varieties with moderate yield reduction after WDV infection. | [147] |
| 2005 | Screening: Description of the Czech winter wheat varieties ‘Banquet’ and ‘Svitava’ with reduced virus titer, moderate susceptibility, and yield reduction. | [148] |
| 2010 | Screening: Description of the Hungarian winter wheat varieties ‘Mv Dalma’ and ‘Mv Vekni’) as partially resistant varieties. | [289] |
| 2015 | Screening: Proof of WDV tolerance of accessions of the species Aeg. Tauschii, Aeg. Cylindrical, Aeg. Searsii, and T. spelta. | [277] |
| 2022 | Screening: Identification of 19 sources of WDV resistance with lower infection rates than ‘MV Vekni,’ including di-, tetra-, and hexaploid genebank wheat varieties as well as the winter wheat variety ‘Fisht.’ | [275] |
| 2022 | Genome-wide association study: Detection of 35 putative QTL for partial WDV resistance on chromosomes 1B, 1D, 2B, 3A, 3B, 4A, 4B, 5A, 6A, 7A, and 7B. | [275] |
| 2022 | QTL analysis: Identification of two significant QTL on chromosome 6A in the variety ‘Mv Vekni.’ | [292] |
| 2023 | Transcriptome analysis: A study of changes in resistant wheat genotypes ‘Svitava’ and ‘Fengyou 3’ compared to susceptible cultivar ‘Akteur’ after WDV infection. | [294] |
| 2023 | QTL study: Identification of a QTL on chromosome 6A in the Dutch experimental line SVP-75360 and a QTL on chromosome 1B of line P1361. | [314] |
| 2024 | QTL study: Identification of QTL in the winter wheat variety Fisht. |
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Buck, K.W. Geminiviruses (Geminiviridae). In Encyclopedia of Virology; Granoff, A., Webster, R.G., Eds.; Academic Press: Cambridge, MA, USA, 1999; pp. 597–606. [Google Scholar] [CrossRef]
- Canto, T.; Aranda, M.A.; Fereres, A. Climate change effects on physiology and population processes of hosts and vectors that influence the spread of hemipteran-borne plant viruses. Glob. Chang. Biol. 2009, 15, 1884–1894. [Google Scholar] [CrossRef]
- Habekuß, A.; Riedel, C.; Schliephake, E.; Ordon, F. Breeding for resistance to insect-transmitted viruses in barley—An emerging challenge due to global warming. J. Für Kult. 2009, 61, 53–61. [Google Scholar] [CrossRef]
- Roos, J.; Hopkins, R.; Kvarnheden, A.; Dixelius, C. The impact of global warming on plant diseases and insect vectors in Sweden. Eur. J. Plant Pathol. 2011, 129, 9–19. [Google Scholar] [CrossRef]
- Ziesche, T.M.; Bell, J.; Ordon, F.; Schliephake, E.; Will, T. Long-term monitoring of insects in agricultural landscapes. Mitteilungen Der DGaaE 2020, 22, 101–106. [Google Scholar]
- Barnett, O.W.; Main, C.E. Plant Virus Disease—Economic Aspects. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 1999; pp. 1318–1326. [Google Scholar] [CrossRef]
- Waterworth, H.E.; Hadidi, A. Economic Losses due to Plant Viruses. In Plant Virus Disease control; APS Press: St. Paul, MN, USA, 1998. [Google Scholar]
- Fraser, R.S.S. Plant Resistance to Viruses|Natural Resistance. In Encyclopedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 1300–1307. [Google Scholar] [CrossRef]
- van Regenmortel, M.H.; Fauquet, C.M.; Bishop, D.H.; Carstens, E.B.; Estes, M.K.; Lemon, S.M.; Maniloff, J.; Mayo, M.A.; McGeoch, D.J.; Pringle, C.R.; et al. Virus Taxonomy: Classification and Nomenclature of Viruses. In Seventh Report of the International Committee on Taxonomy of Viruses; Academic Press: New York, NY, USA, 2000. [Google Scholar]
- Vacke, J. Wheat dwarf virus disease. Biol. Plant 1961, 3, 228–233. [Google Scholar] [CrossRef]
- Mehner, S.; Manurung, B.; Gruntzig, M.; Habekuss, A.; Witsack, W.; Fuchs, E. Investigations into the ecology of the Wheat dwarf virus (WDV) in Saxony-Anhalt, Germany. J. Plant Dis. Prot. 2003, 110, 313–323. [Google Scholar]
- Manurung, B.; Witsack, W.; Mehner, S.; Gruntzig, M.; Fuchs, E. Studies on biology and population dynamics of the leafhopper Psammotettix alienus Dahlb. (Homoptera: Auchenorrhyncha) as vector of Wheat dwarf virus (WDV) in Saxony-Anhalt, Germany. J. Plant Dis. Prot. 2005, 112, 497–507. [Google Scholar]
- Vacke, J. Host plants range and symptoms of wheat dwarf virus. Věd Pr Výz Ust. Rostl Výroby Praha-Ruzyně 1972, 17, 151–162. [Google Scholar]
- MacDowell, S.W.; Macdonald, H.; Hamilton, W.D.O.; Coutts, R.H.A.; Buck, K.W. The nucleotide sequence of cloned wheat dwarf virus DNA. EMBO J. 1985, 4, 2173–2180. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, H.; Coutts, R.H.A.; Buck, K.W. Characterization of a Subgenomic DNA Isolated from Triticum Aestivum Plants Infected with Wheat Dwarf. J. Gen. Virol. 1988, 69, 1339–1344. [Google Scholar] [CrossRef]
- Schalk, H.J.; Matzeit, V.; Schiller, B.; Schell, J.; Gronenborn, B. Wheat dwarf virus, a geminivirus of graminaceous plants needs splicing for replication. EMBO J. 1989, 8, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Lindblad, M.; Waern, P. Correlation of wheat dwarf incidence to winter wheat cultivation practices. Agric. Ecosyst. Environ. 2002, 92, 115–122. [Google Scholar] [CrossRef]
- Lemmetty, A.; Huusela-Veistola, E. First Report of Wheat dwarf virus in Winter Wheat in Finland. Plant Dis. 2005, 89, 912. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guan, Y.; Wu, L.; Guan, X.; Cai, W.; Huang, J.; Dong, W.; Zhang, B. Changing Lengths of the Four Seasons by Global Warming. Geophys. Res. Lett. 2021, 48, e2020GL091753. [Google Scholar] [CrossRef]
- Lindsten, K.; Lindsten, B.; Abdelmoeti, M.; Junti, N. Purification and some properties of wheat dwarf virus. In Proceedings of the 3rd Conference on Virus Diseases of Gramineae in Europe, Rothamsted, UK, 28–30 May 1980; pp. 27–31. [Google Scholar]
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, P.; Golden, M.; Akram, M.; Naimuddin, N.N.; Fernandez, E.; Granier, M.; Rebelo, A.G.; Peterschmitt, M.; Martin, D.P.; Roumagnac, P. Identification and characterisation of a highly divergent geminivirus: Evolutionary and taxonomic implications. Virus Res. 2013, 177, 35–45. [Google Scholar] [CrossRef]
- Varsani, A.; Navas-Castillo, J.; Moriones, E.; Hernández-Zepeda, C.; Idris, A.; Brown, J.K.; Murilo Zerbini, F.; Martin, D.P. Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch. Virol. 2014, 159, 2193–2203. [Google Scholar] [CrossRef]
- Agrios, G.N. Plant Pathology, 3rd ed.; Academic Press: New York, NY, USA, 1988; pp. 3–39. [Google Scholar] [CrossRef]
- Matzeit, V. Wheat Dwarf Virus—Ein Geminivirus Monokotyledoner Pflanzen-DNA-Sequenz, Replikation und Einsatz Seines Genoms zur Amplifikation und Expression Fremder Gene. Ph.D. Thesis, Universität zu Köln, Köln, Germany, 1988. [Google Scholar]
- Zhang, W.; Olson, N.H.; Baker, T.S.; Faulkner, L.; Agbandje-McKenna, M.; Boulton, M.I.; Davies, J.W.; McKenna, R. Structure of the Maize Streak Virus Geminate Particle. Virology 2001, 279, 471–477. [Google Scholar] [CrossRef]
- Boulton, M.I. Functions and interactions of mastrevirus gene products. Physiol. Mol. Plant Pathol. 2002, 60, 243–255. [Google Scholar] [CrossRef]
- Drews, G.; Adam, G.; Heinze, C. Molekulare Pflanzenvirologie; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar] [CrossRef]
- Adejare, G.O.; Coutts, R.H.A. The Isolation and Characterisation of a Virus from Nigerian Cassava Plants Affected by the Cassava Mosaic Disease, and Attempted Transmission of the Disease. J. Phytopathol. 1982, 103, 198–210. [Google Scholar] [CrossRef]
- Harrison, B.D. Advances in Geminivirus Research. Annu. Rev. Phytopathol. 1985, 23, 55–82. [Google Scholar] [CrossRef]
- Damsteegt, V.D.; Igwegbe, E.C.K. Epidemiology and Control of Maize streak disease. In Plant Virus Disease Control; APS Press: St. Paul, MN, USA, 1998; pp. 484–494. [Google Scholar]
- Moffat, A.S. Geminiviruses Emerge as Serious Crop Threat. Science 1999, 286446, 1835. [Google Scholar] [CrossRef]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Lett, J.-M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Virus Taxonomy Profile: Geminiviridae 2021. J. Gen. Virol. 2021, 1022, 001696. [Google Scholar] [CrossRef]
- Family: Geminiviridae. Available online: https://ictv.global/report/chapter/geminiviridae/geminiviridae (accessed on 12 November 2022).
- Fauquet, C.M.; Bisaro, D.M.; Briddon, R.W.; Brown, J.K.; Harrison, B.D.; Rybicki, E.P.; Stenger, D.C.; Stanley, J. Virology division news: Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 2003, 148, 405–420. [Google Scholar] [CrossRef]
- Muhire, B.; Martin, D.P.; Brown, J.K.; Navas-Castillo, J.; Moriones, E.; Zerbini, F.M.; Rivera-Bustamante, R.; Malathi, V.G.; Briddon, R.W.; Varsani, A. A Genome-Wide Pairwise-Identity-Based Proposal for the Classification of Viruses in the Genus Mastrevirus (Family Geminiviridae). Arch. Virol. 2013, 158, 1411–1424. [Google Scholar] [CrossRef]
- Candresse, T.; Filloux, D.; Muhire, B.; Julian, C.; Galzi, S.; Fort, G.; Bernardo, P.; Daugrois, J.H.; Fernandez, E.; Martin, D.P.; et al. Appearances can be deceptive: Revealing a hidden viral infection with deep sequencing in a plant quarantine context. PLoS ONE 2014, 9, e102945. [Google Scholar] [CrossRef]
- NCBI Virus. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=Mastrevirus,%20taxid:11212 (accessed on 22 September 2023).
- Morris, B.A.M.; Richardson, K.A.; Haley, A.; Zhan, X.; Thomas, J.E. The nucleotide sequence of the infectious cloned DNA component of tobacco yellow dwarf virus reveals features of geminiviruses infecting monocotyledonous plants. Virology 1992, 187, 633–642. [Google Scholar] [CrossRef]
- Gutierrez, C. Geminivirus DNA replication. Mol. Life Sci. CMLS 1999, 56, 313–329. [Google Scholar] [CrossRef]
- Thomas, J.E.; Parry, J.N.; Schwinghamer, M.W.; Dann, E.K. Two novel mastreviruses from chickpea (Cicer arietinum) in Australia. Arch. Virol. 2010, 155, 1777–1788. [Google Scholar] [CrossRef]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Gafni, Y.; Epel, B.L. The role of host and viral proteins in intra- and inter-cellular trafficking of geminiviruses. Physiol. Mol. Plant Pathol. 2002, 60, 231–241. [Google Scholar] [CrossRef]
- Ramsell, J.N.E. Genetic Variability of Wheat Dwarf Virus. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2007. [Google Scholar]
- Woolston, C.J.; Barker, R.; Gunn, H.; Boulton, M.I.; Mullineaux, P.M. Agroinfection and nucleotide sequence of cloned wheat dwarf virus DNA. Plant Mol. Biol. 1988, 11, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, M.; Schalk, H.J.; Gronenborn, B. Identification and characterization of wheat dwarf virus from France using a rapid method for geminivirus DNA preparation. Phytopathology 1995, 851, 1449–1455. [Google Scholar] [CrossRef]
- Dickinson, V.J.; Halder, J.; Woolston, C.J. The Product of Maize Streak Virus ORF V1 Is Associated with Secondary Plasmodesmata and Is First Detected with the Onset of Viral Lesions. Virology 1996, 220, 51–59. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gutierrez, C. Geminiviruses and the plant cell cycle. Plant Mol. Biol. 2000, 43, 763–772. [Google Scholar] [CrossRef]
- Gutierrez, C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000, 19, 792–799. [Google Scholar] [CrossRef]
- Gutierrez, C.; Ramirez-Parra, E.; Mar Castellano, M.; Sanz-Burgos, A.P.; Luque, A.; Missich, R. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 2004, 98, 111–119. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Briddon, R.W.; Martin, D.P.; Owor, B.E.; Donaldson, L.; Markham, P.G.; Greber, R.S.; Varsani, A. A novel species of mastrevirus (family Geminiviridae) isolated from Digitaria didactyla grass from Australia. Arch. Virol. 2010, 155, 1529–1534. [Google Scholar] [CrossRef]
- Hofer, J.M.I.; Dekker, E.L.; Reynolds, H.V.; Woolston, C.J.; Cox, B.S.; Mullineaux, P.M. Coordinate Regulation of Replication and Virion Sense Gene Expression in Wheat Dwarf Virus. Plant Cell 1992, 4, 213–223. [Google Scholar] [CrossRef]
- Morris-Krsinich, B.A.M.; Mullineaux, P.M.; Donson, J.; Boulton, M.I.; Markham, P.G.; Short, M.N.; Davies, J.W. Bidirectional transcription of maize streak virus DNA and identification of the coat protein gene. Nucleic Acids Res. 1985, 130, 7237–7256. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.L.; Woolston, C.J.; Xue, Y.; Cox, B.; Mullineaux, P.M. Transcript mapping reveals different expression strategies for the bicistronic RNAs of the geminivirus wheat dwarf virus. Nucleic Acids Res. 1991, 195, 4075–4081. [Google Scholar] [CrossRef]
- Fenoll, C.; Black, D.M.; Howell, S.H. The intergenic region of maize streak virus contains promoter elements involved in rightward transcription of the viral genome. EMBO J. 1988, 7, 1589–1596. [Google Scholar] [CrossRef]
- Accotto, G.P.; Donson, J.; Mullineaux, P.M. Mapping of Digitaria streak virus transcripts reveals different RNA species from the same transcription unit. EMBO J. 1989, 8, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Mullineaux, P.M.; Guerineau, F.; Accotto, G.-P. Processing of complementary sense RNAs of Digitariastreak virus in its host and in transgenic tobacco. Nucleic Acids Res. 1990, 184, 7259–7265. [Google Scholar] [CrossRef]
- Wright, E.A.; Heckel, T.; Groenendijk, J.; Davies, J.W.; Boulton, M.I. Splicing features in maize streak virus virion- and complementary-sense gene expression. Plant J. 1997, 12, 1285–1297. [Google Scholar] [CrossRef]
- Palmer, K.E.; Rybicki, E.P. The Molecular Biology of Mastreviruses. Adv. Virus Res. 1998, 50, 183–234. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, Q.; Liu, W.; Mar, T.; Wei, T.; Liu, Y.; Wang, X. Localization and Distribution of Wheat dwarf virus in Its Vector Leafhopper, Psammotettix alienus. Phytopathology 2014, 104, 897–904. [Google Scholar] [CrossRef]
- Noueiry, A.O.; Lucas, W.J.; Gilbertson, R.L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 1994, 76, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Boulton, M.I.; Oparka, K.J.; Davies, J.W. Interaction of the movement and coat proteins of Maize streak virus: Implications for the transport of viral DNA. J. Gen. Virol. 2001, 82, 35–44. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Andrew, L.P.; Davies, J.W.; Boulton, M.I. A single amino acid change in the coat protein of Maize streak virus abolishes systemic infection, but not interaction with viral DNA or movement protein. Mol. Plant Pathol. 2001, 2, 223–228. [Google Scholar] [CrossRef]
- Noris, E.; Vaira, A.M.; Caciagli, P.; Masenga, V.; Gronenborn, B.; Accotto, G.P. Amino Acids in the Capsid Protein of Tomato Yellow Leaf Curl Virus That Are Crucial for Systemic Infection, Particle Formation, and Insect Transmission. J. Virol. 1998, 722, 10050–10057. [Google Scholar] [CrossRef]
- Liu, H.; Boulton, M.I.; Thomas, C.L.; Prior, D.A.M.; Oparka, K.J.; Davies, J.W. Maize Streak Virus Coat Protein Is Karyophyllic and Facilitates Nuclear Transport of Viral DNA. Mol. Plant Microb. Interact. 1999, 120, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Kotlizky, G.; Boulton, M.I.; Pitaksutheepong, C.; Davies, J.W.; Epel, B.L. Intracellular and Intercellular Movement of Maize Streak Geminivirus V1 and V2 Proteins Transiently Expressed as Green Fluorescent Protein Fusions. Virology 2000, 274, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Sunter, G.; Bisaro, D.M. Transactivation of Geminivirus AR1 and BR1 Gene Expression by the Viral AL2 Gene Product Occurs at the Level of Transcription. Plant Cell 1992, 40, 1321–1331. [Google Scholar] [CrossRef][Green Version]
- Hong, Y.; Saunders, K.; Hartley, M.R.; Stanley, J. Resistance to Geminivirus Infection by Virus-Induced Expression of Dianthin in Transgenic Plants. Virology 1996, 220, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O.; Pinto, Y.M.; Baulcombe, D.C. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 1999, 964, 14147–14152. [Google Scholar] [CrossRef]
- Shivaprasad, P.V.; Akbergenov, R.; Trinks, D.; Rajeswaran, R.; Veluthambi, K.; Hohn, T.; Pooggin, M.M. Promoters, Transcripts, and Regulatory Proteins of Mungbean Yellow Mosaic Geminivirus. J. Virol. 2005, 793, 8149–8163. [Google Scholar] [CrossRef]
- Trinks, D.; Rajeswaran, R.; Shivaprasad, P.V.; Akbergenov, R.; Oakeley, E.J.; Veluthambi, K.; Hohn, T.; Pooggin, M.M. Suppression of RNA Silencing by a Geminivirus Nuclear Protein, AC2, Correlates with Transactivation of Host Genes. J. Virol. 2005, 79, 2517–2527. [Google Scholar] [CrossRef]
- Wang, H.; Buckley, K.J.; Yang, X.; Buchmann, R.C.; Bisaro, D.M. Adenosine Kinase Inhibition and Suppression of RNA Silencing by Geminivirus AL2 and L2 Proteins. J. Virol. 2005, 792, 7410–7418. [Google Scholar] [CrossRef] [PubMed]
- Chowda-Reddy, R.V.; Dong, W.; Felton, C.; Ryman, D.; Ballard, K.; Fondong, V.N. Characterization of the cassava geminivirus transcription activation protein putative nuclear localization signal. Virus Res. 2009, 145, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Castillo-González, C.; Liu, X.; Huang, C.; Zhao, C.; Ma, Z.; Hu, T.; Sun, F.; Zhou, X.; Wang, X.J.; Zhang, X. Geminivirus-Encoded TrAP Suppressor Inhibits the Histone Methyltransferase SUVH4/KYP to Counter Host Defense. eLife 2015, 4, e06671. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Mishra, S.K.; Rahman, J.; Taneja, J.; Sundaresan, G.; Mishra, N.S.; Mukherjee, S.K. Mungbean yellow mosaic Indian virus encoded AC2 protein suppresses RNA silencing by inhibiting Arabidopsis RDR6 and AGO1 activities. Virology 2015, 486, 158–172. [Google Scholar] [CrossRef]
- Kvarnheden, A.; Lindblad, M.; Lindsten, K.; Valkonen, J.P.T. Genetic diversity of Wheat dwarf virus. Arch. Virol. 2002, 147, 205–216. [Google Scholar] [CrossRef]
- Koch, C. Die Bestimmung der DNA-Sequenz des Geminivirus WDV-ER Genoms und Versuche zur Übertragung des Virus auf Gerste mit Agrobacterium tumefaciens. Ph.D. Thesis, Universität Köln, Köln, Germany, 1990. [Google Scholar]
- Schubert, J.; Habekuß, A.; Rabenstein, F. Investigation of differences between wheat and barley forms of Wheat dwarf virus and their distribution in host plants. Plant Prot. Sci. Prague 2003, 38, 43–48. [Google Scholar] [CrossRef]
- Jeske, H. Geminiviruses. In TT Viruses. Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 185–226. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 111, 777–788. [Google Scholar] [CrossRef]
- Wu, B.; Shang, X.; Schubert, J.; Habekuß, A.; Elena, S.F.; Wang, X. Global-scale computational analysis of genomic sequences reveals the recombination pattern and coevolution dynamics of cereal-infecting geminiviruses. Sci. Rep. 2015, 5, 8153. [Google Scholar] [CrossRef]
- Van Bel, A.J.E. The phloem, a miracle of ingenuity. Plant Cell Environ. 2003, 26, 125–149. [Google Scholar] [CrossRef]
- Kammann, M.; Schalk, H.-J.; Matzeit, V.; Schaefer, S.; Schell, J.; Gronenborn, B. DNA replication of wheat dwarf virus, a geminivirus, requires two cis-acting signals. Virology 1991, 184, 786–790. [Google Scholar] [CrossRef]
- Heyraud, F.; Matzeit, V.; Schaefer, S.; Schell, J.; Gronenborn, B. The conserved nonanucleotide motif of the geminivirus stem-loop sequence promotes replicational release of virus molecules from redundant copies. Biochimie 1993, 75, 605–615. [Google Scholar] [CrossRef]
- Laufs, J.; Jupin, I.; David, C.; Schumacher, S.; Heyraud-Nitschke, F.; Gronenborn, B. Geminivirus replication: Genetic and biochemical characterization of Rep protein function, a review. Biochimie 1995, 770, 765–773. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for Plant DNA Replication, Transcription, and Cell Cycle Regulation. Crit. Rev. Plant Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Bosque-Pérez, N.A. Eight decades of maize streak virus research. Virus Res. 2000, 71, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Astier, S.; Albouy, J.; Maury, Y.; Robaglia, C.; Lecoq, H. Principles of Plant Virology: Genome, Pathogenicity, Virus Ecology; Institut National de la Recherche Agronomique: Paris, France, 2007. [Google Scholar]
- Tomenius, K.; Oxelfelt, P. Preliminary Observations of Viruslike Particles in Nuclei in Cells of Wheat Infected with the Wheat Dwarf Disease. J. Phytopathol. 1981, 101, 163–167. [Google Scholar] [CrossRef]
- Huth, W.; Lesemann, D.-E. Nachweis des wheat dwarf virus in Deutschland. Nachrichtenblatt Dtsch. Pflanzenschutzdienstes 1994, 46, 105–106. [Google Scholar]
- Hehnle, S.; Wege, C.; Jeske, H. Interaction of DNA with the Movement Proteins of Geminiviruses Revisited. J. Virol. 2004, 784, 7698–7706. [Google Scholar] [CrossRef]
- Evert, R.F.; Russin, W.A.; Botha, C.E.J. Distribution and frequency of plasmodesmata in relation to photoassimilate pathways and phloem loading in the barley leaf. Planta 1996, 198, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Aoki, N.; Scofield, G.N.; Wang, X.-D.; Patrick, J.W.; Offler, C.E.; Furbank, R.T. Expression and localisation analysis of the wheat sucrose transporter TaSUT1 in vegetative tissues. Planta 2004, 219, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Crawford, K.M.; Zambryski, P.C. Non-Targeted and Targeted Protein Movement through Plasmodesmata in Leaves in Different Developmental and Physiological States. Plant Physiol. 2001, 125, 1802–1812. [Google Scholar] [CrossRef] [PubMed]
- Peterschmitt, M.; Quiot, J.B.; Reynaud, B.; Baudin, P. Detection of maize streak virus antigens over time in different parts of maize plants of a sensitive and a so-called tolerant cultivar by ELISA. Ann. Appl. Biol. 1992, 121, 641–653. [Google Scholar] [CrossRef]
- Mariano, A.C.; Andrade, M.O.; Santos, A.A.; Carolino, S.M.B.; Oliveira, M.L.; Baracat-Pereira, M.C.; Brommonshenkel, S.H.; Fontes, E.P.B. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology 2004, 318, 24–31. [Google Scholar] [CrossRef]
- Maule, A.; Leh, V.; Lederer, C. The dialogue between viruses and hosts in compatible interactions. Curr. Opin. Plant Biol. 2002, 5, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Plant Resistance to Geminiviruses. Available online: https://biblio.iita.org/documents/S20InbkPatilPlantNothomDev.pdf-c1c85057a36d00c1bca8600d973d2cdc.pdf (accessed on 23 September 2023).
- Mehner, S. Zur Ökologie des Wheat Dwarf Virus (WDV) in Sachsen-Anhalt. Ph.D. Thesis, Martin-Luther-Universität Halle-Wittenberg, Halle, Germany, 2005. [Google Scholar]
- Commandeur, U.; Huth, W. Differentiation of strains of Wheat dwarf virus in infected wheat and barley plants by means of polymerase chain reaction. J. Plant Dis. Prot. 1999, 106, 550–552. [Google Scholar]
- Lindsten, K.; Vacke, J. A possible barley adapted strain of wheat dwarf virus (WDV). Acta Phytopathol. Entomol. Hung. 1991, 26, 175–180. [Google Scholar]
- Schubert, J.; Habekuß, A.; Kazmaier, K.; Jeske, H. Surveying cereal-infecting geminiviruses in Germany—Diagnostics and direct sequencing using rolling circle amplification. Virus Res. 2007, 127, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Melcher, U.; Guo, X.; Wang, X.; Fan, L.; Zhou, G. Assessment of codivergence of Mastreviruses with their plant hosts. BMC Evol. Biol. 2008, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Mishchenko, L.T.; Dunich, A.A.; Mishchenko, I.A.; Dashchenko, A.V.; Kozub, N.O.; Kyslykh, T.M.; Molodchenkova, O.O. Wheat dwarf virus in Ukraine: Occurrence, molecular characterization and impact on the yield. J. Plant Dis. Prot. 2022, 129, 107–116. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; McGivern, D.R.; Boulton, M.I.; Thomson, J.A.; Rybicki, E.P. A three-nucleotide mutation altering the Maize streak virus Rep pRBR-interaction motif reduces symptom severity in maize and partially reverts at high frequency without restoring pRBR–Rep binding. J. Gen. Virol. 2005, 86, 803–813. [Google Scholar] [CrossRef]
- Schubert, J.; Habekuß, A.; Wu, B.; Thieme, T.; Wang, X. Analysis of complete genomes of isolates of the Wheat dwarf virus from new geographical locations and descriptions of their defective forms. Virus Genes 2014, 48, 133–139. [Google Scholar] [CrossRef]
- Köklü, G.; Ramsell, J.N.E.; Kvarnheden, A. The complete genome sequence for a Turkish isolate of Wheat dwarf virus (WDV) from barley confirms the presence of two distinct WDV strains. Virus Genes 2007, 34, 359–366. [Google Scholar] [CrossRef]
- Ramsell, J.N.E.; Boulton, M.I.; Martin, D.P.; Valkonen, J.P.T.; Kvarnheden, A. Studies on the host range of the barley strain of Wheat dwarf virus using an agroinfectious viral clone. Plant Pathol. 2009, 58, 1161–1169. [Google Scholar] [CrossRef]
- Wu, X.; Weigel, D.; Wigge, P.A. Signaling in plants by intercellular RNA and protein movement. Genes Dev. 2002, 16, 151–158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Owor, B.E.; Shepherd, D.N.; Taylor, N.J.; Edema, R.; Monjane, A.L.; Thomson, J.A.; Martin, D.P.; Varsani, A. Successful application of FTA® Classic Card technology and use of bacteriophage ϕ29 DNA polymerase for large-scale field sampling and cloning of complete maize streak virus genomes. J. Virol. Methods 2007, 140, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Jungner, J. Die Zwergzikade (Cicadula sexnotata Fall.) und ihre Bekämpfung; Deutsche landwirtschafts-gesellschaft: Berlin, Germany, 1906. [Google Scholar]
- Tullgren, A. Zur Morphologie und Systematik der Hemipteren I. Entomol. Tidskr. Entomol. Föreningen I Stockh. 1918, 1918, 113–133. [Google Scholar] [CrossRef]
- Lindsten, K.; Vacke, J.; Gerhardson, B. A preliminary report on three cereal virus diseases new to Sweden spread by Macrosteles and Psammotettix leafhoppers. Medd. Fran Statens Vaxtskyddsanst. 1970, 1423, 285–297. [Google Scholar]
- Gaborjanyi, R.; Vacke, J.; Bisztray, G. Wheat Dwarf Virus: A New Cereal Pathogen in Hungary; Novenytermeles: Debrecen, Hungary, 1988. [Google Scholar]
- Lapierre, H.; Cousin, M.T.; Della Giustina, W.; Moreau, J.P.; Khogali, M.; Roux, J. Nanisme blé: Agent pathogéne et vecteur. Description, biologie, interaction. Phytoma 1991, 432, 26–28. [Google Scholar]
- Conti, M. Leafhopper-borne plant viruses in Italy. Mem. Della Soc. Entomol. Ital. 1993, 72, 541–547. [Google Scholar]
- Jilaveanu, A.; Vacke, J. Isolation and identification of wheat dwarf virus (WDV) in Romania. Probl. Prot. Plantelor. 1995, 23, 51–62. [Google Scholar]
- Najar, A.; Makkouk, K.M.; Boudhir, H.; Kumari, S.G.; Zarouk, R.; Bessai, R.; Othman, F.B. Viral Diseases of Cultivated Legume and Cereal Crops in Tunisia; Firenze University Press: Florence, France, 2000; pp. 1000–1010. [Google Scholar]
- Lindsten, K.; Lindsten, B. Wheat dwarf—An old disease with new outbreaks in Sweden. J. Plant Dis. Prot. 1999, 106, 325–332. [Google Scholar]
- Sandgren, M.; Lindblad, M. Field studies of Wheat dwarf virus. In Proceedings of the 7th International Congress of Plant Pathology, Edinburgh, UK, 9–16 August 1998. [Google Scholar]
- Lindblad, M. What happened to the wheat dwarf disease. Växtskyddsnotiser 2000, 64, 11–13. [Google Scholar]
- Lindsten, K. Wheat dwarf—An old disease caused by a unique and earlier unknown virus. Vaextskyddsnotiser 1980. [Google Scholar]
- Dlabola, J. Zur Schädlichkeit der Zikaden in Getreidefeldern. Nachrichtenblatt Dtsch. Pflanzenschutzd. 1961, 14, 120–122. [Google Scholar]
- Moreau, J.-P.; Lapierre, H.; Navarro, D.; Debray, P.; Fohrer, F.; Lebrun, I. Distinction des effets du nanisme et de la jaunisse sur le blé. Phytoma Défense Végétaux 1992, 443, 21–25. [Google Scholar]
- Lindsten, K.; Lindsten, B. Occurrence and transmission of Wheat dwarf virus (WDV) in France. In Proceedings of the Third International Conference on Pest in Agriculture, Montpellier, France, 7–9 December 1993; pp. 7–9. [Google Scholar]
- Giustina, W.D.; Lebrun, I.; Lapierre, H.; Lochon, S.; Groupe de Travail „Biologie et Écologie de, P. alienus “. Distribution géographique du vecteur et du virus. Phytoma Défense Végétaux 1991, 432, 30–34. [Google Scholar]
- Anonym. New Knowledges about wheat dwarf virus. Phytoma Défense Végétaux 1992, 443, 17–20. [Google Scholar]
- Vacher, C.; Felix, I.; Bonnand, E. Lutte contre Psammotettix alienus, Cicadelle vectrice de la maladie des pieds chétifs. Perspect. Agric. 1991, 162, 86–89. [Google Scholar]
- Bisztray, G.; Gaborjanyi, R.; Vacke, J. Isolation and characterization of wheat dwarf virus found for the first time in Hungary. J. Plant Dis. Prot. 1989, 96, 449–454. [Google Scholar]
- Jezewska, J. First report of Wheat dwarf virus occurring in Poland. Phytopathol. Pol. 2001, 21, 93–100. [Google Scholar]
- Achon, M.A.; Serrano, L.; Ratti, C.; Rubies-Autonell, C. First Detection of Wheat dwarf virus in Barley in Spain Associated with an Outbreak of Barley Yellow Dwarf. Plant Dis. 2006, 90, 970. [Google Scholar] [CrossRef]
- Viršček Marn, M.; Mavrič Pleško, I. First Report of the Occurrence of Wheat dwarf virus Infecting Wheat in Slovenia. Plant Dis. 2017, 101, 1336. [Google Scholar] [CrossRef]
- Behjatnia, S.A.A.; Afsharifar, A.R.; Tahan, V.; Motlagh, M.H.A.; Gandomani, O.E.; Niazi, A.; Izadpanah, K. Widespread occurrence and molecular characterization of Wheat dwarf virus in Iran. Australas. Plant Pathol. 2011, 40, 12–19. [Google Scholar] [CrossRef]
- Kapooria, R.G.; Ndunguru, J. Occurrence of viruses in irrigated wheat in Zambia. EPPO Bull. 2004, 34, 413–419. [Google Scholar] [CrossRef]
- Ekzayez, A.M.; Kumari, S.G.; Ismail, I. First Report of Wheat dwarf virus and Its Vector (Psammotettix provincialis) Affecting Wheat and Barley Crops in Syria. Plant Dis. 2011, 95, 76. [Google Scholar] [CrossRef]
- Xie, J.; Wang, X.; Liu, Y.; Peng, Y.; Zhou, G. First Report of the Occurrence of Wheat dwarf virus in Wheat in China. Plant Dis. 2007, 91, 111. [Google Scholar] [CrossRef]
- Wang, X.; Wu, B.; Wang, J.F. First report of Wheat dwarf virus infecting barley in Yunnan, China. J. Plant Pathol. 2008, 90, 400. [Google Scholar]
- Bivand, R.; Lewin-Koh, N.; Pebesma, E.; Archer, E.; Baddeley, A.; Bearman, N.; Golicher, D. Package ‘maptools’, Version 1.1–4. 2022.
- Felix, I.; Larcher, J.M.; Maraby, J.; Philippeau, G.; Vinatier, K. Risques d’attaques de cicadelles et conditions d’efficacité des insecticides. Perspect. Agric. 1992, 173, 98–106. [Google Scholar]
- ICTV. Report Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Ramsell, J.N.E.; Lemmetty, A.; Jonasson, J.; Andersson, A.; Sigvald, R.; Kvarnheden, A. Sequence analyses of Wheat dwarf virus isolates from different hosts reveal low genetic diversity within the wheat strain. Plant Pathol. 2008, 57, 834–841. [Google Scholar] [CrossRef]
- Vacke, J.; Cibulka, R. Silky bent grass (Apera spica-venti [L.] Beauv.)—A new host and reservoir of wheat dwarf virus. Plant Prot. Sci. 1999, 35, 47–50. [Google Scholar] [CrossRef]
- Brunt, A.; Crabtree, K.; Dallwitz, M.; Gibbs, A.; Watson, L. Viruses of Plants; CAB International: Oxfordshire, UK, 1996. [Google Scholar] [CrossRef]
- Fohrer, F.; Lebrun, I.; Lapierre, E.H. Acquisitions recéntes sur le virus du nanisme du blé. Phytoma Défense Végétaux 1992, 443, 18–20. [Google Scholar]
- Vacke, J.; Cibulka, R. Response of selected winter wheat varieties to wheat dwarf virus infection at an early growth stage. Czech J. Genet. Plant Breed. 2000, 36, 1–4. [Google Scholar]
- Manurung, B.; Witsack, W.; Mehner, S.; Gruntzig, M.; Fuchs, E. The epidemiology of Wheat dwarf virus in relation to occurrence of the leafhopper Psammotettix alienus in Middle-Germany. Virus Res. 2004, 100, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Širlová, L.; Vacke, J.; Chaloupková, M. Reaction of selected winter wheat varieties to autumnal infection with Wheat dwarf virus. Plant Prot. Sci 2005, 41, 1–7. [Google Scholar] [CrossRef]
- Jones, R.A.C. Global Plant Virus Disease Pandemics and Epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Huth, W. Weizenverzwergung—Bisher übersehen? Pflanzenschutz Prax. 1994, 4, 37–39. [Google Scholar]
- Áy, Z.; Kerényi, Z.; Takács, A.; Papp, M.; Petróczi, I.; Gáborjányi, R.; Silhavy, D.; Pauk, J.; Kertész, Z. Detection of cereal viruses in wheat (Triticum aestivum L.) by serological and molecular methods. Cereal Res. Commun. 2008, 36, 215–224. [Google Scholar] [CrossRef]
- Lindblad, M.; Arenö, P. Temporal and spatial population dynamics of Psammotettix alienus, a vector of wheat dwarf virus. Int. J. Pest Manag. 2002, 48, 233–238. [Google Scholar] [CrossRef]
- Nault, L.R.; Ammar, E.D. Leafhopper and Planthopper Transmission of Plant Viruses. Annu. Rev. Entomol. 1989, 34, 503–529. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479, 278–289. [Google Scholar] [CrossRef]
- Dietzgen, R.G.; Kondo, H.; Goodin, M.M.; Kurath, G.; Vasilakis, N. The family Rhabdoviridae: Mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017, 227, 158–170. [Google Scholar] [CrossRef]
- Liu, B.; Yuan, R.; Liang, Z.; Zhang, T.; Zhu, M.; Zhang, X.; Geng, W.; Fang, P.; Jiang, M.; Wang, Z.; et al. Comprehensive analysis of circRNA expression pattern and circRNA–mRNA–miRNA network in Ctenopharyngodon idellus kidney (CIK) cells after grass carp reovirus (GCRV) infection. Aquaculture 2019, 512, 734349. [Google Scholar] [CrossRef]
- Du, Z.; Fu, Y.; Liu, Y.; Wang, X. Transmission Characteristics of Wheat Yellow Striate Virus by its Leafhopper Vector Psammotettix alienus. Plant Dis. 2020, 104, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, Z.; Wang, H.; Zhang, S.; Cao, M.; Wang, X. Identification and Characterization of Wheat Yellow Striate Virus, a Novel Leafhopper-Transmitted Nucleorhabdovirus Infecting Wheat. Front. Microbiol. 2018, 9, 468. [Google Scholar] [CrossRef] [PubMed]
- Lundsgaard, T. Filovirus-like particles detected in the leafhopper Psammotettix alienus. Virus Res. 1997, 48, 5–40. [Google Scholar] [CrossRef] [PubMed]
- Bock, J.O.; Lundsgaard, T.; Pedersen, P.A.; Christensen, L.S. Identification and partial characterization of Taastrup virus: A newly identified member species of the Mononegavirales. Virology 2004, 319, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Liu, W.; Cao, M.; Wang, X. Full genome sequence of a novel iflavirus from the leafhopper Psammotettix alienus. Arch. Virol. 2019, 164, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Liu, W.; Cao, M.; Wang, X. Sequence analysis and genomic organization of a novel chuvirus, Tàiyuán leafhopper virus. Arch. Virol. 2019, 164, 617–620. [Google Scholar] [CrossRef]
- Han, X.; Wang, H.; Wu, N.; Liu, W.; Cao, M.; Wang, X. Leafhopper Psammotettix alienus hosts chuviruses with different genomic structures. Virus Res. 2020, 285, 197–992. [Google Scholar] [CrossRef]
- Fu, Y.; Cao, M.; Wang, H.; Du, Z.; Liu, Y.; Wang, X. Discovery and characterization of a novel insect-specific reovirus isolated from Psammotettix alienus. J. Gen. Virol. 2020, 101, 884–892. [Google Scholar] [CrossRef]
- Harding, R.M.; Burns, P.; Geijskes, R.J.; McQualter, R.M.; Dale, J.L.; Smith, G.R. Molecular Analysis of Fiji Disease Virus Segments 2, 4 and 7 Completes the Genome Sequence. Virus Genes 2006, 32, 43–47. [Google Scholar] [CrossRef]
- Lot, H.; Delecolle, B.; Boccardo, G.; Marzachi, C.; Milne, R.G. Partial characterization of reovirus-like particles associated with garlic dwarf disease. Plant Pathol. 1994, 43, 537–546. [Google Scholar] [CrossRef]
- Xie, L.; LV, M.-F.; Yang, J.; Chen, J.-P.; Zhang, H.-M. Genomic and phylogenetic evidence that Maize rough dwarf and Rice black-streaked dwarf fijiviruses should be classified as different geographic strains of a single species. Acta Virol. 2017, 61, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Distéfano, A.J.; Conci, L.R.; Muñoz Hidalgo, M.; Guzmán, F.A.; Hopp, H.E.; del Vas, M. Sequence and phylogenetic analysis of genome segments S1, S2, S3 and S6 of Mal de Río Cuarto virus, a newly accepted Fijivirus species. Virus Res. 2003, 92, 113–121. [Google Scholar] [CrossRef]
- Isogai, M.; Lindsten, K.; Uyeda, I. Taxonomic characteristics of fijiviruses based on nucleotide sequences of the oat sterile dwarf virus genome. J. Gen. Virol. 1998, 79, 1479–1485. [Google Scholar] [CrossRef]
- Teakle, D.; Hicks, S.; Karan, M.; Hacker, J.; Greber, R.; Donaldson, J. Host range and geographic distribution of pangola stunt virus and its planthopper vectors in Australia. Aust. J. Agric. Res. 1991, 42, 819–826. [Google Scholar] [CrossRef]
- Zhou, G.; Wen, J.; Cai, D.; Li, P.; Xu, D.; Zhang, S. Southern rice black-streaked dwarf virus: A new proposed Fijivirus species in the family Reoviridae. Chin. Sci. Bull. 2008, 533, 3677–3685. [Google Scholar] [CrossRef]
- Nakashima, N.; Koizumi, M.; Watanabe, H.; Noda, H. Complete nucleotide sequence of the Nilaparvata lugens reovirus: A putative member of the genus Fijivirus. J. Gen. Virol. 1996, 77, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, G.; Wang, X. Detection of wheat dwarf virus (WDV) in wheat and vector leafhopper (Psammotettix alienus Dahlb.) by real-time PCR. J. Virol. Methods 2010, 169, 416–419. [Google Scholar] [CrossRef]
- Le Roux, J.J.; Rubinoff, D. Molecular data reveals California as the potential source of an invasive leafhopper species, Macrosteles sp. nr. severini, transmitting the aster yellows phytoplasma in Hawaii. Ann. Appl. Biol. 2009, 154, 419–427. [Google Scholar] [CrossRef]
- Derlink, M.; Pavlovčič, P.; Stewart, A.J.A.; Virant-Doberlet, M. Mate recognition in duetting species: The role of male and female vibrational signals. Anim. Behav. 2014, 90, 181–193. [Google Scholar] [CrossRef]
- Abt, I.; Jacquot, E. Wheat Dwarf. In Virus Diseases of Tropical and Subtropical Crops; Tennant, P., Fermin, R., Eds.; Plant Protection Series; CAB International: Boston, MA, USA, 2015; pp. 27–41. [Google Scholar] [CrossRef]
- Vilbaste, J. Preliminary key for the identification of the nymphs of North European Homoptera, Cicadinea. II. Cicadelloidea. Ann. Zool. Fenn. 1982, 19, 1–20. [Google Scholar]
- Della Giustina, W. Homoptères Cicadellidae 3: Compléments Faune de France 73, INRA ed.; Fédération Française des Sociétés de Sciences Naturelles: Paris, France, 1989. [Google Scholar]
- Biedermann, R.; Niedrighaus, R. Die Zikaden Deutschlands. In Bestimmungstafeln für alle Arten; Wissenschaftlich Akademischer Buchvertrieb-Fründ: Scheeßel, Germany, 2004. [Google Scholar]
- Kunz, G.; Nickel, H.; Niedrighaus, R. Photographic Atlas of the Planthoppers and Leafhoppers of Germany; Wissenschaftlich Akademischer Buchvertrieb-Fründ: Scheeßel, Germany, 2011. [Google Scholar]
- Tishechkin, D.Y. The variability of acoustic signals and some morphological characters in Psammotettix striatus (Homoptera, Cicadellidae) from Russia and adjacent countries. Зooлoгический журнал 1999, 78. [Google Scholar]
- Biedermann, R.; Niedringhaus, R. The Plant- and Leafhoppers of Germany: Identification Key to all Species; Wabv Fründ: New York, NY, USA, 2009. [Google Scholar]
- Heady, S.E.; Nault, L.R.; Shambaugh, G.F.; Fairchild, L. Acoustic and Mating Behavior of Dalbulus Leaf hoppers (Homoptera: Cicadellidae). Ann. Entomol. Soc. Am. 1986, 79, 727–736. [Google Scholar] [CrossRef]
- Gillham, M.C. Variation in acoustic signals within and among leafhopper species of the genus Alebra (Homoptera, Cicadellidae). Biol. J. Linn. Soc. 1992, 45, 1–15. [Google Scholar] [CrossRef]
- Tishechkin, D.Y. Vibrational communication in Aphrodinae leafhoppers (Deltocephalinae auct.; Homoptera: Cicadellidae) and related groups with notes on classification of higher taxa. Russ. Entomol. J. 2000, 9, 1–66. [Google Scholar]
- Percy, D.M.; Boyd, E.A.; Hoddle, M.S. Observations of acoustic signaling in three sharpshooters: Homalodisca vitripennis, Homalodisca liturata, and Graphocephala atropunctata (Hemiptera: Cicadellidae). Ann. Entomol. Soc. Am. 2008, 101, 253–259. [Google Scholar] [CrossRef]
- Tishechkin, D.Y. The use of bioacoustic characters for distinguishing between cryptic species in insects: Potentials, restrictions, and prospects. Entomol. Rev. 2014, 94, 289–309. [Google Scholar] [CrossRef]
- Derlink, M.; Abt, I.; Mabon, M.; Julian, C.; Virant-Doberlet, M.; Jacquot, E. Mating behaviour of Psammotettix alienus Dahlbom (Hemiptera: Cicadellidae). Insect Sci. 2018, 25, 148–160. [Google Scholar] [CrossRef]
- Schlick-Steiner, B.C.; Steiner, F.M.; Seifert, B.; Stauffer, C.; Christian, E.; Crozier, R.H. Integrative Taxonomy: A Multisource Approach to Exploring Biodiversity. Annu. Rev. Entomol. 2010, 55, 421–438. [Google Scholar] [CrossRef]
- Bluemel, J.K.; Derlink, M.; Pavlovcic, P.; Russio, I.-R.M.; Andrew King, R.; Corbett, E.; Sherrard-Smith, E.; Blejec, A.; Wilson, M.R.; Stewart, A.J.A.; et al. Integrating vibrational signals, mitochondrial DNA and morphology for species determination in the genus Aphrodes (Hemiptera: Cicadellidae). Syst. Entomol. 2014, 39, 304–324. [Google Scholar] [CrossRef]
- Kamitani, S. DNA Barcodes of Japanese Leafhoppers. ESAKIA 2011, 50, 81–88. [Google Scholar] [CrossRef]
- Gwiazdowski, R.A.; Foottit, R.G.; Maw, H.E.L.; Hebert, P.D.N. The Hemiptera (Insecta) of Canada: Constructing a Reference Library of DNA Barcodes. PLoS ONE 2015, 10, e0125635. [Google Scholar] [CrossRef]
- Abt, I.; Derlink, M.; Mabon, R.; Virant-Doberlet, M.; Jacquot, E. Integrating multiple criteria for the characterization of Psammotettix populations in European cereal fields. Bull. Entomol. Res. 2018, 108, 185–202. [Google Scholar] [CrossRef] [PubMed]
- Guglielmino, A.; Virla, E.G. Postembryonic development and biology of Psammotettix alienus (Dahlbom) (Homoptera, Cicadellidae) under laboratory conditions. Boll. Zool. Agrar. Bachic. 1997, 29, 65–80. [Google Scholar]
- Manurung, B. Untersuchungen zur Biologie und Ökologie der Zwergzikade Psammotettix alienus Dahlb. (Auchenorrhyncha) und zu ihrer Bedeutung als Vektor des Wheat Dwarf Virus (Weizenverzwergungs-Virus, WDV). Ph.D. Thesis, Martin–Luther-Universität Halle-Wittenberg, Halle, Germany, 2002. [Google Scholar]
- Schiemenz, H.; Emmrich, R.; Witsack, W. Beiträge zur Insektenfauna Ost- deutschlands: Homoptera—Auchenorrhyncha (Cicadina) (Insecta) Teil IV: Unterfamilie Deltocephalinae. Faun. Abh. 1996, 20, 153–258. [Google Scholar]
- Alla, S.; Moreau, J.P.; Frerot, B. Effects of the aphid Rhopalosiphum padi on the leafhopper Psammotettix alienus under laboratory conditions. Entomol. Exp. Appl. 2001, 98, 203–209. [Google Scholar] [CrossRef]
- Van Nieuwenhove, G.A.; Frías, E.A.; Virla, E.G. Effects of temperature on the development, performance and fitness of the corn leafhopper Dalbulus maidis (DeLong) (Hemiptera: Cicadellidae): Implications on its distribution under climate change. Agric. For. Entomol. 2016, 18, 1–10. [Google Scholar] [CrossRef]
- Schiemenz, H. Die Zikadenfauna mitteleuropäischer Trockenrasen (Homoptera, Auchenorrhyncha). Entomol. Abh. Staatl. Mus. Für Tierkd. Dresd. 1969, 36, 201–280. [Google Scholar]
- Ghodoum Parizipour, M.H.; Behjatnia, S.A.A.; Afsharifar, A.; Izadpanah, K. Natural hosts and efficiency of leafhopper vector in transmission of Wheat dwarf virus. J. Plant Pathol. 2016, 483–492. [Google Scholar]
- Lindblad, M.; Sigvald, R. Temporal spread of wheat dwarf virus and mature plant resistance in winter wheat. Crop Prot. 2004, 23, 229–234. [Google Scholar] [CrossRef]
- Backus, E.A. Sensory systems and behaviours which mediate hemipteran plant-feeding: A taxonomic overview. J. Insect Physiol. 1988, 34, 151–165. [Google Scholar] [CrossRef]
- Zhao, L.; Dai, W.; Zhang, C.; Zhang, Y. Morphological characterization of the mouthparts of the vector leafhopper Psammotettix striatus (L.) (Hemiptera: Cicadellidae). Micron 2010, 41, 754–759. [Google Scholar] [CrossRef]
- Hewer, A.; Becker, A.; van Bel, A.J.E. An aphid’s Odyssey—The cortical quest for the vascular bundle. J. Exp. Biol. 2011, 2142, 3868–3879. [Google Scholar] [CrossRef]
- Storey, H.H. Transmission studies of maize streak disease. Ann. Appl. Biol. 1928, 15, 1–25. [Google Scholar] [CrossRef]
- Wang, Y.; Dang, M.; Hou, H.; Mei, Y.; Qian, Y.; Zhou, X. Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 2014, 95, 2082–2088. [Google Scholar] [CrossRef]
- Reynaud, B.; Peterschmitt, M. A study of the mode of transmission of maize streak virus by Cicadulina mbila using an enzyme-linked immunosorbent assay. Ann. Appl. Biol. 1992, 121, 85–94. [Google Scholar] [CrossRef]
- Tholt, G.; Samu, F.; Kiss, B. Feeding behaviour of a virus-vector leafhopper on host and non-host plants characterised by electrical penetration graphs. Entomol. Exp. Appl. 2015, 155, 123–136. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [PubMed]
- Brault, V.; Uzest, M.; Monsion, B.; Jacquot, E.; Blanc, S. Aphids as transport devices for plant viruses. C. R. Biol. 2010, 333, 524–538. [Google Scholar] [CrossRef]
- Yazdkhasti, E.; Hopkins, R.J.; Kvarnheden, A. Reservoirs of plant virus disease: Occurrence of wheat dwarf virus and barley/cereal yellow dwarf viruses in Sweden. Plant Pathol. 2021, 70, 1552–1561. [Google Scholar] [CrossRef]
- Nickel, H.; Remane, R. Artenliste der Zikaden Deutschands, (Checklist of the planthoppers and leafhoppers of Germany, with notes on food plants, diet width, life cycles, geographic range and conservation status). Beiträge Zikadenkunde 2002, 5, 27–64. [Google Scholar]
- Ossiannilsson, F. The Auchenorrhyncha (Homoptera) of Fennoscandia and Denmark. Part 2: The Families Cicadidae, Cercopidae, Membracidae, and Cicadellidae (excl. Deltocephalinae). Scand. Sci. Press 1981, 7, 223–593. [Google Scholar] [CrossRef]
- Koppányi, T. A lucernásban kalakuló Cicadinea együttes évszakos és állományok korával járó változásának vizsgálata. A study into the seasonal and crop age related changes of Cicadinea assemblages in alfalfa.). Debreceni Agrártudományi Egy. Tudományos Közleményei 1976, 18, 27–60. [Google Scholar]
- Drobnjakovic, T.; Peric, P.; Marcic, D.; Picciau, L.; Alma, A.; Mitrovic, J.; Duduk, B.; Bertaccini, A. Leafhoppers and cixiids in phytoplasma-infected carrot fields: Species composition and potential phytoplasma vectors. Pestic. I Fitomed. 2010, 25, 311–318. [Google Scholar] [CrossRef]
- Kiss, B.; Redei, D.; Koczor, S. Occurrence and feeding of hemipterans on common ragweed (Ambrosia artemisiifolia) in Hungary. Bull. Insectol. 2008, 61, 195–196. [Google Scholar]
- Riedle-Bauer, M.; Tiefenbrunner, W.; Otreba, J.; Hanak, K.; Schildberger, B.; Regner, F. Epidemiological observations on Bois Noir in Austrian vineyards. Mitteilungen Klosterneubg. 2006, 56, 177–181. [Google Scholar]
- Landi, L.; Isidoro, N.; Riolo, P. Natural Phytoplasma Infection of Four Phloem-Feeding Auchenorrhyncha Across Vineyard Agroecosystems in Central-Eastern Italy. J. Econ. Entomol. 2013, 106, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Tholt, G.; Kiss, B. Host range of Psammotettix alienus (Dahlbom). Növényvédelem 2011, 47, 229–235. [Google Scholar]
- Stafford, C.A.; Walker, G.P. Characterization and correlation of DC electrical penetration graph waveforms with feeding behavior of beet leafhopper, Circulifer tenellus. Entomol. Exp. Appl. 2009, 130, 113–129. [Google Scholar] [CrossRef]
- Alvarez, A.E.; Tjallingii, W.F.; Garzo, E.; Vleeshouwers, V.; Dicke, M.; Vosman, B. Location of resistance factors in the leaves of potato and wild tuber-bearing Solanum species to the aphid Myzus persicae. Entomol. Exp. Appl. 2006, 121, 145–157. [Google Scholar] [CrossRef]
- Lei, H.; Lenteren, J.C.; Tjallingii, W.F. Analysis of resistance in tomato and sweet pepper against the greenhouse whitefly using electrically monitored and visually observed probing and feeding behaviour. Entomol. Exp. Appl. 1999, 92, 299–309. [Google Scholar] [CrossRef]
- Lei, H.; Van Lenteren, J.C.; Xu, R.M. Effects of plant tissue factors on the acceptance of four greenhouse vegetable host plants by the greenhouse whitefly: An Electrical Penetration Graph (EPG) study. Eur. J. Entomol. 2001, 98, 31–36. [Google Scholar] [CrossRef]
- McLean, D.L.; Kinsey, M.G. A Technique for Electronically Recording Aphid Feeding and Salivation. Nature 1964, 202939, 1358–1359. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Electronic recording of penetration behaviour by aphids. Entomol. Exp. Appl. 1978, 24, 721–730. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Electrical nature of recorded signals during stylet penetration by aphids. Entomol. Exp. Appl. 1985, 38, 177–186. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Electrical Recording of Stylet Penetration Activities. In Aphids, Their Biology, Natural Enemies and Control; Elsevier: Amsterdam, The Netherlands, 1988; pp. 95–108. [Google Scholar]
- Prado, E.; Tjallingii, W.F. Aphid activities during sieve element punctures. Entomol. Exp. Appl. 1994, 72, 157–165. [Google Scholar] [CrossRef]
- Harrewijn, P.; Kayser, H. Pymetrozine, a Fast-Acting and Selective Inhibitor of Aphid Feeding. In-situStudies with Electronic Monitoring of Feeding Behaviour. Pestic. Sci. 1997, 49, 130–140. [Google Scholar] [CrossRef]
- Mesfin, T.; Bosque-Pérez, N.A. Feeding behavior of Cicadulina storeyi China (Homoptera: Cicadellidae) on maize varieties susceptible or resistant to maize streak virus. Afr. Entomol. 1998, 6, 185–191. [Google Scholar]
- Helden, M.; Tjallingii, W.F. Experimental Design and Analysis in EPG Experiments with Emphasis on Plant Resistance Research. In Principles and Applications of Electronic Monitoring and Other Techniques in the Study of Homopteran Feeding Behavior; Entomological Society of America: Annapolis, MD, USA, 2000; Volume 144. [Google Scholar] [CrossRef]
- Liu, X.D.; Zhai, B.P.; Zhang, X.X.; Zong, J.M. Impact of transgenic cotton plants on a non-target pest, Aphis gossypii Glover. Ecol. Entomol. 2005, 30, 307–315. [Google Scholar] [CrossRef]
- Prado, E.; Fred Tjallingii, W. Behavioral Evidence for Local Reduction of Aphid-Induced Resistance. J. Insect Sci. 2007, 7, 48. [Google Scholar] [CrossRef]
- Martin, B.; Fereres, A.; Tjallingii, W.F.; Collar, J.L. Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J. Gen. Virol. 1997, 780, 2701–2705. [Google Scholar] [CrossRef]
- Sauge, M.H.; Lambert, P.; Pascal, T. Co-localisation of host plant resistance QTLs affecting the performance and feeding behaviour of the aphid Myzus persicae in the peach tree. Heredity 2012, 108, 292–301. [Google Scholar] [CrossRef]
- Giordanengo, P. EPG-Calc: A PHP-based script to calculate electrical penetration graph (EPG) parameters. Arthropod Plant Interact. 2014, 8, 163–169. [Google Scholar] [CrossRef]
- Tjallingii, W.F.; Esch, T.H.H. Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol. Entomol. 1993, 18, 317–328. [Google Scholar] [CrossRef]
- Khan, Z.R.; Saxena, R.C. Probing Behavior of Three Biotypes of Nilaparvata lugens (Homoptera: Delphacidae) on Different Resistant and Susceptible Rice Varieties. J. Econ. Entomol. 1988, 81, 1338–1345. [Google Scholar] [CrossRef]
- Kimmins, F.M. Electrical penetration graphs from Nilaparvata lugens on resistant and susceptible rice varieties. Entomol. Exp. Appl. 1989, 50, 69–79. [Google Scholar] [CrossRef]
- Powell, K.S.; Gatehouse, J.A. Mechanism of Mannose-Binding Snowdrop Lectin for Use against Brown Planthopper in Rice. In Rice Genetics III: (In 2 Parts); World Scientific: Singapore, 1996; pp. 753–758. [Google Scholar] [CrossRef]
- Seo, B.Y.; Kwon, Y.-H.; Jung, J.K.; Kim, G.-H. Electrical penetration graphic waveforms in relation to the actual positions of the stylet tips of Nilaparvata lugens in rice tissue. J. Asia Pac. Entomol. 2009, 12, 89–95. [Google Scholar] [CrossRef]
- Calatayud, P.-A.; Seligmann, C.D.; Polania, M.A.; Bellotti, A.C. Influence of parasitism by encyrtid parasitoids on the feeding behaviour of the cassava mealybug Phenacoccus herreni. Entomol. Exp. Appl. 2001, 98, 271–278. [Google Scholar] [CrossRef]
- Harrewijn, P.; Piron, P.G.M.; Ponsen, M.B. Evolution of vascular feeding in aphids: An electrophysiological study. Entomol. Exp. Appl. 1998, 9, 29–34. [Google Scholar]
- Kingston, K.B. Digestive and Feeding Physiology of Grape Phylloxera (Daktulosphaira vitifoliae Fitch). Ph.D. Thesis, Australian National University, Canberra, Australia, 2007. [Google Scholar]
- Joost, P.H.; Riley, D.G. Imidacloprid effects on probing and settling behavior of Frankliniella fusca and Frankliniella occidentalis (Thysanoptera: Thripidae) in tomato. J. Econ. Entomol. 2005, 98, 1622–1629. [Google Scholar] [CrossRef]
- Kindt, F.; Joosten, N.N.; Tjallingii, W.F. Electrical penetration graphs of thrips revised: Combining DC- and AC-EPG signals. J. Insect Physiol. 2006, 52, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.P.; Janssen, J.A.M. Electronic Recording of Whitefly (Homoptera: Aleyrodidae) Feeding and Oviposition Behavior. In Principles and Applications of Electronic Monitoring and Other Techniques in the Study of Homopteran Feeding Behavior; Entomological Society of America: Annapolis, MD, USA, 2000; p. 172. [Google Scholar] [CrossRef]
- Kimmins, F.M.; Bosque-Perez, N.A. Electrical Penetration Graphs from Cicadulina spp. and the Inoculation of a Persistent Virus into Maize. In Proceedings of the 9th International Symposium on Insect-Plant Relationships; Springer: Berlin/Heidelberg, Germany, 1996; pp. 46–49. [Google Scholar] [CrossRef]
- Lett, J.-M.; Granier, M.; Grondin, M.; Turpin, P.; Molinaro, F.; Chiroleu, F.; Peterschmitt, M.; Reynaud, B. Electrical penetration graphs from Cicadulina mbila on maize, the fine structure of its stylet pathways and consequences for virus transmission efficiency. Entomol. Exp. Appl. 2001, 101, 93–109. [Google Scholar] [CrossRef]
- Miranda, M.P.; Fereres, A.; Appezzato-da-Gloria, B.; Lopes, J.R.S. Characterization of electrical penetration graphs of Bucephalogonia xanthophis, a vector of Xylella fastidiosain citrus. Entomol. Exp. Appl. 2010, 134, 35–49. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 2006, 57, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Wayadande, A.C. Leafhopper Probing Behavior Associated with Maize Chlorotic Dwarf Virus Transmission to Maize. Phytopathology 1993, 83, 522–526. [Google Scholar] [CrossRef]
- Backus, E.A.; Holmes, W.J.; Schreiber, F.; Reardon, B.J.; Walker, G.P. Sharpshooter X Wave: Correlation of an Electrical Penetration Graph Waveform with Xylem Penetration Supports a Hypothesized Mechanism for Xylella fastidiosa Inoculation. Ann. Entomol. Soc. Am. 2009, 102, 847–867. [Google Scholar] [CrossRef]
- Pillon, O. Importance en Champagne de deux parasitoides de la cicadelle vectrice du nanisme du ble. In Proceedings of the ANPP—3rd International Conference on Pests in Agriculture, Montpellier, France, 7–9 December 1993. [Google Scholar]
- Samu, F.; Beleznai, O.; Tholt, G. A potential spider natural enemy against virus vector leafhoppers in agricultural mosaic landscapes—Corroborating ecological and behavioral evidence. Biol. Control 2013, 67, 390–396. [Google Scholar] [CrossRef]
- Kogan, M. Integrated Pest Management: Historical Perspectives and Contemporary Developments. Annu. Rev. Entomol. 1998, 43, 243–270. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; van der Walt, E.; Dent, K.; Varsani, A.; Rybicki, E.P. Maize streak virus: An old and complex ‘emerging’ 62 pathogen. Mol. Plant Pathol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Yazdkhasti, E. Epidemiology of Wheat Dwarf Virus. Ph.D. Thesis, Acta Universitatis Agriculturae Sueciae, Uppsala, Sweden, 2022. [Google Scholar]
- Eweida, M.; Oxelfelt, P.; Tomenius, K. Concentration of virus and ultrastructural changes in oats at various stages of barley yellow dwarf virus infection. Ann. Appl. Biol. 1988, 112, 313–321. [Google Scholar] [CrossRef]
- Aranda, M.A.; Freitas-Astúa, J. Ecology and diversity of plant viruses, and epidemiology of plant virus-induced diseases. Ann. Appl. Biol. 2017, 171, 1–4. [Google Scholar] [CrossRef]
- Weibull, A.-C.; Östman, Ö. Species composition in agroecosystems: The effect of landscape, habitat, and farm management. Basic Appl. Ecol. 2003, 4, 349–361. [Google Scholar] [CrossRef]
- Ansari, M.S.; Moraiet, M.A.; Ahmad, S. Insecticides: Impact on the Environment and Human Health. In Environmental Deterioration and Human Health; Springer: Berlin/Heidelberg, Germany, 2014; pp. 99–123. [Google Scholar] [CrossRef]
- Biondi, A.; Desneux, N.; Siscaro, G.; Zappalà, L. Using organic-certified rather than synthetic pesticides may not be safer for biological control agents: Selectivity and side effects of 14 pesticides on the predator Orius laevigatus. Chemosphere 2012, 87, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Dudley, N.; Attwood, S.J.; Goulson, D.; Jarvis, D.; Bharucha, Z.P.; Pretty, J. How should conservationists respond to pesticides as a driver of biodiversity loss in agroecosystems? Biol. Conserv. 2017, 209, 449–453. [Google Scholar] [CrossRef]
- Guimarães-Cestaro, L.; Martins, M.F.; Martínez, L.C.; Alves, M.L.T.M.F.; Guidugli-Lazzarini, K.R.; Nocelli, R.C.F.; Malaspina, O.; Serrão, J.E.; Teixeira, É.W. Occurrence of virus, microsporidia, and pesticide residues in three species of stingless bees (Apidae: Meliponini) in the field. Sci. Nat. 2020, 107, 1–14. [Google Scholar] [CrossRef]
- Staskawicz, B.J. Genetics of Plant-Pathogen Interactions Specifying Plant Disease Resistance. Plant Physiol. 2001, 125, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, L.; Shung, C.-Y.; Sunter, G.; Bisaro, D.M. Adenosine Kinase Is Inactivated by Geminivirus AL2 and L2 Proteins. Plant Cell 2003, 152, 3020–3032. [Google Scholar] [CrossRef]
- Leke, W.N. Molecular Epidemiology of Begomoviruses That Infect Vegetable Crops in Southwestern Cameroon. Ph.D. Thesis, Acta Universitatis Agriculturae Sueciae, Uppsala, Sweden, 2010. [Google Scholar]
- Wulff, B.B.H.; Moscou, M.J. Strategies for transferring resistance into wheat: From wide crosses to GM cassettes. Front. Plant Sci. 2014, 5, 692. [Google Scholar] [CrossRef]
- Kanyuka, K.; Lovell, D.J.; Mitrofanova, O.P.; Hammond-Kosack, K.; Adams, M.J. A controlled environment test for resistance to Soil-borne cereal mosaic virus (SBCMV) and its use to determine the mode of inheritance of resistance in wheat cv. Cadenza and for screening Triticum monococcum genotypes for sources of SBCMV resistance. Plant Pathol. 2004, 53, 154–160. [Google Scholar] [CrossRef]
- Ward, E.; Kanyuka, K.; Motteram, J.; Kornyukhin, D.; Adams, M.J. The use of conventional and quantitative real-time PCR assays for Polymyxa graminis to examine host plant resistance, inoculum levels and intraspecific variation. New Phytol. 2005, 875–885. [Google Scholar] [CrossRef]
- Hall, M.D.; Brown-Guedira, G.; Klatt, A.; Fritz, A.K. Genetic analysis of resistance to soil-borne wheat mosaic virus derived from Aegilops tauschii. Euphytica 2009, 169, 169–176. [Google Scholar] [CrossRef]
- Zaharieva, M.; Monneveux, P.; Henry, M.; Rivoal, R.; Valkoun, J.; Nachit, M.M. Evaluation of a Collection of Wild Wheat Relative Aegilops Geniculata Roth and Identification of Potential Sources for Useful Traits. Dev. Plant Breed. 2001, 739–746. [Google Scholar] [CrossRef]
- Pfrieme, A.-K.; Ruckwied, B.; Habekuß, A.; Will, T.; Stahl, A.; Pillen, K.; Ordon, F. Identification and Validation of Quantitative Trait Loci for Wheat Dwarf Virus Resistance in Wheat (Triticum spp.). Front. Plant Sci. 2022, 13, 828639. [Google Scholar] [CrossRef] [PubMed]
- Pfrieme, A.K.; Stahl, A.; Pillen, K.; Will, T. Comparison of two different experimental environments for resistance screenings for leafhopper-transmitted wheat dwarf virus in wheat. J. Plant Dis. Prot. 2023; submitted. [Google Scholar]
- Nygren, J.; Shad, N.; Kvarnheden, A.; Westerbergh, A. Variation in Susceptibility to Wheat dwarf virus among Wild and Domesticated Wheat. PLoS ONE 2015, 10, e0121580. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.; Habekuß, A.; Schliephake, E.; Niks, R.; Broer, I.; Ordon, F. Pyramiding of Ryd2 and Ryd3 conferring tolerance to a German isolate of Barley yellow dwarf virus-PAV (BYDV-PAV-ASL-1) leads to quantitative resistance against this isolate. Theor. Appl. Genet. 2011, 123, 69–76. [Google Scholar] [CrossRef]
- Scheurer, K.S.; Friedt, W.; Huth, W.; Waugh, R.; Ordon, F. QTL analysis of tolerance to a German strain of BYDV-PAV in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2001, 103, 1074–1083. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the Microplate Method of Enzyme-Linked Immunosorbent Assay for the Detection of Plant Viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.J.; Macdonald, H.; Coutts, R.H.A.; Buck, K.W. Agroinfection of Triticum aestivum with Cloned DNA of Wheat Dwarf Virus. J. Gen. Virol. 1988, 69, 891–896. [Google Scholar] [CrossRef]
- Boulton, M.I. Agrobacterium-Mediated Transfer of Geminiviruses to Plant Tissues. In Plant Gene Transfer and Expression Protocols; Springer: Berlin/Heidelberg, Germany, 1995; pp. 77–93. [Google Scholar] [CrossRef]
- Boulton, M.I. Construction of Infectious Clones for DNA Viruses: Mastreviruses. In Plant Virology Protocols: From Viral Sequence to Protein Function; Springer: Berlin/Heidelberg, Germany, 2008; pp. 503–523. [Google Scholar] [CrossRef]
- Bukvayová, N.; Henselová, M.; Vajcíková, V.; Kormanová, T. Occurrence of dwarf virus of winter wheat and barley in several regions of Slovakiaduring the growing seasons 2001–2004. Plant Soil Environ. 2006, 52, 392. [Google Scholar] [CrossRef]
- Rabenstein, F.; Sukhacheva, E.; Habekuß, A.; Schubert, J. Differentiation of Wheat dwarf virus isolates from wheat, triticale, and barley by means of a monoclonal antibody. In Proceedings of the X Conference on Viral Diseases of Gramineae in Europe, Louvain-la-Neuve, Belgium, 12–14 September 2005; p. 60. [Google Scholar]
- Kundu, J.K.; Gadiou, S.; Červená, G. Discrimination and genetic diversity of Wheat dwarf virus in the Czech Republic. Virus Genes 2009, 38, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Glais, L.; Jacquot, E. Detection and Characterization of Viral Species/Subspecies Using Isothermal Recombinase Polymerase Amplification (RPA) Assays. In Plant Pathology: Techniques and Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 207–225. [Google Scholar] [CrossRef]
- Thresh, J.M. Cropping Practices and Virus Spread. Annu. Rev. Phytopathol. 1982, 20, 193–216. [Google Scholar] [CrossRef]
- Benkovics, A.H.; Vida, G.; Nelson, D.; Veisz, O.; Bedford, I.; Silhavy, D.; Boulton, M.I. Partial resistance to Wheat dwarf virus in winter wheat cultivars. Plant Pathol. 2010, 59, 1144–1151. [Google Scholar] [CrossRef]
- Schneider, A.; Molnár-Láng, M. Detection of the 1RS chromosome arm in Martonvásár wheat genotypes containing 1Bl.1Rs or 1Al.1Rs translocations using SSR and STS markers. Acta Agron. Hung. 2009, 57, 409–416. [Google Scholar] [CrossRef]
- Schlegel, R. Current List of Wheats with Rye and Alien Introgression. Available online: http://www.ryegene-map.de/rye-introgression/html/entry_m.html. (accessed on 22 May 2020).
- Ruckwied, B. Sources of Resistance and Identification of QTLs for Wheat Dwarf Virus (WDV) Resistance in Wheat (Triticum spp.) and Wild Relatives; Julius Kühn-Institut, Bundesforschungsinstitut für Kulturpflanzen: Quedlinburg, Germany, 2022. [Google Scholar]
- Kumar, R.V. Plant Antiviral Immunity Against Geminiviruses and Viral Counter-Defense for Survival. Front. Microbiol. 2019, 10, 1460. [Google Scholar] [CrossRef]
- Sharaf, A.; Nuc, P.; Ripl, J.; Alquicer, G.; Ibrahim, E.; Wang, X.; Maruthi, M.N.; Kundu, J.K. Transcriptome Dynamics in Triticum aestivum Genotypes Associated with Resistance against the Wheat Dwarf Virus. Viruses 2023, 15, 689. [Google Scholar] [CrossRef] [PubMed]
- Schoelz, J.E.; Harries, P.A.; Nelson, R.S. Intracellular Transport of Plant Viruses: Finding the Door out of the Cell. Mol. Plant 2011, 4, 813–831. [Google Scholar] [CrossRef]
- Frederickson Matika, D.E.; Loake, G.J. Redox Regulation in Plant Immune Function. Antioxid. Redox Signal. 2014, 21, 1373–1388. [Google Scholar] [CrossRef]
- Mayer, M.P. Recruitment of Hsp70 chaperones: A crucial part of viral survival strategies. In Reviews of Physiology, Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–46. [Google Scholar] [CrossRef]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef]
- Yuan, W.; Jiang, T.; Du, K.; Chen, H.; Cao, Y.; Xie, J.; Li, M.; Carr, J.P.; Wu, B.; Fan, Z.; et al. Maize phenylalanine ammonia-lyases contribute to resistance toSugarcane mosaic virusinfection, most likely through positive regulation of salicylic acid accumulation. Mol. Plant Pathol. 2019, 200, 1365–1378. [Google Scholar] [CrossRef]
- Zhu, F.; Yuan, S.; Wang, S.-D.; Xi, D.-H.; Lin, H.-H. The higher expression levels of dehydroascorbate reductase and glutathione reductase in salicylic acid-deficient plants may contribute to their alleviated symptom infected with RNA viruses. Plant Signal. Behav. 2011, 6, 1402–1404. [Google Scholar] [CrossRef]
- Selway, J.W. Antiviral activity of flavones and flavans. Prog. Clin. Biol. Res. 1986, 213, 521–536. [Google Scholar] [PubMed]
- Hrmova, M.; Hussain, S.S. Plant Transcription Factors Involved in Drought and Associated Stresses. Int. J. Mol. Sci. 2021, 221, 5662. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Khan, M.K.; Hamurcu, M.; Brestic, M.; Topal, A.; Gezgin, S. Insight into the Root Transcriptome of a Boron-Tolerant Triticum zhukovskyi Genotype Grown under Boron Toxicity. Agronomy 2022, 120, 2421. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Tong, G.; Xi, C.; Liu, J.; Zhao, H.; Wang, Y.; Ren, D.; Han, S. MPK3/MPK6-mediated phosphorylation of ERF72 positively regulates resistance to Botrytis cinerea through directly and indirectly activating the transcription of camalexin biosynthesis enzymes. J. Exp. Bot. 2022, 73, 413–428. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, W.; Wang, L.; Wang, X. Replication-associated proteins encoded by Wheat dwarf virus act as RNA silencing suppressors. Virus Res. 2014, 190, 34–39. [Google Scholar] [CrossRef]
- Chellappan, P.; Vanitharani, R.; Pita, J.; Fauquet, C.M. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 2004, 784, 7465–7477. [Google Scholar] [CrossRef]
- Li, F.; Wang, Y.; Zhou, X. SGS3 Cooperates with RDR6 in Triggering Geminivirus-Induced Gene Silencing and in Suppressing Geminivirus Infection in Nicotiana Benthamiana. Viruses 2017, 9, 247. [Google Scholar] [CrossRef]
- Ding, S.-W.; Li, H.; Lu, R.; Li, F.; Li, W.-X. RNA silencing: A conserved antiviral immunity of plants and animals. Virus Res. 2004, 102, 109–115. [Google Scholar] [CrossRef]
- Shen, Q.; Bao, M.; Zhou, X. A plant kinase plays roles in defense response against geminivirus by phosphorylation of a viral pathogenesis protein. Plant Signal. Behav. 2012, 7, 888–892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Bai, G.; Hunger, R.M.; Bockus, W.W.; Yu, J.; Carver, B.F.; Brown-Guedira, G. Association Study of Resistance to Soilborne wheat mosaic virus in U.S. Winter Wheat. Phytopathology 2011, 1011, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yang, X.; Zhang, D.; Bai, G.; Chao, S.; Bockus, W. Genome-wide association analysis identified SNPs closely linked to a gene resistant to Soil-borne wheat mosaic virus. Theor. Appl. Genet. 2014, 127, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Hourcade, D.; Bogard, M.; Bonnefoy, M.; Savignard, F.; Mohamadi, F.; Lafarge, S.; Du Cheyron, P.; Mangel, N.; Cohan, J.P. Genome-wide association analysis of resistance to wheat spindle streak mosaic virus in bread wheat. Plant Pathol. 2019, 68, 609–616. [Google Scholar] [CrossRef]
- Buerstmayr, M.; Buerstmayr, H. Two major quantitative trait loci control wheat dwarf virus resistance in four related winter wheat populations. Theor. Appl. Genet. 2023, 136, 103. [Google Scholar] [CrossRef]
- Li, H.; Conner, R.L.; Liu, Z.; Li, Y.; Chen, Y.; Zhou, Y.; Duan, X.; Shen, T.; Chen, Q.; Graf, R.J.; et al. Characterization of Wheat-Triticale Lines Resistant to Powdery Mildew, Stem Rust, Stripe Rust, Wheat Curl Mite, and Limitation on Spread of WSMV. Plant Dis. 2007, 91, 368–374. [Google Scholar] [CrossRef]
- Parlevliet, J.E. Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 2002, 124, 147–156. [Google Scholar] [CrossRef]
- Palloix, A.; Ayme, V.; Moury, B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 2009, 183, 190–199. [Google Scholar] [CrossRef]
- Brown, J.K.; Idris, A.M.; Ostrow, K.M.; Goldberg, N.; French, R.; Stenger, D.C. Genetic and phenotypic variation of the Pepper golden mosaic virus Complex. Phytopathology 2005, 950, 1217–1224. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Riedel, C. Pyramidisierung von QTL im Hinblick auf eine Verbesserung der Barley Yellow Dwarf Virus Toleranz der Gerste und Genetische Analyse der Toleranz Gegenüber Wheat Dwarf Virus. Ph.D. Thesis, University of Rostock, Rostock, Germany, 2013. [Google Scholar]
- Ayalew, H.; Tsang, P.W.; Chu, C.; Wang, J.; Liu, S.; Chen, C.; Ma, X.F. Comparison of TaqMan, KASP and rhAmp SNP genotyping platforms in hexaploid wheat. PLoS ONE 2019, 14, e0217222. [Google Scholar] [CrossRef]
- Karlstedt, F. Identification and Mapping of QTL for Resistance against Zymoseptoria tritici in the Winter Wheat Accession HTRI1410 (Triticum aestivum L. subsp. Spelta); Julius Kühn-Institut, Bundesforschungsinstitut für Kulturpflanzen: Quedlinburg, Germany, 2020. [Google Scholar]
- Soleimani, B.; Lehnert, H.; Trebing, S.; Habekuß, A.; Ordon, F.; Stahl, A.; Will, T. Identification of Markers Associated with Wheat Dwarf Virus (WDV) Tolerance/Resistance in Barley (Hordeum vulgare ssp. vulgare) Using Genome-Wide Association Studies. Viruses 2023, 15, 1568. [Google Scholar] [CrossRef]
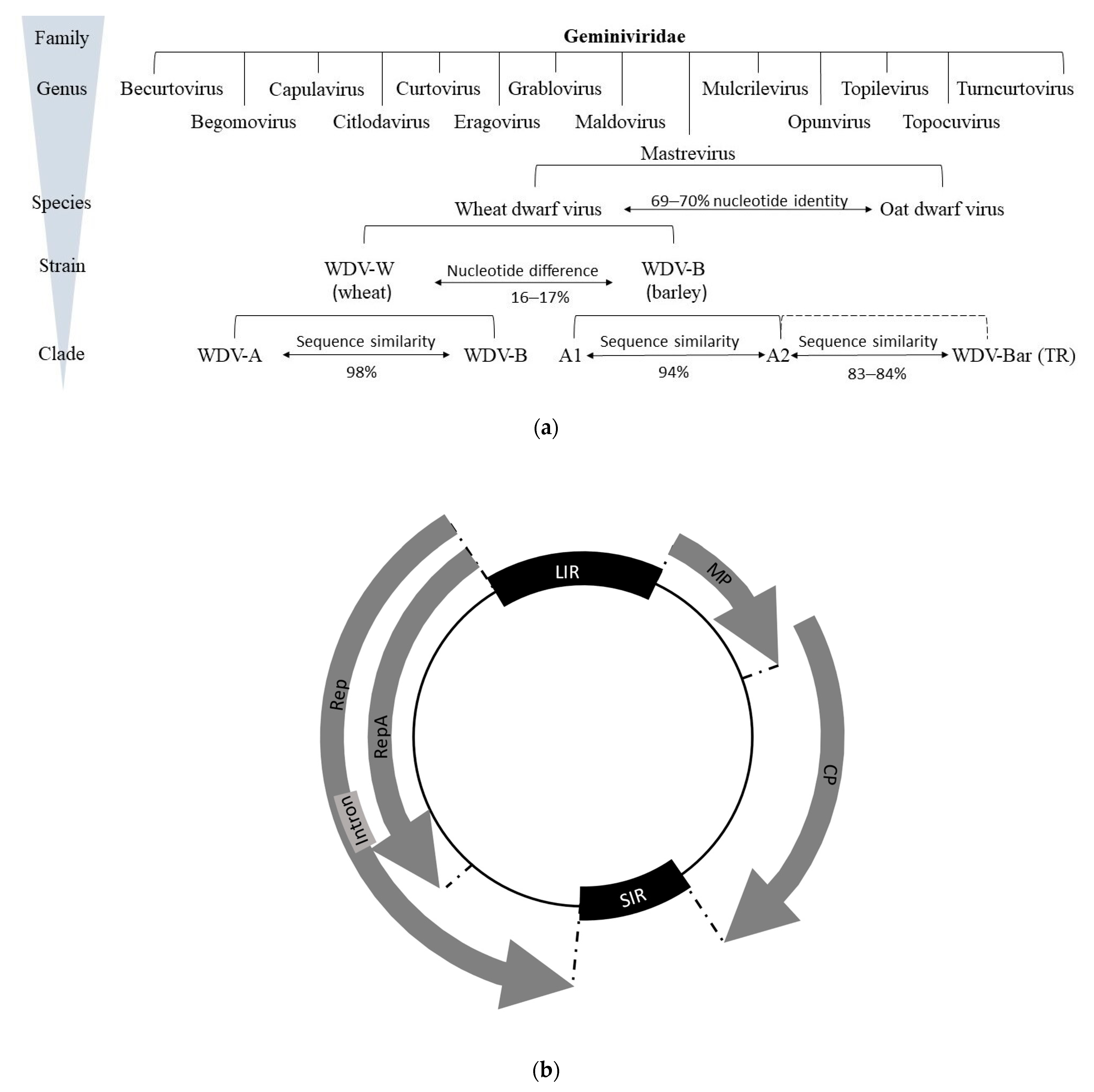
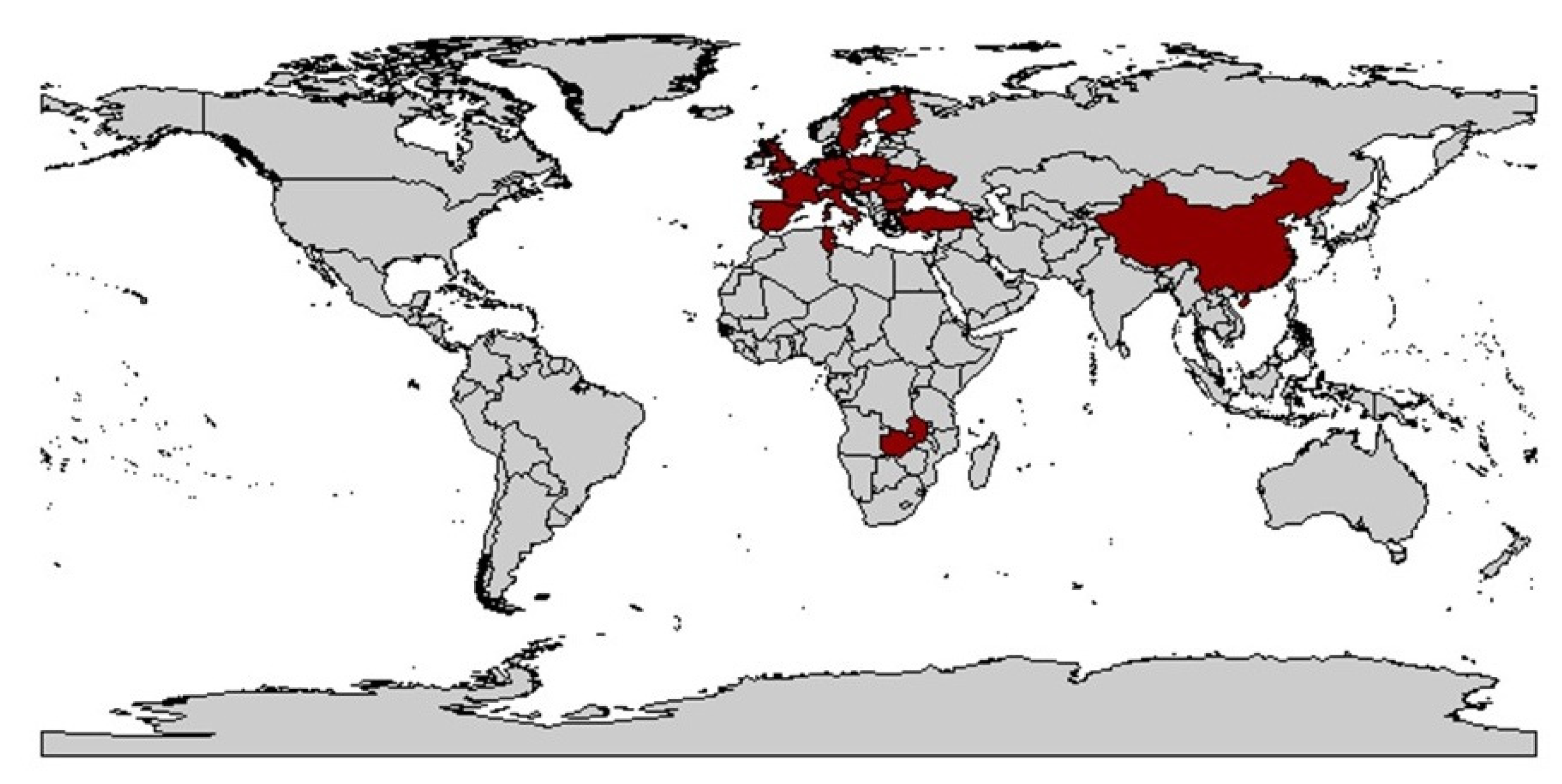

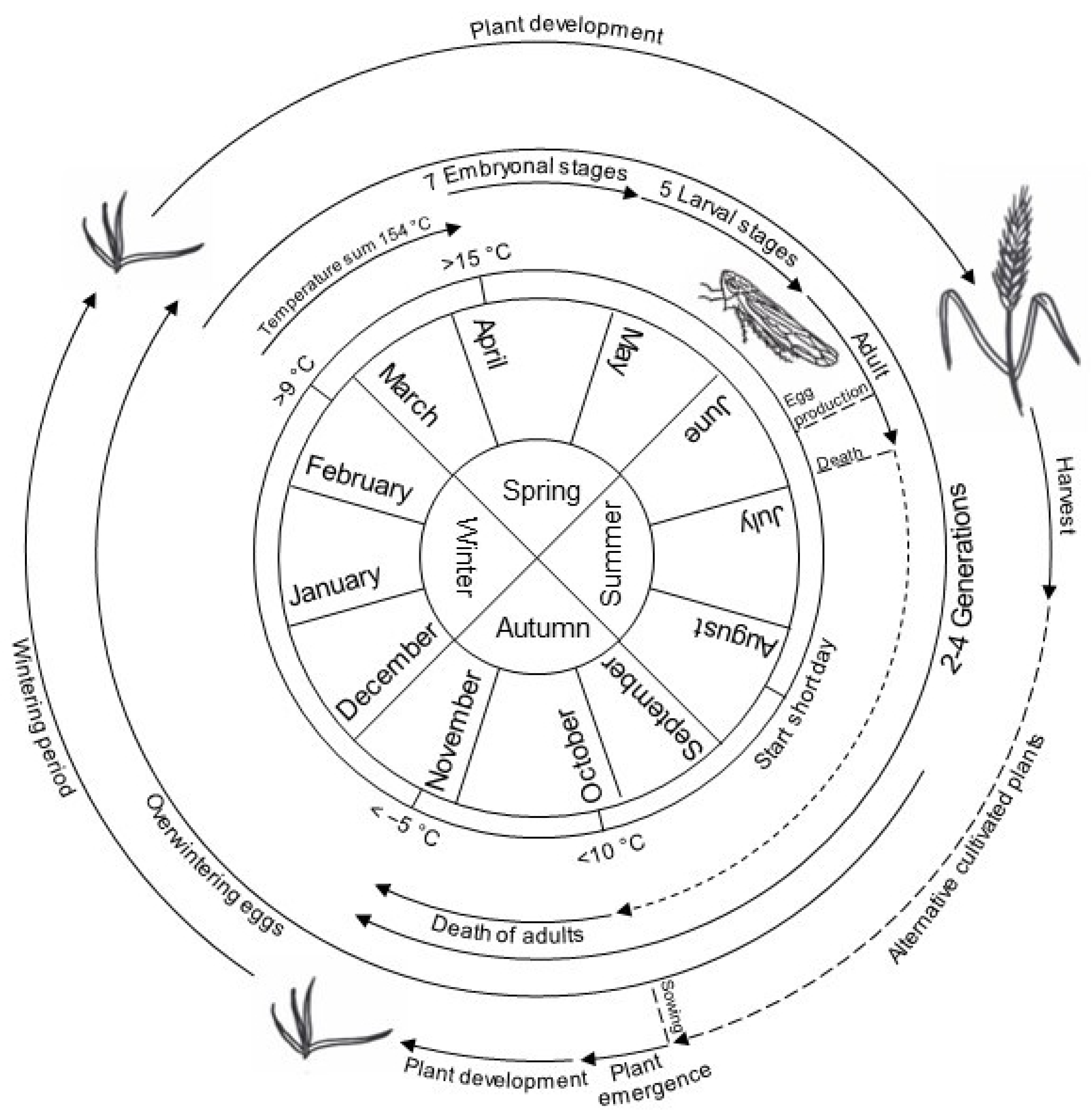
| Time | Event | Reference |
|---|---|---|
| Early 20th century | The first observed dwarfing symptoms of wheat, called slidsjuka | [114,115] |
| Early 20th century | Relatively low field prevalence of WDV; only a few symptoms of dwarfing have been described in scientific literature | [116,117,118,119,120] |
| Early 1950s | Less undersowing in wheat; increased use of combine harvesters | [124] |
| Around 1950 | Decline of slidsjuka due to changes in agricultural practices | [121,122,123,124] |
| 1950–1980/1990 | Slidsjuka occurred sporadically | [121,122,123] |
| 1961 | The first report of a direct relationship between virus, vector, and symptoms; no virus particle detected | [10,125] |
| 1980 | Increased incidence of disease in European countries | [124] |
| 1980 | Identification and taxonomic classification of WDV | [124] |
| 1981 | Leafhopper P. alienus was made responsible for WDV occurence | [114] |
| Late 1980s | A new disease (pieds chétifs) occurred in France in association with P. alienus; the disease was identified as WDV | [126,127] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfrieme, A.-K.; Will, T.; Pillen, K.; Stahl, A. The Past, Present, and Future of Wheat Dwarf Virus Management—A Review. Plants 2023, 12, 3633. https://doi.org/10.3390/plants12203633
Pfrieme A-K, Will T, Pillen K, Stahl A. The Past, Present, and Future of Wheat Dwarf Virus Management—A Review. Plants. 2023; 12(20):3633. https://doi.org/10.3390/plants12203633
Chicago/Turabian StylePfrieme, Anne-Kathrin, Torsten Will, Klaus Pillen, and Andreas Stahl. 2023. "The Past, Present, and Future of Wheat Dwarf Virus Management—A Review" Plants 12, no. 20: 3633. https://doi.org/10.3390/plants12203633
APA StylePfrieme, A.-K., Will, T., Pillen, K., & Stahl, A. (2023). The Past, Present, and Future of Wheat Dwarf Virus Management—A Review. Plants, 12(20), 3633. https://doi.org/10.3390/plants12203633






