The Effects of Rhizophagus irregularis Inoculation on Transcriptome of Medicago lupulina Leaves at Early Vegetative and Flowering Stages of Plant Development
Abstract
1. Introduction
2. Results
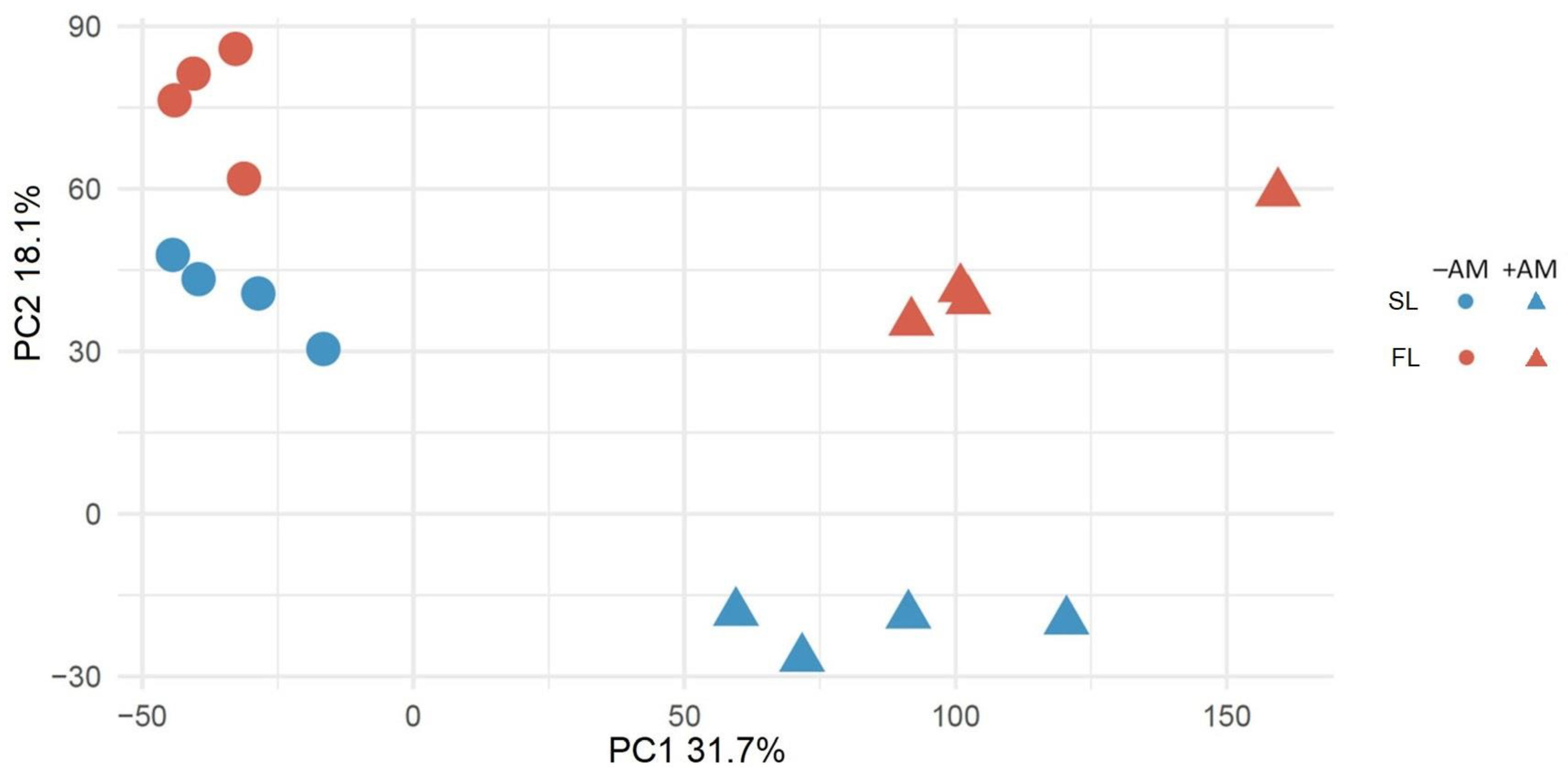
2.1. The Effect of AM Fungus on Medicago Lupulina at Second Leaf and Flowering Stages of Plant Development
2.2. Differential Gene Expression Determined by the Interaction of Medicago Lupulina with Rhizophagus irregularis
3. Discussion
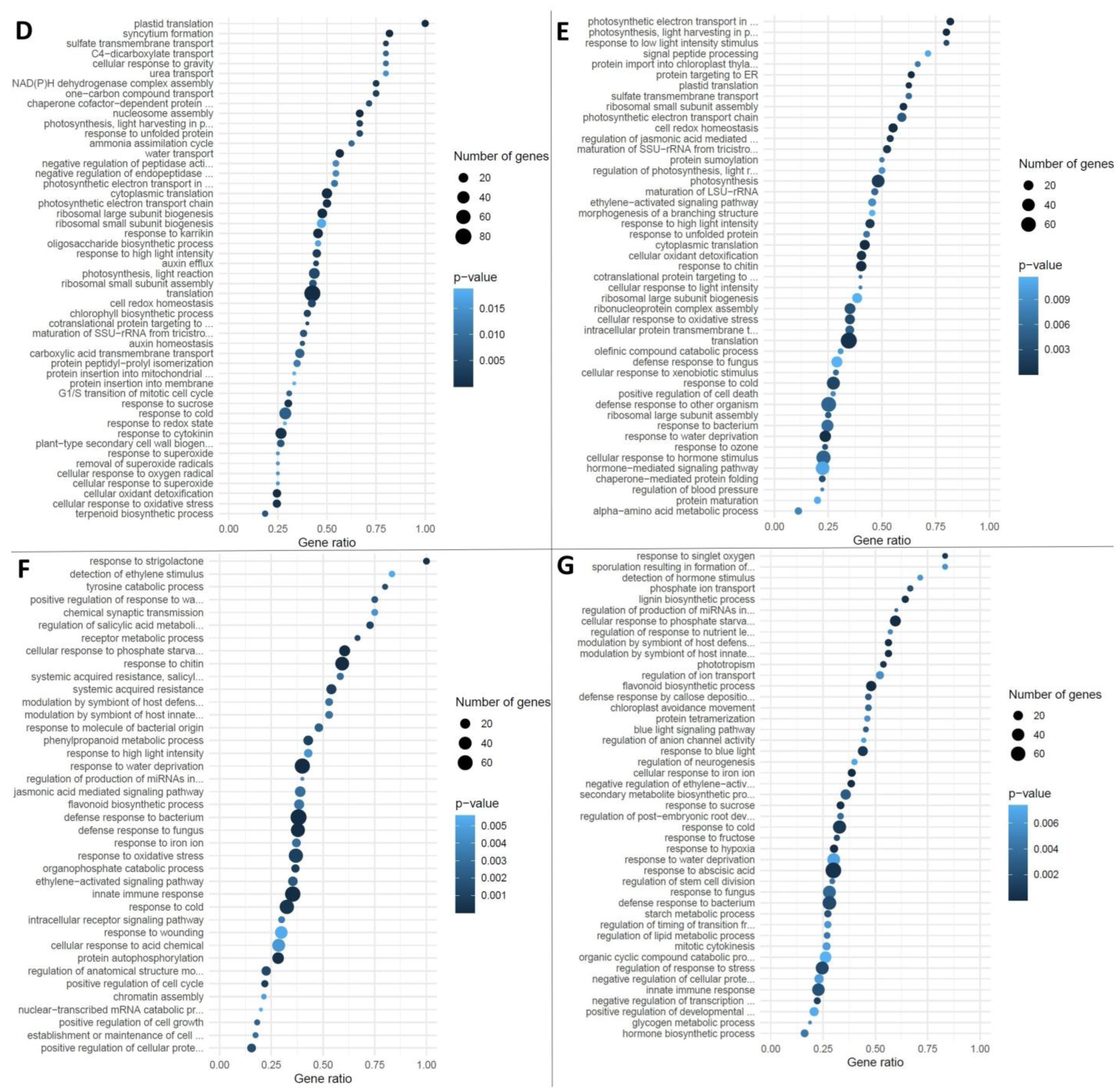
3.1. Diversity in the Effects of R. irregularis on M. lupulina Leaf Transcriptome Is Defined by High Response of MlS-1 Plants to AM and Adaptation to Phosphate Starvation
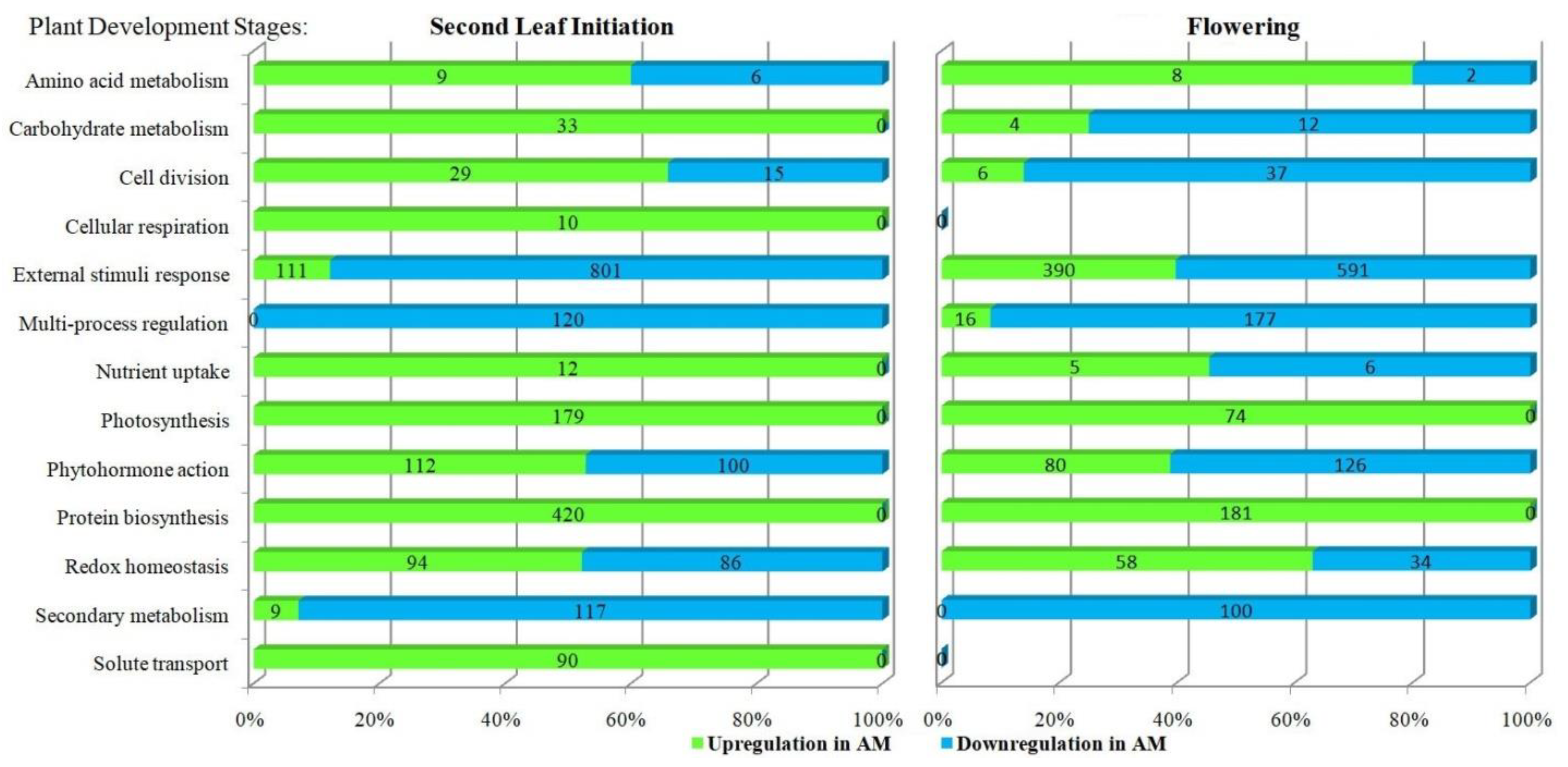
3.2. Functional Categories of Up- and Downregulated Genes by AM in M. lupulina Leaves
4. Materials and Methods
4.1. Plant and Fungus Biomaterials
4.2. Experimental Design and Plant Growth Conditions
4.3. RNA Extraction, Library Preparation, and Sequencing
4.4. Bioinformatic Analysis
4.5. Data Availability
4.6. Evaluation of Mycorrhization Parameters
4.7. Evaluation of Mycorrhizal Growth Response—AM Symbiotic Efficiency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Küster, H.; Vieweg, M.F.; Manthey, K.; Baier, M.C.; Hohnjec, N.; Perlick, A.M. Identification and expression regulation of symbiotically activated legume genes. Phytochemistry 2007, 68, 8–18. [Google Scholar] [CrossRef]
- Kaur, S.; Campbell, B.J.; Suseela, V. Root metabolome of plant-arbuscular mycorrhizal symbiosis mirrors the mutualistic or parasitic mycorrhizal phenotype. New Phytol. 2022, 234, 672–687. [Google Scholar] [CrossRef]
- Kaur, S.; Suseela, V. Unraveling arbuscular mycorrhiza-induced changes in plant primary and secondary metabolome. Metabolites 2020, 10, 335. [Google Scholar] [CrossRef]
- Schweiger, R.; Baier, M.C.; Persicke, M.; Müller, C. High specificity in plant leaf metabolic responses to arbuscular mycorrhiza. Nat. Commun. 2014, 5, 3886. [Google Scholar] [CrossRef]
- Rivero, J.; Álvarez, D.; Flors, V.; Azcón-Aguilar, C.; Pozo, M.J. Root metabolic plasticity underlies functional diversity in mycorrhiza-enhanced stress tolerance in tomato. New Phytol. 2018, 220, 1322–1336. [Google Scholar] [CrossRef] [PubMed]
- Watts-Williams, S.J.; Emmett, B.D.; Levesque-Tremblay, V.; MacLean, A.M.; Sun, X.; Satterlee, J.W.; Fei, Z.; Harrison, M.J. Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ. 2019, 42, 1758–1774. [Google Scholar] [CrossRef]
- Goddard, M.-L.; Belval, L.; Martin, I.R.; Roth, L.; Laloue, H.; Deglène-Benbrahim, L.; Valat, L.; Bertsch, C.; Chong, J. Arbuscular mycorrhizal symbiosis triggers major changes in primary metabolism together with modification of defense responses and signaling in both roots and leaves of Vitis vinifera. Front. Plant Sci. 2021, 12, 721614. [Google Scholar] [CrossRef] [PubMed]
- Blažková, A.; Jansa, J.; Püschel, D.; Vosatka, M.; Janousková, M. Is mycorrhiza functioning influenced by the quantitative composition of the mycorrhizal fungal community? Soil Biol. Biochem. 2021, 157, 108249. [Google Scholar] [CrossRef]
- Fester, T.; Fetzer, I.; Buchert, S.; Lucas, R.; Rillig, M.C.; Härtig, C. Towards a systemic metabolic signature of the arbuscular mycorrhizal interaction. Oecologia 2011, 167, 913–924. [Google Scholar] [CrossRef]
- Hill, E.M.; Robinson, L.A.; Abdul-Sada, A.; Vanbergen, A.J.; Hodge, A.; Hartley, S.E. Arbuscular mycorrhizal fungi and plant chemical defence: Effects of colonisation on above ground and below ground metabolomes. J. Chem. Ecol. 2018, 44, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Tavarini, S.; Passera, B.; Martini, A.; Avio, L.; Sbrana, C.; Giovannetti, M.; Angelini, L.G. Plant growth, steviol glycosides and nutrient uptake as affected by arbuscular mycorrhizal fungi and phosphorous fertilization in Stevia rebaudiana Bert. Ind. Crops Prod. 2018, 111, 899–907. [Google Scholar] [CrossRef]
- Dreher, D.; Baldermann, S.; Schreiner, M.; Hause, B. An arbuscular mycorrhizal fungus and a root pathogen induce different volatiles emitted by Medicago truncatula roots. J. Adv. Res. 2019, 19, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Vo, A.T.; Haddidi, I.; Daood, H.; Mayer, Z.; Posta, K. Impact of arbuscular mycorrhizal inoculation and growth substrate on biomass and content of polyphenols in Eclipta prostrata. Hortic. Sci. 2019, 54, 1976–1983. [Google Scholar] [CrossRef]
- Casarrubias-Castillo, K.; Montero-Vargas, J.M.; Dabdoub-González, N.; Winkler, R.; Martínez-Gallardo, N.A.; Avilés-Arnaut, H.; Délano-Frier, J.P. Distinct gene expression and secondary metabolite profiles in suppressor of prosystemin-mediated responses2 (spr2) tomato mutants having impaired mycorrhizal colonization. PeerJ 2020, 16, e8888. [Google Scholar] [CrossRef]
- Johny, L.; Cahill, D.M.; Adholeya, A. AMF enhance secondary metabolite production in ashwagandha, licorice, and marigold in a fungi-host specific manner. Rhizosphere 2021, 17, 100314. [Google Scholar] [CrossRef]
- Rasouli, F.; Amini, T.; Asadi, M.; Hassanpouraghdam, M.B.; Aazami, M.A.; Ercisli, S.; Skrovankova, S.; Mlcek, J. Growth and antioxidant responses of lettuce (Lactuca sativa L.) to arbuscular mycorrhiza inoculation and seaweed extract foliar application. Agronomy 2022, 12, 401. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, W.; Geng, Y.; Liu, K.; Shao, X. Cooperation with arbuscular mycorrhizal fungi increases plant nutrient uptake and improves defenses against insects. Front. Ecol. Evol. 2022, 23, 833389. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, J.; Yang, F.; Tao, S.; Yan, X.; Zhou, Z.; Zhang, Y. Arbuscular mycorrhizal fungi improve the growth and performance in the seedlings of Leymus chinensis under alkali and drought stresses. PeerJ 2022, 10, e12890. [Google Scholar] [CrossRef]
- Kashif, M.; Sang, Y.; Mo, S.; Rehman, S.; Khan, S.; Khan, M.R.; He, S.; Jiang, C. Deciphering the biodesulfurization pathway employing marine mangrove Bacillus aryabhattai strain NM1-A2 according to whole genome sequencing and transcriptome analyses. Genomics 2023, 115, 110635. [Google Scholar] [CrossRef]
- Liaquat, F.; Munis, M.F.H.; Arif, S.; Haroon, U.; Shi, J.; Saqib, S.; Zaman, W.; Che, S.; Liu, Q. PacBio single-molecule long-read sequencing reveals genes tolerating manganese stress in Schima superba saplings. Front. Genet. 2021, 12, 635043. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.K.; Maurya, A.K.; Irvin, L.; Kelly, M.P.; Schoenherr, A.P.; Huguet-Tapia, J.C.; Bombarely, A. A snapshot of the transcriptome of Medicago truncatula (Fabales: Fabaceae) shoots and roots in response to an arbuscular mycorrhizal fungus and the pea aphid (Acyrthosiphon pisum) (Hemiptera: Aphididae). Environ. Entomol. 2023, 52, 667–680. [Google Scholar] [CrossRef]
- Jing, S.; Li, Y.; Zhu, L.; Su, J.; Yang, T.; Liu, B.; Ma, B.; Ma, F.; Li, M.; Zhang, M. Transcriptomics and metabolomics reveal effect of arbuscular mycorrhizal fungi on growth and development of apple plants. Front. Plant Sci. 2022, 13, 1052464. [Google Scholar] [CrossRef] [PubMed]
- Mamontova, T.; Afonin, A.M.; Ihling, C.; Soboleva, A.; Lukasheva, E.; Sulima, A.S.; Shtark, O.Y.; Akhtemova, G.A.; Povydysh, M.N.; Sinz, A.; et al. Profiling of seed proteome in pea (Pisum sativum L.) lines characterized with high and low responsivity to combined inoculation with nodule bacteria and arbuscular mycorrhizal fungi. Molecules 2019, 24, 1603. [Google Scholar] [CrossRef] [PubMed]
- Watson, B.S.; Asirvatham, V.S.; Wang, L.; Sumner, L.W. Mapping of the proteome of barrel medic (Medicago truncatula). Plant Physiol. 2003, 131, 1104–1123. [Google Scholar] [CrossRef] [PubMed]
- Valot, B.; Negroni, L.; Zivy, M.; Gianinazzi, S.; Dumas-Gaudot, E. A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics 2006, 6, S145–S155. [Google Scholar] [CrossRef]
- Desalegn, G.; Turetschek, R.; Kaul, H.-P.; Wienkoop, S. Microbial symbionts affect Pisum sativum proteome and metabolome under Didymella pinodes infection. J. Proteomics 2016, 143, 173–187. [Google Scholar] [CrossRef]
- Aloui, A.; Recorbet, G.; Lemaître-Guillier, C.; Mounier, A.; Balliau, T.; Zivy, M.; Wipf, D.; Dumas-Gaudot, E. The plasma membrane proteome of Medicago truncatula roots as modified by arbuscular mycorrhizal symbiosis. Mycorrhiza 2018, 281, 1–16. [Google Scholar] [CrossRef]
- Schliemann, W.; Ammer, C.; Strack, D. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 2008, 69, 112–146. [Google Scholar] [CrossRef]
- Yurkov, A.P.; Jacobi, L.M.; Kryukov, A.A.; Perevedentseva, L.G.; Shishova, M.F. Brief overview of arbuscular mycorrhiza research conducted in Russia over the XXth century. Revista GEINTEC 2021, 11, 71–92. [Google Scholar] [CrossRef]
- Yurkov, A.; Puzanskiy, R.; Kryukov, A.; Gorbunova, A.; Kudriashova, T.; Jacobi, L.; Kozhemyakov, A.; Romanyuk, D.; Aronova, E.; Avdeeva, G.; et al. The role of Medicago lupulina interaction with Rhizophagus irregularis in determination of root metabolome at early stages of AM symbiosis. Plants 2022, 11, 2338. [Google Scholar] [CrossRef]
- Shtark, O.Y.; Puzanskiy, R.K.; Avdeeva, G.S.; Yurkov, A.P.; Smolikova, G.N.; Yemelyanov, V.V.; Kliukova, M.S.; Shavarda, A.L.; Kirpichnikova, A.A.; Zhernakov, A.I.; et al. Metabolic alterations in pea leaves during arbuscular mycorrhiza development. PeerJ 2019, 7, e7495. [Google Scholar] [CrossRef]
- Shtark, O.; Puzanskiy, R.; Avdeeva, G.; Yemelyanov, V.; Shavarda, A.; Romanyuk, D.; Kliukova, M.; Kirpichnikova, A.; Tikhonovich, I.; Zhukov, V.; et al. Metabolic alterations in Pisum sativum roots during plant growth and arbuscular mycorrhiza development. Plants 2021, 10, 1033. [Google Scholar] [CrossRef]
- Grunwald, U.; Guo, W.; Fischer, K.; Isayenkov, S.; Ludwig-Müller, J.; Hause, B.; Yan, X.; Küster, H.; Franken, P. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 2009, 229, 1023–1034. [Google Scholar] [CrossRef]
- Wang, M.; Schäfer, M.; Li, D.; Halitschke, R.; Dong, C.; McGale, E.; Paetz, C.; Song, Y.; Li, S.; Dong, J.; et al. Blumenols as shoot markers of root symbiosis with arbuscular mycorrhizal fungi. eLife 2018, 7, e37093. [Google Scholar] [CrossRef]
- Afonin, A.M.; Leppyanen, I.V.; Kulaeva, O.A.; Shtark, O.Y.; Tikhonovich, I.A.; Dolgikh, E.A.; Zhukov, V.A. A high coverage reference transcriptome assembly of pea (Pisum sativum L.) mycorrhizal roots. Vavilovskii Zhurnal Genet. Sel. 2020, 24, 331–339. [Google Scholar] [CrossRef]
- Zhernakov, A.; Rotter, B.; Winter, P.; Borisov, A.; Tikhonovich, I.; Zhukov, V. Massive Analysis of cDNA Ends (MACE) for transcript-based marker design in pea (Pisum sativum L.). Genom. Data 2016, 11, 75–76. [Google Scholar] [CrossRef]
- Zawada, A.M.; Rogacev, K.S.; Müller, S.; Rotter, B.; Winter, P.; Fliser, D.; Heine, G.H. Massive analysis of cDNA Ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease. Epigenetics 2014, 9, 161–172. [Google Scholar] [CrossRef]
- Zhernakov, A.I.; Shtark, O.Y.; Kulaeva, O.A.; Fedorina, J.V.; Afonin, A.M.; Kitaeva, A.B.; Tsyganov, V.E.; Afonso-Grunz, F.; Hoffmeier, K.; Rotter, B.; et al. Mapping-by-sequencing using NGS-based 3′-MACE-Seq reveals a new mutant allele of the essential nodulation gene Sym33 (IPD3) in pea (Pisum sativum L.). PeerJ 2019, 2, e6662. [Google Scholar] [CrossRef]
- Bojahr, J.; Nhengiwa, O.; Krezdorn, N.; Rotter, B.; Saal, B.; Ruge-Wehling, B.; Struck, C.; Winter, P. Massive analysis of cDNA ends (MACE) reveals a co-segregating candidate gene for LpPg1 stem rust resistance in perennial ryegrass (Lolium perenne). Theor. Appl. Genet. 2016, 129, 1915–1932. [Google Scholar] [CrossRef]
- Fischer, K.; Dieterich, R.; Nelson, M.N.; Kamphuis, L.G.; Singh, K.B.; Rotter, B.; Krezdorn, N.; Winter, P.; Wehling, P.; Ruge-Wehling, B. Characterization and mapping of LanrBo: A locus conferring anthracnose resistance in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 2015, 128, 2121–2130. [Google Scholar] [CrossRef]
- Afonin, A.M.; Gribchenko, E.S.; Zorin, E.A.; Sulima, A.S.; Romanyuk, D.A.; Zhernakov, A.I.; Shtark, O.Y.; Akhtemova, G.A.; Zhukov, V.A. Unique transcriptome features of pea (Pisum sativum L.) lines with differing responses to beneficial soil microorganisms. Ecol. Genet. 2021, 19, 131–141. [Google Scholar] [CrossRef]
- Keymer, A.; Pimprikar, P.; Wewer, V.; Huber, C.; Brands, M.; Bucerius, S.L.; Delaux, P.-M.; Klingl, V.; von Röpenack-Lahaye, E.; Wang, T.L.; et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. eLife 2017, 6, e29107. [Google Scholar] [CrossRef]
- Bravo, A.; Brands, M.; Wewer, V.; Dormann, P.; Harrison, M.J. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 2017, 214, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Shtark, O.Y.; Borisov, A.Y.; Zhukov, V.A.; Tikhonovich, I.A. Mutually beneficial legume symbioses with soil microbes and their potential for plant production. Symbiosis 2012, 58, 51–62. [Google Scholar] [CrossRef]
- Shtark, O.Y.; Danilova, T.N.; Naumkina, T.S.; Vasilchikov, A.G.; Chebotar, V.K.; Kazakov, A.E.; Zhernakov, A.I.; Nemankin, T.A.; Prilepskaya, N.A.; Borisov, A.Y.; et al. Analysis of pea (Pisum sativum L.) source material for breeding of cultivars with high symbiotic potential and choice of criteria for its evaluation. Ecol. Genet. 2006, 4, 22–28. [Google Scholar] [CrossRef]
- Yurkov, A.P.; Jacobi, L.M.; Gapeeva, N.E.; Stepanova, G.V.; Shishova, M.F. Development of arbuscular mycorrhiza in highly responsive and mycotrophic host plant—Black medick (Medicago lupulina L.). Russ. J. Dev. Biol. 2015, 46, 263–275. [Google Scholar] [CrossRef]
- Yurkov, A.P.; Veselova, S.V.; Jacobi, L.M.; Stepanova, G.V.; Yemelyanov, V.V.; Kudoyarova, G.R.; Shishova, M.F. The effect of inoculation with arbuscular mycorrhizal fungus Rhizophagus irregularis on cytokinin content in highly mycotrophic Medicago lupulina line under low phosphorus level in soil. Plant Soil Environ. 2017, 63, 519–524. [Google Scholar] [CrossRef]
- Yurkov, A.P.; Veselova, S.V.; Jacobi, L.M.; Stepanova, G.V.; Kudoyarova, G.R.; Shishova, M.F. Effect of inoculation with arbuscular mycorrhizal fungus Rhizophagus irregularis on auxin content in highly mycotrophic black medick under low phosphorus in soil. Sel’skokhozyaistvennaya Biol. 2017, 52, 830–838. [Google Scholar] [CrossRef]
- Yurkov, A.P.; Puzanskiy, R.K.; Avdeeva, G.S.; Jacobi, L.M.; Gorbunova, A.O.; Kryukov, A.A.; Kozhemyakov, A.P.; Laktionov, Y.V.; Kosulnikov, Y.V.; Romanyuk, D.A.; et al. Mycorrhiza-induced alterations in metabolome of Medicago lupulina leaves during symbiosis development. Plants 2021, 10, 2506. [Google Scholar] [CrossRef] [PubMed]
- Yurkov, A.; Kryukov, A.; Gorbunova, A.; Sherbakov, A.; Dobryakova, K.; Mikhaylova, Y.; Afonin, A.; Shishova, M. AM-induced alteration in the expression of genes, encoding phosphorus transporters and enzymes of carbohydrate metabolism in Medicago lupulina. Plants 2020, 9, 486. [Google Scholar] [CrossRef]
- Pecrix, Y.; Staton, S.E.; Sallet, E.; Lelandais-Brière, C.; Moreau, S.; Carrère, S.; Blein, T.; Jardinaud, M.; Latrasse, D.; Zouine, M.; et al. Medicago Truncatula A17 r5.0 Genome Portal. 2022. Available online: https://medicago.toulouse.inra.fr/MtrunA17r5.0-ANR/ (accessed on 31 July 2023).
- Pecrix, Y.; Staton, S.E.; Sallet, E.; Lelandais-Brière, C.; Moreau, S.; Carrère, S.; Blein, T.; Jardinaud, M.; Latrasse, D.; Zouine, M.; et al. Whole-genome landscape of Medicago truncatula symbiotic genes. Nat. Plants 2018, 4, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef]
- Tran, B.; Cavagnaro, T.; Jewell, N.; Brien, C.; Berger, B.; Watts-Williams, S. High-throughput phenotyping reveals growth of Medicago truncatula is positively affected by arbuscular mycorrhizal fungi even at high soil phosphorus availability. Plants People Planet 2020, 3, 600–613. [Google Scholar] [CrossRef]
- Schroeder, M.S.; Janos, D.P. Plant growth, phosphorus nutrition, and root morphological responses to arbuscular mycorrhizas, phosphorus fertilization, and intraspecific density. Mycorrhiza 2005, 15, 203–216. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: San Diego, CA, USA, 2008. [Google Scholar]
- Adolfsson, L.; Nziengui, H.; Abreu, I.N.; Šimura, J.; Beebo, A.; Herdean, A.; Aboalizadeh, J.; Široká, J.; Moritz, T.; Novák, O.; et al. Enhanced secondary- and hormone metabolism in leaves of arbuscular mycorrhizal Medicago truncatula. Plant Physiol. 2017, 175, 392–411. [Google Scholar] [CrossRef]
- Cope, K.R.; Kafle, A.; Yakha, J.K.; Pfeffer, P.E.; Strahan, G.D.; Garcia, K.; Subramanian, S.; Bücking, H. Physiological and transcriptomic response of Medicago truncatula to colonization by high- or low-benefit arbuscular mycorrhizal fungi. Mycorrhiza 2022, 32, 281–303. [Google Scholar] [CrossRef]
- Cesaro, P.; Massa, N.; Cantamessa, S.; Todeschini, V.; Bona, E.; Berta, G.; Barbato, R.; Lingua, G. Tomato responses to Funneliformis mosseae during the early stages of arbuscular mycorrhizal symbiosis. Mycorrhiza 2020, 30, 601–610. [Google Scholar] [CrossRef]
- Yuttavanichakul, W.; Teamtisong, K.; Teaumroong, N.; Boonkerd, N.; Tittabutr, P. Brevibacillus sp. promotes maize root colonization by Acaulospora tuberculata and the alteration of associated plant protein responses. J. Plant Interact. 2018, 13, 543–554. [Google Scholar] [CrossRef]
- Aleandri, M.P.; Martignoni, D.; Reda, R.; Chilosi, G. Effects of preconditioning through mycorrhizal inoculation on the control of melon root rot and vine decline caused by Monosporascus cannonballus. J. Phytopathol. 2015, 163, 898–907. [Google Scholar] [CrossRef]
- Tan, Z.; Hu, Y.; Lin, Z. Expression of SYMRK affects the development of arbuscular mycorrhiza in tobacco roots. Acta Physiol. Plant. 2013, 35, 85–94. [Google Scholar] [CrossRef]
- Kim, G.-B.; Son, S.-U.; Yu, H.-J.; Mun, J.-H. MtGA2ox10 encoding C20-GA2-oxidase regulates rhizobial infection and nodule development in Medicago truncatula. Sci. Rep. 2019, 9, 5952. [Google Scholar] [CrossRef]
- Gao, H.; Ma, K.; Ji, G.; Pan, L.; Zhou, Q. Lipid transfer proteins involved in plant-pathogen interactions and their molecular mechanisms. Mol. Plant Pathol. 2022, 23, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Karlo, M.; Boschiero, C.; Landerslev, K.G.; Blanco, G.S.; Wen, J.; Mysore, K.S.; Dai, X.; Zhao, P.X.; de Bang, T.C. The CLE53-SUNN genetic pathway negatively regulates arbuscular mycorrhiza root colonization in Medicago truncatula. J. Exp. Bot. 2020, 71, 4972–4984. [Google Scholar] [CrossRef]
- Ren, C.-G.; Kong, C.-C.; Yan, K.; Xie, Z.-H. Transcriptome analysis reveals the impact of arbuscular mycorrhizal symbiosis on Sesbania cannabina expose to high salinity. Sci. Rep. 2019, 9, 2780. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Y.; Zhao, J.; Zhen, X.; Guo, C.; Shu, Y. Genome-wide identification and characterization of R2R3 MYB genes in Medicago truncatula. Genet. Mol. Biol. 2019, 42, 611–623. [Google Scholar] [CrossRef]
- Wang, P.; Snijders, R.; Kohlen, W.; Liu, J.; Bisseling, T.; Limpens, E. Medicago SPX1 and SPX3 regulate phosphate homeostasis, mycorrhizal colonization, and arbuscule degradation. Plant Cell 2021, 33, 3470–3486. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; Molinero-Rosales, N.; Flores, D.F.; Díaz, M.V.; García-Garrido, J.M. Identification and expression analysis of GRAS transcription factor genes involved in the control of arbuscular mycorrhizal development in tomato. Front. Plant Sci. 2019, 10, 268. [Google Scholar] [CrossRef]
- Aloui, A.; Recorbet, G.; Robert, F.; Schoefs, B.; Bertrand, M.; Henry, C.; Gianinazzi-Pearson, V.; Dumas-Gaudot, E.; Aschi-Smiti, S. Arbuscular mycorrhizal symbiosis elicits shoot proteome changes that are modified during cadmium stress alleviation in Medicago truncatula. BMC Plant Biol. 2011, 11, 75. [Google Scholar] [CrossRef]
- Sharma, N.; Aggarwal, A.; Yadav, K. Arbuscular mycorrhizal fungi enhance growth, physiological parameters and yield of salt-stressed Phaseolus mungo (L.) Hepper. Eur. J. Environ. Sci. 2017, 7, 5–13. [Google Scholar] [CrossRef][Green Version]
- Schweiger, R.; Baier, M.C.; Müller, C. Arbuscular mycorrhiza-induced shifts in foliar metabolism and photosynthesis mirror the developmental stage of the symbiosis and are only partly driven by improved phosphate uptake. MPMI 2014, 27, 1403–1412. [Google Scholar] [CrossRef]
- Machiani, A.M.; Javanmard, A.; Machiani, H.R.; Sadeghpour, A. Arbuscular mycorrhizal fungi and changes in primary and secondary metabolites. Plants 2022, 11, 2183. [Google Scholar] [CrossRef]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar] [CrossRef] [PubMed]
- Wipf, D.; Mongelard, G.; van Tuinen, D.; Gutierrez, L.; Casieri, L. Transcriptional responses of Medicago truncatula upon sulfur deficiency stress and arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2014, 5, 680. [Google Scholar] [CrossRef] [PubMed]
- Quiroga, G.; Erice, G.; Aroca, R.; Chaumont, F.; Ruiz-Lozano, J.M. Enhanced drought stress tolerance by the arbuscular mycorrhizal symbiosis in a drought-sensitive maize cultivar is related to a broader and differential regulation of host plant aquaporins than in a drought-tolerant cultivar. Front. Plant Sci. 2017, 19, 1056. [Google Scholar] [CrossRef]
- Hsieh, C.; Chen, Y.-H.; Chang, K.-C.; Yang, S.-Y. Transcriptome analysis reveals the mechanisms for mycorrhiza-enhanced salt tolerance in rice. Front. Plant Sci. 2022, 13, 1072171. [Google Scholar] [CrossRef]
- Wang, T.; Zhao, M.; Zhang, X.; Liu, M.; Yang, C.; Chen, Y.; Chen, R.; Wen, J.; Mysore, K.S.; Zhang, W.H. Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 2017, 68, 5937–5948. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martın, A.C.; Iglesias, J.; Leyva, A.; PazAres, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Develop. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.; Ramirez, M.; Valdés-López, O.; Lara, M.; Tesfaye, M.; Vance, C. Identification of candidate phosphorus stress induced genes in Phaseolus vulgaris through clustering analysis across several plant species. Func. Plant Biol. 2006, 33, 789–797. [Google Scholar] [CrossRef]
- Liu, J.; Maldonado-Mendoza, I.; Lopez-Meyer, M.; Cheung, F.; Town, C.D.; Harrison, M.J. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 2007, 50, 529–544. [Google Scholar] [CrossRef]
- Arosio, P.; Ingrassia, R.; Cavadini, P. Ferritins: A family ofmolecules for iron storage, antioxidation and more. Biochim. Biophys. Acta 2009, 1790, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Ali, S.; Al Azzawi, T.N.I.; Saqib, S.; Ullah, F.; Ayaz, A.; Zaman, W. The key roles of ROS and RNS as a signaling molecule in plant-microbe interactions. Antioxidants 2023, 12, 268. [Google Scholar] [CrossRef] [PubMed]
- Belmondo, S.; Calcagno, C.; Genre, A.; Puppo, A.; Pauly, N.; Lanfranco, L. NADPH oxidases in the arbuscular mycorrhizal symbiosis. Plant Signal. Behav. 2016, 11, e1165379. [Google Scholar] [CrossRef]
- Kapoor, R.; Singh, N. Arbuscular mycorrhiza and reactive oxygen species. In Arbuscular Mycorrhizas and Stress Tolerance of Plants; Wu, Q.S., Ed.; Springer: Singapore, 2017; pp. 225–243. [Google Scholar] [CrossRef]
- Liu, A.; Chen, S.; Wang, M.; Liu, D.; Chang, R.; Wang, Z.; Lin, X.; Bai, B.; Ahammed, G.J. Arbuscular mycorrhizal fungus alleviates chilling stress by boosting redox poise and antioxidant potential of tomato seedlings. J. Plant Growth. Regul. 2016, 35, 109–120. [Google Scholar] [CrossRef]
- Saroy, K.; Garg, N. Relative effectiveness of arbuscular mycorrhiza and polyamines in modulating ROS generation and ascorbate-glutathione cycle in Cajanus cajan under nickel stress. Environ. Sci. Pollut. Res. Int. 2021, 28, 48872–48889. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, F.; Tang, M. Transcriptome analysis of arbuscular mycorrhizal Casuarina glauca in damage mitigation of roots on NaCl stress. Microorganisms 2022, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Pedranzani, H.E.; Gutiérrez, M.H.; Arias, S.M.; Zapico, M.G.; Ruiz-Lozano, J.M. Arbuscular mycorrhiza interaction with Medicago sativa plants: Study of abiotic stress tolerance in sustainable agriculture. AIA Av. Investig. Agropecu 2021, 25, 26–40. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Liu, C.; Gao, Y.; Han, L.; Chu, H. Arbuscular mycorrhizal fungi contribute to reactive oxygen species homeostasis of Bombax ceiba L. under drought stress. Front. Microbiol. 2022, 13, 991781. [Google Scholar] [CrossRef]
- González-Guerrero, M.; Cano, C.; Azcón-Aguilar, C.; Ferrol, N. GintMT1 encodes a functional metallothionein in Glomus intraradices that responds to oxidative stress. Mycorrhiza 2007, 17, 327–335. [Google Scholar] [CrossRef]
- Benabdellah, K.; Merlos, M.A.; Azcón-Aguilar, C.; Ferrol, N. GintGRX1, the first characterized glomeromycotan glutaredoxin, is a multifunctional enzyme that responds to oxidative stress. Fungal Genet. Biol. 2009, 46, 94–103. [Google Scholar] [CrossRef]
- Nordberg, J.; Arnér, E.S. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 2001, 31, 1287–1312. [Google Scholar] [CrossRef]
- Song, F.; Li, J.; Fan, X.; Zhang, Q.; Chang, W.; Yang, F.; Geng, G. Transcriptome analysis of Glomus mosseae/Medicago sativa mycorrhiza on atrazine stress. Sci. Rep. 2016, 6, 20245. [Google Scholar] [CrossRef] [PubMed]
- Kirpichnikov, N.A.; Volkov, A.A.; Chernyshkova, L.B.; Yurkov, A.P.; Yakobi, L.M.; Kozhemyakov, A.P.; Zavalin, A.A. Effect of phosphorus fertilizers, lime materials, and biopreparations in barley and clover plants in a mixed plantation. Agrohimia 2012, 11, 16–27. [Google Scholar]
- Efimova, I.L.; Yurkov, A.P. New methods of agroecology for improving the quality of apple planting material. Proc. Kuban State Univ. 2015, 4, 73–77. [Google Scholar]
- Yurkov, A.P.; Stepanova, G.V.; Yakobi, L.M.; Kozhemyakov, A.P.; Sergaliev, N.H.; Amenova, R.K.; Djaparov, R.S.; Volodin, M.A.; Tlepov, A.S.; Baymukanov, E.N. Productivity of spring and winter wheat in drought conditions dependent on the application of arbuscular mycorrhizal fungus Glomus intraradices. Fodd. Prod. 2012, 12, 18–24. [Google Scholar]
- Kryukov, A.A.; Yurkov, A.P. Optimization procedures for molecular-genetic identification of arbuscular mycorrhizal fungi in symbiotic phase on the example of two closely kindred strains. Mikol. I Fitopatol. 2018, 52, 38–48. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2023. Available online: https://github.com/s-andrews/FastQC (accessed on 31 July 2023).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Ponce-Soto, G.Y.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.E.; Usadel, B. MapMan4: A refined protein classification and annotation framework applicable to multi-omics data analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer New York: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods-a bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158–161. [Google Scholar] [CrossRef]
- Trouvelot, A.; Kough, J.L.; Gianinazzi-Pearson, V. Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes ayant une signification fonctionnelle. In Physiological and Genetical Aspects of Mycorrhizae; Gianinazzi-Pearson, V., Gianinazzi, S., Eds.; INRA-Press: Paris, France, 1986; pp. 217–221. [Google Scholar]
- Vorobyev, N.I.; Yurkov, A.P.; Provorov, N.A. Certificate N2010612112 about the Registration of the Computer Program “Program for Calculating the Mycorrhization Indices of Plant Roots” (Dated 2 December 2016); The Federal Service for Intellectual Property: Moscow, Russia, 2016. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yurkov, A.P.; Afonin, A.M.; Kryukov, A.A.; Gorbunova, A.O.; Kudryashova, T.R.; Kovalchuk, A.I.; Gorenkova, A.I.; Bogdanova, E.M.; Kosulnikov, Y.V.; Laktionov, Y.V.; et al. The Effects of Rhizophagus irregularis Inoculation on Transcriptome of Medicago lupulina Leaves at Early Vegetative and Flowering Stages of Plant Development. Plants 2023, 12, 3580. https://doi.org/10.3390/plants12203580
Yurkov AP, Afonin AM, Kryukov AA, Gorbunova AO, Kudryashova TR, Kovalchuk AI, Gorenkova AI, Bogdanova EM, Kosulnikov YV, Laktionov YV, et al. The Effects of Rhizophagus irregularis Inoculation on Transcriptome of Medicago lupulina Leaves at Early Vegetative and Flowering Stages of Plant Development. Plants. 2023; 12(20):3580. https://doi.org/10.3390/plants12203580
Chicago/Turabian StyleYurkov, Andrey P., Alexey M. Afonin, Alexey A. Kryukov, Anastasia O. Gorbunova, Tatyana R. Kudryashova, Anastasia I. Kovalchuk, Anastasia I. Gorenkova, Ekaterina M. Bogdanova, Yuri V. Kosulnikov, Yuri V. Laktionov, and et al. 2023. "The Effects of Rhizophagus irregularis Inoculation on Transcriptome of Medicago lupulina Leaves at Early Vegetative and Flowering Stages of Plant Development" Plants 12, no. 20: 3580. https://doi.org/10.3390/plants12203580
APA StyleYurkov, A. P., Afonin, A. M., Kryukov, A. A., Gorbunova, A. O., Kudryashova, T. R., Kovalchuk, A. I., Gorenkova, A. I., Bogdanova, E. M., Kosulnikov, Y. V., Laktionov, Y. V., Kozhemyakov, A. P., Romanyuk, D. A., Zhukov, V. A., Puzanskiy, R. K., Mikhailova, Y. V., Yemelyanov, V. V., & Shishova, M. F. (2023). The Effects of Rhizophagus irregularis Inoculation on Transcriptome of Medicago lupulina Leaves at Early Vegetative and Flowering Stages of Plant Development. Plants, 12(20), 3580. https://doi.org/10.3390/plants12203580





