Transcriptomic Time-Course Sequencing: Insights into the Cell Wall Macromolecule-Mediated Fruit Dehiscence during Ripening in Camellia oleifera
Abstract
:1. Introduction
2. Results
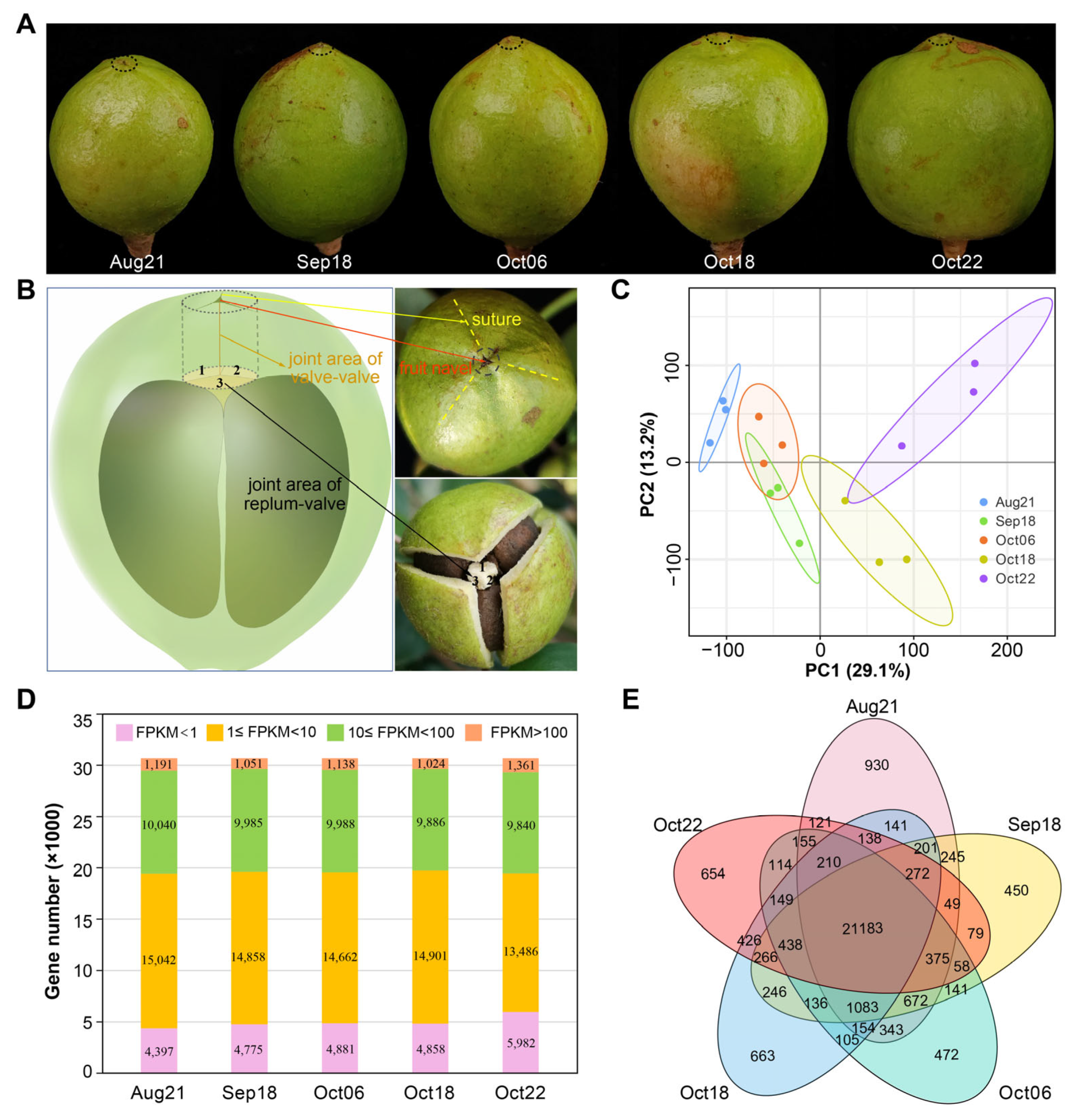
2.1. Overview of the Transcriptomic Analysis of Dissected Fruit Tissues
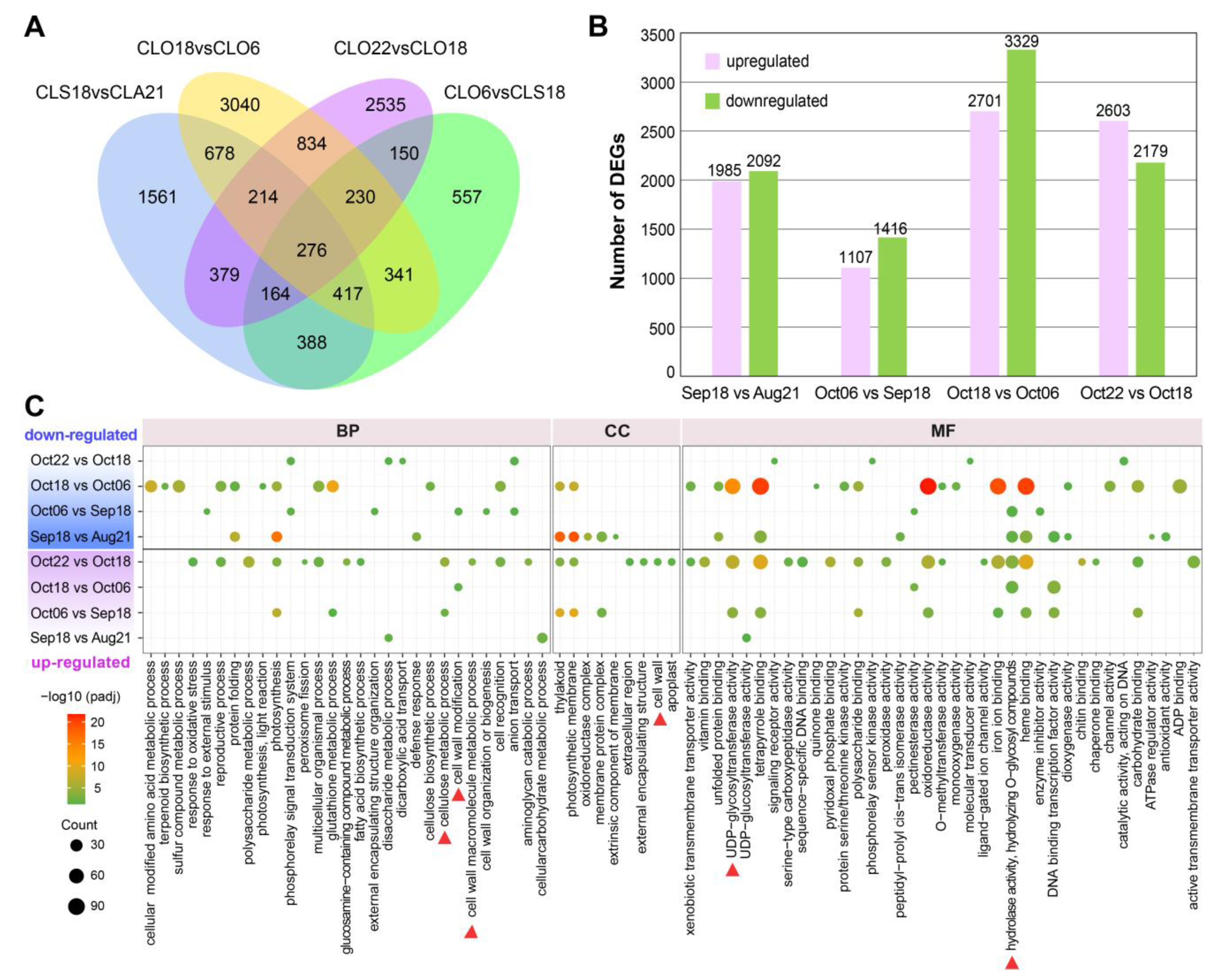
2.2. Analysis of DEGs Suggested Drastic Transcriptional Changes Occurred in the Final Stages of Pericarp–Replum Tissue
2.3. Time-Course Analysis of Pericarp–Replum Tissues Showed Strongly Induced Cell Wall-Related Transcriptional Changes in the Last Two Stages
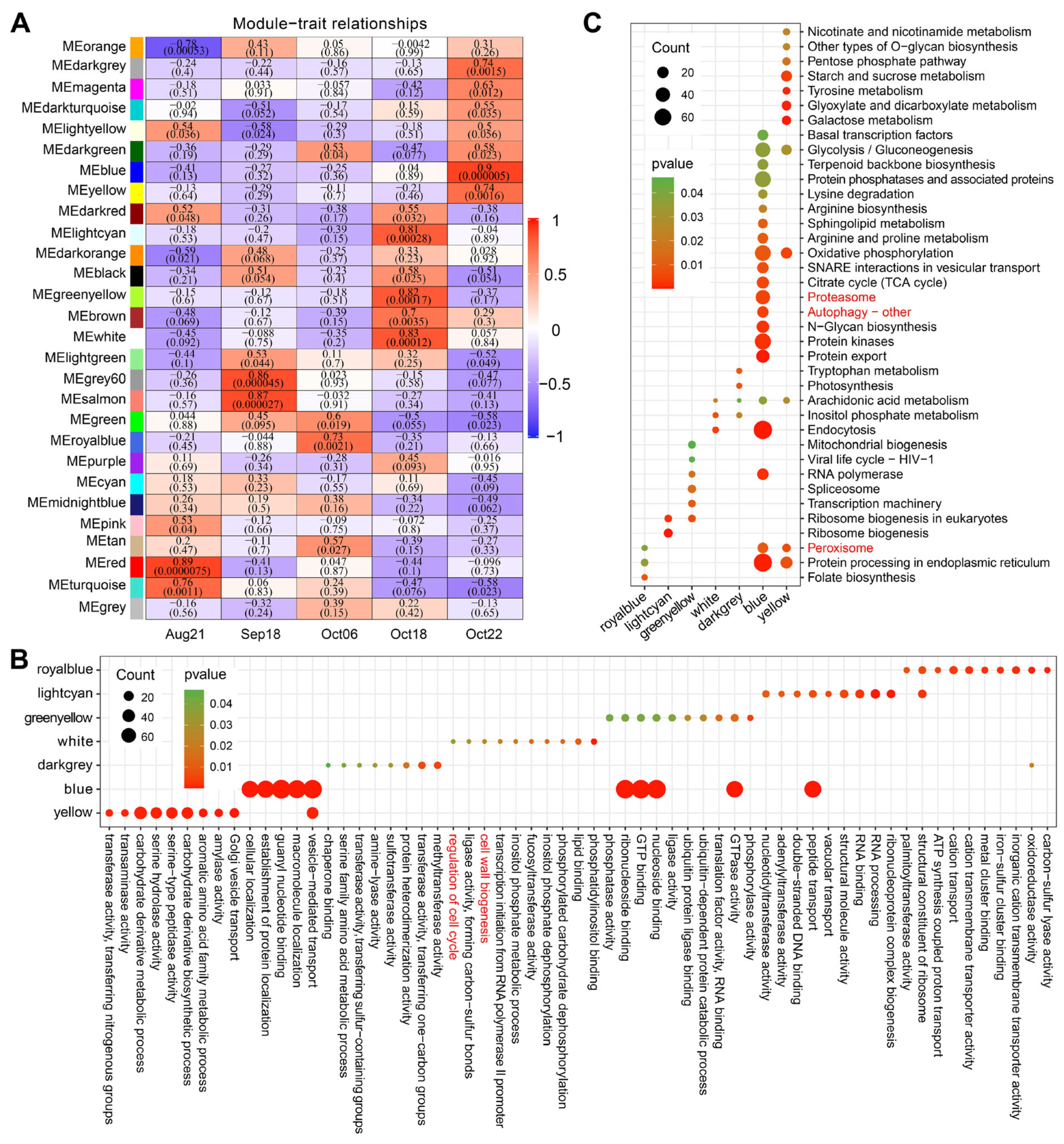
2.4. Co-Expression Network Analysis Revealed Relationships between Developmental Stages and DEGs
2.5. Analysis of Cell Wall-Related Hub Genes in Co-Expressed Modules That Were Highly Associated with the Final Developmental Stage of C. oleifera Fruit Dehiscence
2.6. Comprehensive Search for Potential Regulatory Factors Related to Fruit Dehiscence in C. oleifera
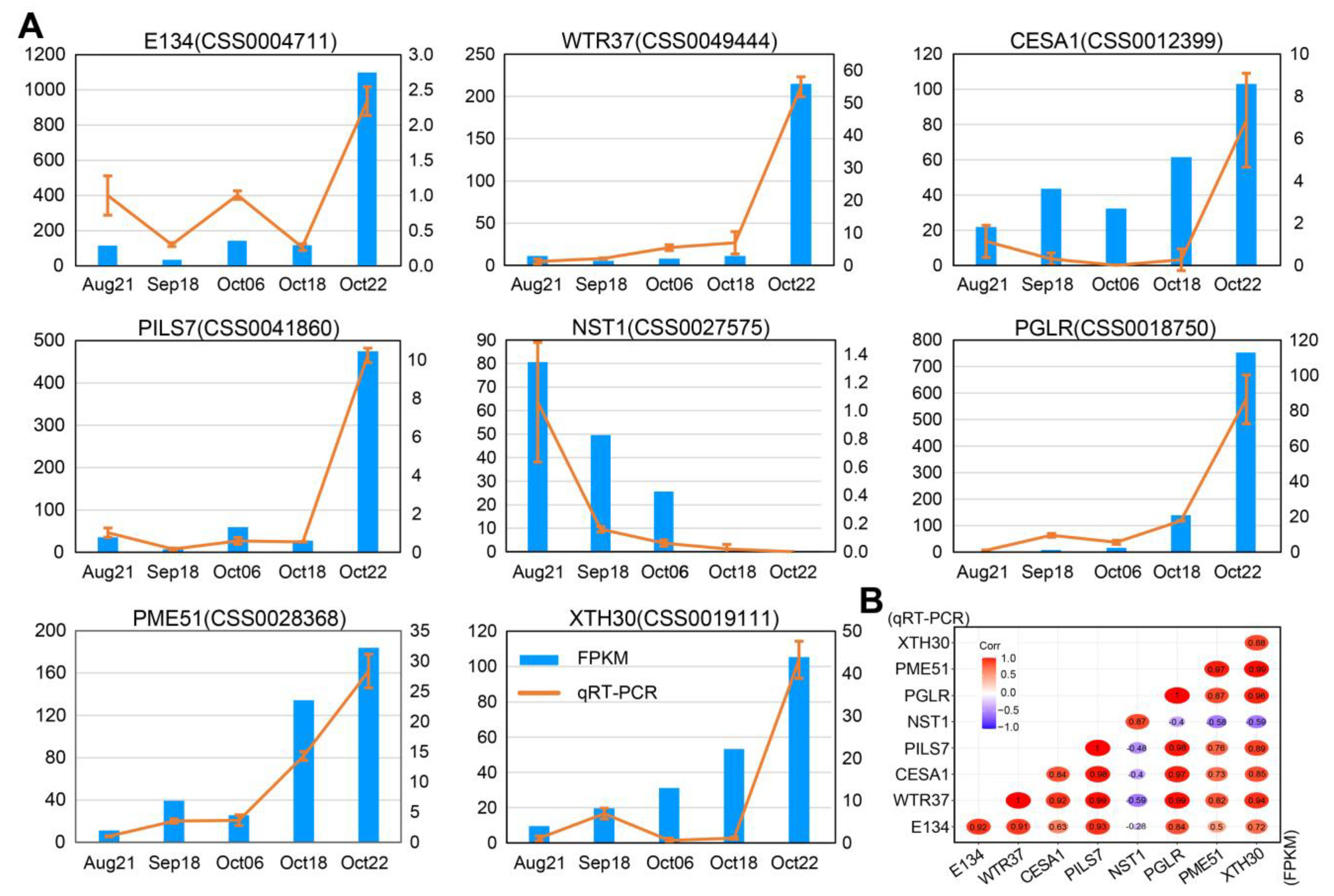
2.7. Quantitative Real-Time PCR (qRT-PCR) Assays of DEGs
3. Discussion
3.1. Tissues Located in the Fruit Navel Region Are Likely to Be the Most Critical Structural and Functional Basis for the Dehiscence of C. oleifera Fruit
3.2. Dehiscing Control in the Later Stage of C. oleifera Fruit Development Points to Cell Wall Activity
3.3. Hydrolases Targeting the Cell Wall Macromolecular Components Are Important Factors in the Dehiscence of C. oleifera Fruit
4. Methods
4.1. Sample Collection and Histological Analysis
4.2. High-Throughput Transcriptome Sequencing
4.3. Sequence Assembly, Functional Annotation, and Classification
4.4. Gene Clustering and Weighted Gene Co-Expression Network Analysis (WGCNA)
4.5. Quantitative RT-PCR (qPCR) Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Funatsuki, H.; Suzuki, M.; Hirose, A.; Inaba, H.; Yamada, T.; Hajika, M.; Komatsu, K.; Katayama, T.; Sayama, T.; Ishimoto, M.; et al. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Natl. Acad. Sci. USA 2014, 111, 17797–17802. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-K.; Li, Y.-L.; Ding, L.-N.; Sarwar, R.; Zhao, F.-Y.; Tan, X.-L. Mechanism and Regulation of Silique Dehiscence, Which Affects Oil Seed Production. Front. Plant Sci. 2020, 11, 580. [Google Scholar] [CrossRef] [PubMed]
- Lenser, T.; TheiBen, G. Conservation of fruit dehiscence pathways between Lepidium campestre and Arabidopsis thaliana sheds light on the regulation of INDEHISCENT. Plant J. 2013, 76, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Benitez, D.; Casimiro-Soriguer, I.; Torres, A.M. First approach to pod dehiscence in faba bean: Genetic and histological analyses. Sci. Rep. 2020, 10, 17678. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.; Liu, J.; Cheng, H.; Li, C.; Fu, L.; Wang, W.; Wang, H.; Hao, M.; Mei, D.; Liu, K.; et al. A lignified-layer bridge controlled by a single recessive gene is associated with high pod-shatter resistance in Brassica napus L. Crop. J. 2022, 10, 638–646. [Google Scholar] [CrossRef]
- Ogutcen, E.; Pandey, A.; Khan, M.K.; Marques, E.; Penmetsa, R.V.; Kahraman, A.; Von Wettberg, E.J.B. Pod Shattering: A Homologous Series of Variation Underlying Domestication and an Avenue for Crop Improvement. Agronomy 2018, 8, 137. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Pelaz, S.; Yanofsky, M.F. Control of Carpel and Fruit Development in Arabidopsis. Annu. Rev. Biochem. 1999, 68, 321–354. [Google Scholar] [CrossRef]
- Ballester, P.; Ferrándiz, C. Shattering fruits: Variations on a dehiscent theme. Curr. Opin. Plant Biol. 2017, 35, 68–75. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Weigel, D.; Yanofsky, M.F. A genetic framework for fruit patterning in Arabidopsis thaliana. Development 2005, 132, 4687–4697. [Google Scholar] [CrossRef]
- Ostergaard, L.; Kempin, S.A.; Bies, D.; Klee, H.J.; Yanofsky, M.F. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol. J. 2006, 4, 45–51. [Google Scholar] [CrossRef]
- Sander, L.; Child, R.; Ulvskov, P.; Albrechtsen, M.; Borkhardt, B. Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: Evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol. Biol. 2001, 46, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kay, P.; Swain, W.S.M. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Bai, M.; Tong, P.; Hu, Y.; Yang, M.; Wu, H. CELLULASE6 and MANNANASE7 Affect Cell Differentiation and Silique Dehiscence. Plant Physiol. 2018, 176, 2186–2201. [Google Scholar] [CrossRef]
- Changfu, Z.; Xiaohua, Y.; Pin, L.; Jin, L. Growth characteristics and dynamic analysis of water and oil content on oil-tea camellia fruit. J. Yangzhou Univ. Agric. Life Sci. Ed. 2013, 34, 49–53. [Google Scholar]
- Ping, L.; Kailiang, W.; Changfu, Z.; Yunhai, X.; Xiaohua, Y.; Hengfu, Y. Seed transcriptomics analysis in Camellia oleifera uncovers genes associated with oil content and fatty acid composition. Int. J. Mol. Sci. 2018, 19, 118. [Google Scholar]
- Zhang, F.; Li, Z.; Zhou, J.; Gu, Y.; Tan, X. Comparative study on fruit development and oil synthesis in two cultivars of Camellia oleifera. BMC Plant Biol. 2021, 21, 348. [Google Scholar] [CrossRef]
- Luo, F.; Fei, X.; Fang, X.; Wang, J.; Wang, Y. Effects of process methods on physicochemical property and nutrient content of Camellia seed oil. Acta Agric. Univ. Jiangxiensis 2012, 34, 87–92. [Google Scholar]
- Tan, X. Advances in the molecular breeding of Camellia oleifera. J. Cent. South Univ. For. Technol. 2023, 43, 1–24. [Google Scholar]
- Yao, X.; Ren, H. Oil-Tea Camellia Genetic Resource in China; Science Press: Beijing, China, 2020. [Google Scholar]
- Xia, Y.; Yao, X. Fruit size classification and cracking characteristics analysis of Camellia oleifera clones. J. Cent. South Univ. For. Technol. 2019, 39, 24–33. [Google Scholar]
- Zhao, S. Identification of a homologue to the Arabidopsis thaliana INDEHISCENT gene in Camellia oleifera. South China For. Sci. 2020, 48, 32–34. [Google Scholar]
- Gan, S.Z.R. Identification of SPATULA/ALCATRAZ genes related to fruit development in Camellia oleifera. South China For. Sci. 2020, 48, 7–11. [Google Scholar]
- Wang, H.; Schippers, J.H.M. The Role and Regulation of Autophagy and the Proteasome During Aging and Senescence in Plants. Genes 2019, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.P.; Bhatia, V.S. Characters of Pod Anatomy Associated with Resistance to Pod-Shattering in Soybean. Ann. Bot. 1995, 76, 483–485. [Google Scholar] [CrossRef]
- Matilla, A.J. How is the silique fruit dismantled over its maturation. Funct. Plant Sci. Biotechnol. 2007, 1, 85–93. [Google Scholar]
- Zhang, Q.; Tu, B.; Liu, C.; Liu, X. Pod anatomy, morphology and dehiscing forces in pod dehiscence of soybean (Glycine max (L.) Merrill). Flora 2018, 248, 48–53. [Google Scholar] [CrossRef]
- Parker, T.A.; Lo, S.; Gepts, P. Pod shattering in grain legumes: Emerging genetic and environment-related patterns. Plant Cell 2021, 33, 179–199. [Google Scholar] [CrossRef]
- Thomas, B.R.; Inouhe, M.; Simmons, C.R.; Nevins, D.J. Endo-1,3;1,4-beta-glucanase from coleoptiles of rice and maize: Role in the regulation of plant growth. Int. J. Biol. Macromol. 2000, 27, 145–149. [Google Scholar] [CrossRef]
- Ranocha, P.; Denancé, N.; Vanholme, R.; Freydier, A.; Martinez, Y.; Hoffmann, L.; Köhler, L.; Pouzet, C.; Renou, J.-P.; Sundberg, B.; et al. Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010, 63, 469–483. [Google Scholar] [CrossRef]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef]
- Xu, S.L.; Rahman, A.; Baskin, T.I.; Kieber, J.J. Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 2008, 20, 3065–3079. [Google Scholar] [CrossRef] [PubMed]
- Louvet, R.; Rayon, C.; Domon, J.-M.; Rusterucci, C.; Fournet, F.; Leaustic, A.; Crépeau, M.-J.; Ralet, M.-C.; Rihouey, C.; Bardor, M.; et al. Major changes in the cell wall during silique development in Arabidopsis thaliana. Phytochemistry 2011, 72, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Shi, Y.; Huang, K.; Zhong, Y.; Chen, J.; Sun, Z.; Luan, M.; Chen, J. Integrative transcriptome and proteome analyses provide new insights into different stages of Akebia trifoliata fruit cracking during ripening. Biotechnol. Biofuels 2020, 13, 149. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-C.; Wu, J.-Y.; Zhang, H.-N.; Shi, S.-Y.; Liu, L.-Q.; Shu, B.; Liang, Q.-Z.; Xie, J.-H.; Wei, Y.-Z. De Novo Assembly and Characterization of Pericarp Transcriptome and Identification of Candidate Genes Mediating Fruit Cracking in Litchi chinensis Sonn. Int. J. Mol. Sci. 2014, 15, 17667–17685. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef]
- Lewis, M.W.; E Leslie, M.; Liljegren, S.J. Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 2006, 9, 59–65. [Google Scholar] [CrossRef]
- Srivastava, V.; Mckee, L.S.; Bulone, V. Plant Cell Walls: eLS; John Wiley & Sons Ltd: Chichester, UK, 2017. [Google Scholar]
- Li, Y.-L.; Yu, Y.-K.; Zhu, K.-M.; Ding, L.-N.; Wang, Z.; Yang, Y.-H.; Cao, J.; Xu, L.-Z.; Li, Y.-M.; Tan, X.-L. Down-regulation of MANNANASE7 gene in Brassica napus L. enhances silique dehiscence-resistance. Plant Cell Rep. 2021, 40, 361–374. [Google Scholar] [CrossRef]
- Hongo, S.; Sato, K.; Yokoyama, R.; Nishitani, K. Demethylesterification of the Primary Wall by PECTIN METHYLESTERASE35 Provides Mechanical Support to the Arabidopsis Stem. Plant Cell 2012, 24, 2624–2634. [Google Scholar] [CrossRef]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Maruo, T.; Ishibashi, C.; Ueda, Y.; Kobayashi, N.; Yamagishi, M.; Itamura, H. Expression of genes encoding xyloglucan endotransglycosylase/hydrolase in ‘Saijo’ persimmon fruit during softening after deastringency treatment. Postharvest Biol. Technol. 2011, 62, 89–92. [Google Scholar] [CrossRef]
- Roberts, J.A.; Whitelaw, C.A.; Gonzalez-Carranza, Z.H.; McManus, M.T. Cell separation processes in plants-models, mechanisms and manipulation. Ann. Bot. 2000, 86, 223–235. [Google Scholar] [CrossRef]
- Ferrándiz, C. Regulation of fruit dehiscence in Arabidopsis. J. Exp. Bot. 2002, 53, 2031–2038. [Google Scholar] [CrossRef]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant. 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Wu, M.; Gu, L. TCseq: Time Course Sequencing Data Analysis. R Package Version 1.20.0. Available online: http://mirrors.nju.edu.cn/bioconductor/3.15/bioc/html/TCseq.html (accessed on 2 March 2022).
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.F.; Lin, P.; Yao, X.H.; Wang, K.L.; Chang, J.; Han, X.J. Selection of reference genes for quantitative real-time PCR in six oil-tea Camellia based on RNA-seq. Mol. Biol. 2013, 47, 836–851. [Google Scholar] [CrossRef]
- Long, W.; Yao, X.; Wang, K.; Sheng, Y.; Lv, L. De novo transcriptome assembly of the cotyledon of Camellia oleifera for discovery of genes regulating seed germination. BMC Plant Biol. 2022, 22, 265. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, Y.; Yao, X.; Liu, L.; Yu, C.; Wang, K.; Wang, K.; Chang, J.; Chen, J.; Cao, Y. Transcriptomic Time-Course Sequencing: Insights into the Cell Wall Macromolecule-Mediated Fruit Dehiscence during Ripening in Camellia oleifera. Plants 2023, 12, 3314. https://doi.org/10.3390/plants12183314
Sheng Y, Yao X, Liu L, Yu C, Wang K, Wang K, Chang J, Chen J, Cao Y. Transcriptomic Time-Course Sequencing: Insights into the Cell Wall Macromolecule-Mediated Fruit Dehiscence during Ripening in Camellia oleifera. Plants. 2023; 12(18):3314. https://doi.org/10.3390/plants12183314
Chicago/Turabian StyleSheng, Yu, Xiaohua Yao, Linxiu Liu, Chunlian Yu, Kunxi Wang, Kailiang Wang, Jun Chang, Juanjuan Chen, and Yongqing Cao. 2023. "Transcriptomic Time-Course Sequencing: Insights into the Cell Wall Macromolecule-Mediated Fruit Dehiscence during Ripening in Camellia oleifera" Plants 12, no. 18: 3314. https://doi.org/10.3390/plants12183314
APA StyleSheng, Y., Yao, X., Liu, L., Yu, C., Wang, K., Wang, K., Chang, J., Chen, J., & Cao, Y. (2023). Transcriptomic Time-Course Sequencing: Insights into the Cell Wall Macromolecule-Mediated Fruit Dehiscence during Ripening in Camellia oleifera. Plants, 12(18), 3314. https://doi.org/10.3390/plants12183314





