African Mountain Thistles: Three New Genera in the Carduus-Cirsium Group
Abstract
:1. Introduction
2. Results
2.1. Target Loci Recovery
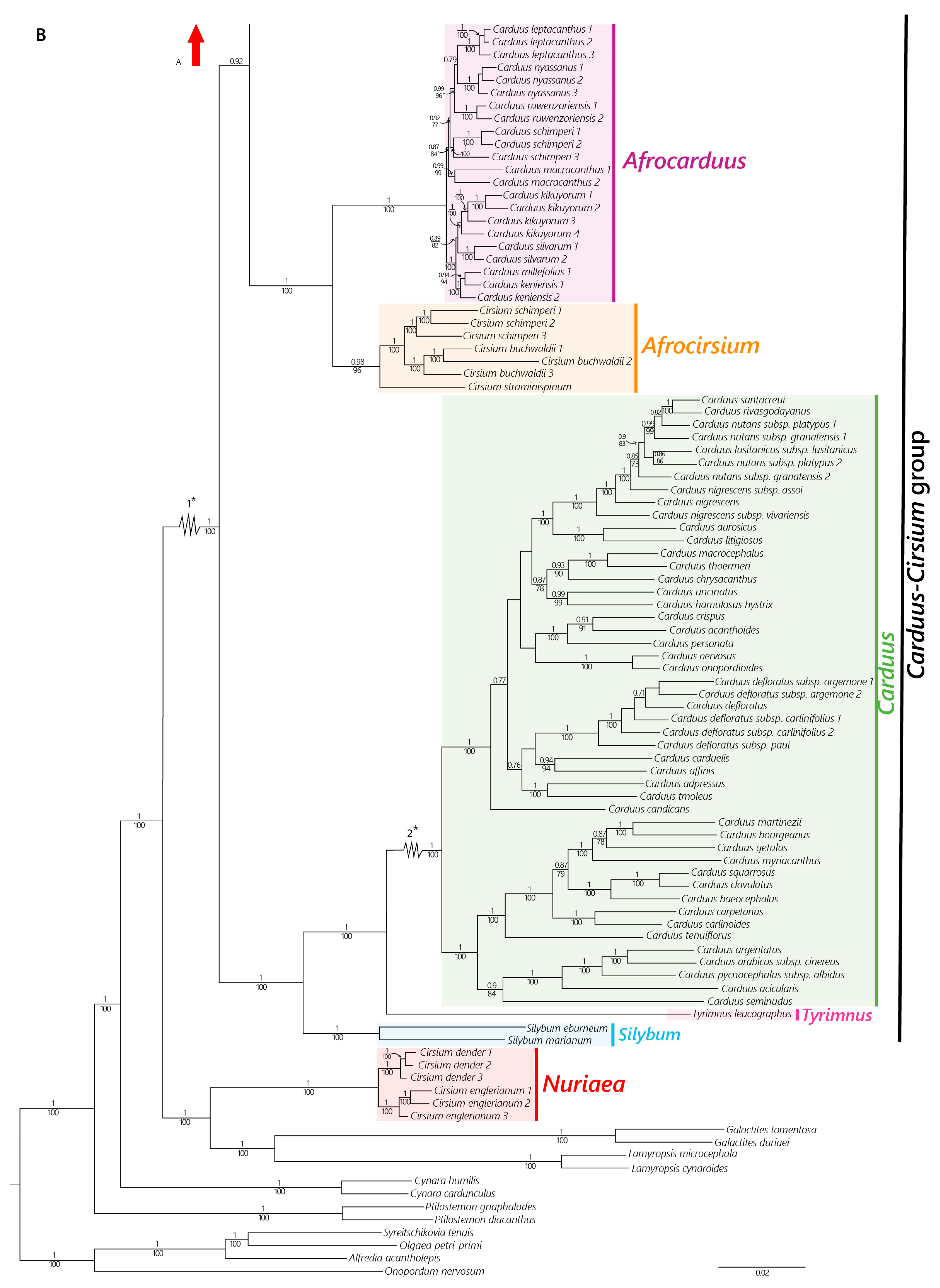
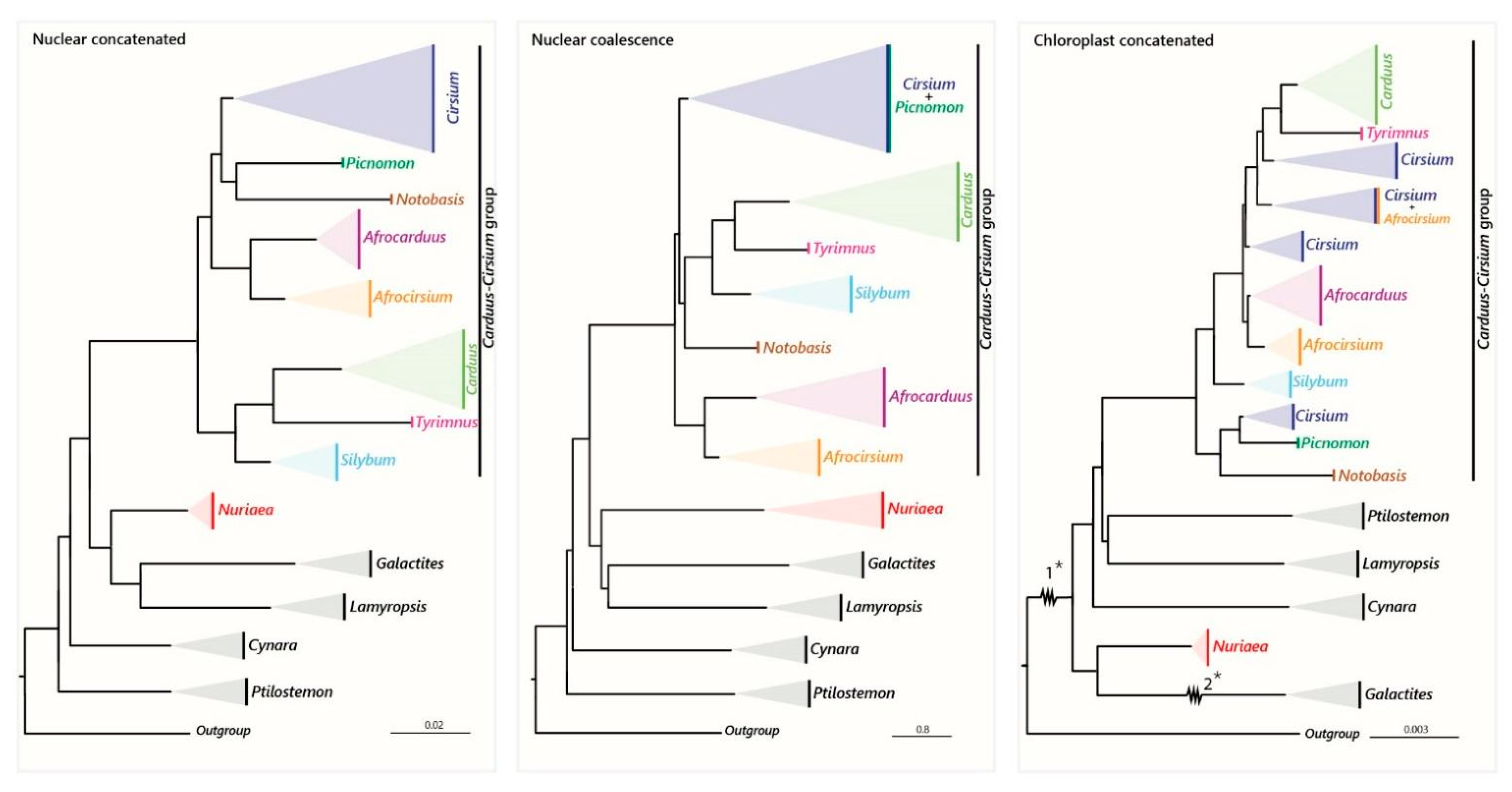
2.2. Phylogenetic Relationships
2.2.1. Nuclear Dataset
2.2.2. Plastid Dataset
3. Materials and Methods
3.1. Taxon Sampling
3.2. DNA Extraction, Library, Capture and Sequencing
3.3. Molecular Data Processing
3.4. Phylogenetic Analyses
3.5. Morphological Examination
4. Discussion
4.1. Utility of Hyb-Seq and Incongruence between the Phylogenies
4.2. The African Carduus and Cirsium
4.3. The Carduus-Cirsium Group
5. Taxonomic Proposal
- Afrocarduus (Kazmi) Garcia-Jacas, Moreyra & Susanna, gen. et stat nov.
- ≡ Carduus subgenus Afrocarduus Kazmi, Mitt. Bot. Staatssamml. München 5: 139 (1963) [basionym].
- Spiny perennial herbs, sometimes rosulate and acaulescent, more often caulescent to 2.5 m high. Stems usually winged or interruptedly winged, fistulose. Leaves very variable, from narrowly lanceolate to oval–lanceolate or elliptic, dentate to pinnatisect, spiny specially on lobes and teeth, glabrous or sparsely arachnoid–pilose above, more densely pilose below especially on nerves, often with septate hairs; nerves prominent. Capitula globose or campanulate, terminal, solitary or many-clustered in the center of rosettes in acaulescent species, homogamous, 0.5–3 cm wide. Involucral bracts glabrous or arachnoid–pilose, sometimes lanuginose; margin minutely denticulate of entire, scariose, glabrous or pilose with septate hairs, without appendages; apical spine always present, 1 to 20 mm long. Florets white, pink or purple; anther filaments papillose with very short papillae to 0.1 mm. Achenes ovate, laterally compressed, glabrous, smooth or somewhat striate, 3–9 mm long, with a short apical coronule; apical caruncle absent. Pappus to 55 mm long; setae scabrid-short barbellate. Type: Afrocarduus leptacanthus (Fresen.) Garcia-Jacas, Moreyra & Susanna.
- Afrocarduus afromontanus (R.E.Fr.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus afromontanus R.E.Fr., Acta Horti Berg. 8: 21 (1925) [basionym].
- Examined material: Kenya: South Kinangop, The Elephant Mt. alt. 2700 m, 6.5.1968, O.M.Mwangangi 1003 (BR0000016058613); Mt. Kenya (versant ouest), alt. 3300 m, 02.1912, C.Alluaud KE204 (MNHN-P-P0010072).
- Afrocarduus keniensis (R.E.Fr.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus keniensis R.E.Fr., Acta Horti Berg. 8: 31 (1925) [basionym].
- Examined material: Ethiopia: Kenya: Mt. Elgon, 06.12.1920, G.Lindblom s.n. (S! [photo]). East side of Mt. Kenya, alt. 3078 m, 05.15.1926, J.P.Chapin 48 (BR0000016058637); Mt. Kenya (Versant Ouest), alt. 3200–3300 m, 02.06.1912, K.E. (?) 214 (MNHN-P-P0010092). Tanzania: Kilimandjaro, alt. 3000–4000 m, P.E.Janssens s.n. (BR0000016058644); Kilimandjaro, alt. 3580 m, 03.07.1934, H.J.E.Schlieben 4912 (BR0000016058675); Kilimandscharo, SO seite, alt. 3580 m, 03.07.1934, H.J.Schilieben 4912 (MNHN-P-P0010088).
- Afrocarduus kikuyorum (R.E.Fr.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus kikuyorum R.E.Fr., Acta Horti Berg. 8: 24 (1925) [basionym] ≡ Carduus nyassanus subsp. kikuyorum (R.E.Fr.) C.Jeffrey, Fl. Trop. E. Africa 49 (1969).
- Examined material: Kenya: Samburu, Mt. Nyiru, Mario Forest Zone, alt. 2500 m, 03.30.1995, B.Bytebier 232 (BR0000016059610). Rwanda: N.-Rwanda, O.-flank van de Muhavura, alt. 3100 m, 02.19.1972, P. N.-Rwanda, O.-flank van de Muhavura 9457 (BR0000021711428); Northern Province, Volcanoes National Park, Muhavura volcano, M.Galbany-Casals 2779, J.A.Calleja & M.Kandziora (BC); Northern Province, Volcanoes National Park, Muhavura volcano, M.Galbany-Casals 2790, J.A.Calleja & M.Kandziora (M.Galbany pers.herb.). Tanzania: Kilimanjaro, alt. 1800 m, 12.16.1933, H.J.E. Schilieben 4361, (BR0000016060241); Ngorongoro conservation area, Empakaai Crater, top of the south rim, alt. 2900 m, Eastern exposure, 02.28.1973, G.W.Frame P21 (BR0000016060265). Uganda: Western Province, Kigezi Dist. Virunga-Kette, Sattel zwischen Muhavura und Mgahinga, alt. 3000 m, 11.14.1954, H.U.Stauffer 771 (BR0000016060197).
- Afrocarduus leptacanthus (Fresen.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus leptacanthus Fresen., Mus. Senckenberg. 3(1): 70 (1839) [basionym]
- = Carduus abyssinicus Sch.Bip., Linnaea 19(3): 332 (1846).
- Examined material: Congo: Parc National Albert, entre Kakalali et Butahu, le long du chemin-limite du P.N.A. (Sousd. du Ruwenz.), Congo Belge, alt. approx. 1650 m, 08.05.1952, H.Fredericq 7857 (BR0000016059047). Ethiopia: sine loc., E.Rüppell s.n., 06.01.1832 (FR! [photo]). Kenya: Landiani. Mau escarpment, alt. 2500 m, 10.1903, C.Alluaud KE80 (MNHN-P-P0010115). Rwanda: Wisumo, Prefecture: Kibuye, alt. 2200 m, 03.16.1973, G.Troupin 14755 (BR0000016059368); N.-Rwanda, terr. Ruhengeri, Kinigi, Rops-plantage, alt. approx. 2300 m, 02.24.1972, P.Van der Veken 9539 (BR0000016059375); Northern Province, Volcanoes National Park, Mount Gahinga, cultivated lands outside the park limit, 02.08.2022, M.Galbany-Casals 2756, J.A.Calleja & M.Kandziora (M.Galbany pers. herb.). Tanzania: Ruvuma region Kitulo, 10.18.1969, J.Prins-Lampert 576 (WAG.1744238).
- Afrocarduus macracanthus (Kazmi) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus macracanthus Kazmi, Mitt. Bot. Staatssamml. München 5: 164 (1963) [basionym].
- Examined material: Ethiopia: Bale prov., Bale Mts National Park, Garba Goracha, in steep slope just S of the camp site, alt. 3950 m, 11.02.1973, K.O.Hedberg 5623 (WAG.1361919); Mt. Boruluccu, along the road to Ticcio, about 30 km SE of Asella, alt. 4000 m, 12.06.1965. W.J.J.O.Wilde & B.E.E. De Wilde-Duyfjes 9187 (BR0000016059559); Mt. Boruluccu, along the road to Ticcio, about 30 km SE of Asella, alt. 4000 m, 12.06.1965, W.J.J.O.De Wilde & B.E.E.Wilde-Duyfjes (MNHN-P-P0010116).
- Afrocarduus millefolius (R.E.Fr.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus millefolius R.E.Fr., Acta Horti Berg. 8: 34 (1925) [basionym].
- Examined material: Kenya: Western slopes of Mt. Kenya, along the trail from West Kenia Forest Station to summit, British East Africa, alt. 3630 m, 09.21–27.1909, E.A. Mearns, (BR0000016059566); Fort Jerusalem, Aberdares National Park on N. Kinangop-Nyeri road, alt. 3200 m, 7.30.1960, E.Polhill 228, (BR0000016059573); Mt. Kinangop, alt. 2800 m, 02.18.1912., C.Alluaud ke 268 (MNHN-P-P0010124).
- Afrocarduus nyassanus (S.Moore) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus leptacanthus var. nyassanus S.Moore, Journ. Linn. Soc. Bot., 37: 326 (1906) [basionym] ≡ Carduus nyassanus (S.Moore) R.E.Fr., Acta Horti Berg. 8: 25 (1925).
- Examined material: Congo: Bord marécageux d’une rivière, Près de Kasiki (Plateau des Marungu, Katanga), alt. 2000 m, 11.26.1969, S.Lisowski, F.Malaisse & J.Symoens 8378 (BR0000016059948). Malawi: Zomba plateau, Chingwe’s Hole area, alt. 1800 m, 11.20.1981, J.D.Chapman & E.J.Tawakali 5998 (BR0000016059641). Rwanda: Northern Province, Volcanoes National Park, Mount Gahinga, 02.08.2022, M.Galbany-Casals, J.A.Calleja & M.Kandziora 2765.
- Afrocarduus ruwenzoriensis (S.Moore) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus ruwenzoriensis S.Moore, J. Linn. Soc., Bot. 35: 364 (1902) [basionym].
- Examined material: Congo: Ruisseau affluent de la Mososa, (à l’Est de Mahungu), alt. 3180 m, 05.29.1953, F. in De Witte 9134 (BR0000005891160).
- Afrocarduus schimperi (Sch.Bip.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus schimperi Sch.Bip., Linnaea 19 (3): 334 (1846) [basionym].
- Examined material: Congo: Mts. S.W. of Lemera, lower Ruzizi Valley, Kivu district, alt. 2830 m, 07.17.1927, J.Chapin 515 (BR0000016059450); Mont Muki, Prairie, alt. 3050 m, 07.27.1955, U.J.Kinet 5 (BR0000016059481); Munt Muhende, 03.1982, H.Scaetta 2424 (BR0000016059504); Mont Muhende, H.Scaetta 2424 (BR0000016059511); Sur les crêtes rocheuses du Mt. Kyanza au Sud Ouest du Lac Lungwe, alt. 2700 m. 08.01.1959, A.Michelson 1078 (BR0000016059498). Ethiopia: Kaffa: SE of Folla, some 15 km N of Ghibe bridge (on Addis-Jimma road), alt. 2000–2100 m, 12.02.1970, I.Friis, A.Hunde, K.Jacobsen 548 (WAG.1361800); Gojam, near Debre Markos, on the road to Elias, alt. 2175 m, 12.22.1972, C.J.P.Seegeler 2968 (WAG.1361869); Gondar, Semiam Mountains, small valley running down to main valley between Geech and Ambaras, alt. 3360 m, 09.19.1969, M.G.G.Gilbert 105 (WAG.1361871); about 10 km SE of Hagere Selam, SE of Wondo, alt. approx. 3000 m, 03.13.1966, W.J.J.O de Wilde & B.E.E.Wilde-Duyfjes 10299 (WAG.1361877); Road Wondo-Agere Selam, 17 km, from Wondo, alt. approx. 2450 m, 11.17.1967, E.Westphal & J.Westphal-Stevels 2675 (BR0000016060616); Mt. Entotto, about 5 km N. of Addis Ababa, alt. approx. 2600 m, 02.12.1966, E.Westphal & J.Westphal-Stevels 9974 (BR0000016060623); Kaffa prov. S.E. of Folla, some 15 km N. of Ghibe bridge, on Addis-Jimma road, alt. 2000–2100 m, 12.02.1970, I.Friis 548 (BR0000016060630); Begemder Prov. Semian Mountains, small valley running to main valley, between Geech and Ambaras, alt. approx. 3360 m, 09.19.1969, J.J.F.E. de Wilde & M.G.Gilbert 105 (BR0000021711459); Begemder Prov. Semian Mountains, small valley running to main valley, between Geech and Ambaras, alt. approx. 3360 m, 09.19.1969, J.J.F.E. de Wilde & M.G.Gilbert 105 (BR0000021711459); Mt Wanchi, near rim of crater, alt. 3100 m, 09.20.2003, C.C.H.Jongkind 5974 (WAG.1362060); Col de Mororo, prov. Bale, alt. 3200 m, 05.16.1970, J.L.Guillaumet 2674 (MNHN-P-P00931227). Kenya: Nanyuki distr. Endarasha ranch near Sirimun R., alt. 2000 m, 08.09.1965, J.B. Gillett 16827 (MNHN-P-P0010076); S. Nyanza, Kisii district (k5), alt. 1930 m, near Ramasha, 01.14.1978, A.C.Plaizier 483 (WAG.1361797); Ol’Pusimoru sawmill about 9 miles from Olokurto, alt. 2600 m, 05.20.1961, P.E.Glover, Gwynne & Samuel 1339 (BR0000016060678). Tanzania: Masai district, 15 km from Lolionto on Marok road, 11.09.1974, J.B. Gillet 16333 (BR0000016060708); on way to Aiteho (illegible), Hubulu dist., alt. 1828 m, 08.31.1932, B.D.Burtt 4268 (BR0000016060685).
- Afrocarduus silvarum (R.E.Fr.) Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Carduus silvarum R.E.Fr., Acta Horti Berg. 8: 22 (1925) [basionym].
- Examined material: Kenya: Upper Imenti Forest, within Meru Municipality boundary, alt. 1740 m, 05.28–29.1974, R.B. & A.J.Faden 74/914 (BR0000016059528); 10 km E of Kieni, alt. 2100 m, 06.17.1986, K.H.J.Beentje 2938 (WAG.1362065). Uganda: British Uganda, Station Lamuru (Buschiges Hochland), alt. 3000, 07.11.1909, G.Schefflet 330 (MNHN-P-P0010145).
- A complete taxonomic and nomenclatural revision of Afrocarduus is in preparation and will be published as a separate work.
- Afrocirsium Calleja, Garcia-Jacas, Moreyra & Susanna, gen. nov.
- Spiny perennial herbs, caulescent, 0.50 to 2.5 m high. Stems unwinged, fistulose. Leaves lanceolate, shortly decurrent, entire, lobate or pinnatisect with spiny lobes, glabrescent above, white–tomentose below, usually with reticulate nerves. Capitula solitary or clustered, campanulate, 1–3 cm wide, homogamous. Involucral bracts ovate or lanceolate; margin scariose, denticulate, the outer ones with a fimbriate, black appendix; apical spine short, to 3 mm. Florets pink or purple; anther filaments papillose with very short papillae to 0.1 mm. Achenes ovate, laterally compressed, glabrous, smooth or somewhat striate, 4–7 mm long, with a short apical coronule; apical caruncle absent. Pappus to 12 mm long; setae plumose. Type: Afrocirsium schimperi (Vatke) Calleja, Garcia-Jacas, Moreyra & Susanna, comb. nov.
- Afrocirsium buchwaldii (O.Hoffm.) Calleja, Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Cirsium buchwaldii O.Hoffm. in Bot. Jahrb. Syst. 38(2): 211 (1906) [basionym].
- Ind. Loc.: [Tanzania] Usambara: Gale; auf nassen Hochgebirgswiesen im Quellgebiet des Kwasindo, um 1200 m, im ganzen Wambugu-Lande häufig (BUCHWALD n. 318)—Blühend und fruchten im Dezember 1895). Nyassaland: Uhehe, Utschungwe-Berge, um 1600 m ü. M. (Frau Hauptmann PRINCE.—Blühend 1899).
- Lectotype (designated here): [Tanzania] Usambara, 06.1900, Buchwald 318 (BM000924815!).
- Notes: O. Hoffmann’s original materials were mainly kept in B herbarium, but most of the collections of this herbarium were destroyed. We designate, as a lectotype, a specimen that, although it is currently kept in BM, was acquired from B, as the label indicates “ex Museo botanico Berolinensi”.
- Examined material: Congo: Kivu, slope of Mt. Visoke facing Mt. Mikeno, alt. 3100, 02.18.1975, W.G. d’Arcy 7948 (MNHN-P-P0010147). Etiopia: Bale Prov., Web river, just W. of Dinshu, about 157 km E. of Shashamane, along the road to Goba, alt. approx. 3000 m, 07.24.1970, J.J.F.E.Wilde 6805 (BR0000016273061). Malawi: N. Prov., Nkhata Bay Dist. Vipya Plateau, 23 mi. SW of Mzuzu, Lwafa drift, alt. approx. 1675 m, 05.15.1976, J.Pawek 11273 (WAG.1322534). Rwanda: Northern Province, Volcanoes National Park, Mount Gahinga, swampy area in a clearing of the bamboo forest, 02.08.2022, M.Galbany-Casals, J.A.Calleja & M.Kandziora 2762. Tanzania: Iringa, Makete District. Kitulo Plateau, 03.01.1991, H.Suleiman, M.J.Fundi 13 (WAG.1322531); Iringa; Kitulo, Kitulo Plateau, 02.10.1969, J.Prins-Lampert 166 (WAG.1534453/4).
- Afrocirsium schimperi (Vatke) Calleja, Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Cnicus schimperi Vatke in Linnaea 39(5): 511 (1875) [basionym] ≡ Cirsium schimperi (Vatke) C.Jeffrey in Cufod., Bull. Jard. Bot. Natl. Belg. 37(3, Suppl.): 1174 (1967).
- Ind. Loc.: [Ethiopia] in rivulorum ripa et in aqua ipsa ad Gaffat 8100′ a. m. Sept. 1863. (n. 1238). Eadem in coll. a. 1853. n. 36.
- Lectotype (designated by Jeffrey, 1968): [Ethiopia] Abyssinia, au Bachufern im wasser 8100′ uber mur Gaffat, 25.09.1863, Schimper 1238 (K000418422!). Isolectotypes: BM 000924814!, E 00611482! [photo], GRA0003185-0! [photo], S10-1686! [photo].
- Examined material: Ethiopia: About 5 km NE of Addis Ababa, behind Italian Embassy, alt. approx. 2600 m, 10.16.1965, W.J.J.O.de Wilde & de Wilde-Duyfjes B.E.E. 8280, (BR0000016273092); Gojam: 4 km N of Debre Tabor along the old, now completely disused road to Addis Zemen, alt. 2550 m, 09.13.2004, I.Friis, G.S.Bidgood, A.Hailu, B.Yitbarek 11553 (WAG.1534438); about 5 km NE. of Addis Abeba. Behind Italian Ambassy, alt. approx. 2600 m, W.J.J.O.de Wilde & B.E.E.Wilde-Duyfjes 8280 (MNHN-P-P0010150); Debr’Eski, alt. 8000 m, 11.02.1851, W.Schimper 36 (MNHN-P-P0010151).
- Afrocirsium straminispinum (C.Jeffrey) Calleja, Garcia-Jacas, Moreyra & Susanna, comb. nov.
- ≡ Cirsium polyacanthum Hochst. ex A.Rich., Tent. Fl. Abyss. 1: 456 (1848) [basionym], nom. illeg., non Kar. & Kit. (1841) ≡ Cirsium straminispinum C.Jeffrey in Cufod., Bull. Jard. Bot. Natl. Belg. 37(3, Suppl.): 1174 (1967).
- Ind. Loc.: [Ethiopia] Crescit prope Tchélatchékanné non procul a convalle fluvii Taccazé, mense Julio (Quartin Dillon) et prope Demerki, in provincia Semiène (Schimper).
- Lectotype (designated here): [Ethiopia] Abyssinie, Demerki, 09.08.1838, Schimper 1147 (P0010166! [photo]). Isolectotypes: P0010167! [photo], P0010168! [photo], P0010169! [photo], BR0000008360465! [photo], LG0000090028557! [photo], K000418441!, S10-1669! [photo]).
- Examined material: Ethiopia, Abyssinie, Tchélatchékanné, 1844, MM.Quartin-Dillon et Petit s.n. (P0010162! [photo], P0010163! [photo], P0010164! [photo], P0010165! [photo], P0010170! [photo]).
- Nuriaea Susanna, Calleja & Moreyra, gen. nov.
- Spiny perennial herbs, (2.5) 3–4 (5) m high. Stems interruptedly spiny-winged, fistulose, sulcate, densely pilose in the grooves. Basal leaves very large, to 1 m long, cauline smaller to 40 cm, bipinnatisect, with basalmost lobes transformed in a stout spine’s 2 mm diam. and 3–4 cm length, sparsely pilose above, white–tomentose below. Capitula terminal in loose panicles, globose–umbilicate, 4–7 cm wide, homogamous. Involucral bracts glabrous; outermost bracts patent or reflexed, with a scariose triangular appendate, purple in N. engleriana, green in N. dender; middle bracts with a more developed appendage, entire in N. engleriana, pinnulate–spiny in N. dender. Florets white, or pink-purple, 5–6 cm long including the ovary; anther filaments papillose. Achenes lanceolate, compressed, glabrous, smooth or somewhat striate, 1 cm long, with a short apical coronule; apical caruncle absent. Pappus to 5 cm long; setae plumose. Type: Nuriaea engleriana (O. Hoffm.) Susanna, Calleja & Moreyra.
- The name of the genus honors our coauthor Núria Garcia-Jacas, who passed away on April the 28th 2023, with this paper almost finished. Rest in peace.
- Nuriaea engleriana (O. Hoffm.) Susanna, Calleja & Moreyra, comb. nov.
- ≡ Cirsium englerianum O. Hoffm. in Bot. Jahrb. Syst. 38(2): 210 (1906) [basionym].
- Ind. Loc.: Gallahochland: im Lande der Arussi-Galla, im Uferwald am Awala-See (Dr. ELLENBECK n. 1715.—Blühend im Dezember 1900).
- Neotype (designated by Friis in Norweg. J. Bot. 22 [3]: 203 (1975)): Ethiopia, Kaffa Prov. S.E. of Folla, some 15 km N og Ghibe bridge on Addis-Jimma road, 7°52′ N, 37°11′ E, 2000–2100 m a.s.l., in shrub grassland on edge of path, 12.02.1970, Danish Botanical Expedition to Ethiopia 1970 (I.Friis, A.Hounde & K.Jacobsen) 551 (C100000339! [photo]). Isoneotypes (designated by Friis in Norweg. J. Bot. 22 [3]: 203 (1975)): BR0000005537143! (photo), ETH000000309! (photo), WAG 0258805! (photo).
- Notes: O. Hoffmann’s original materials were originally kept in B but were destroyed. Due to the lack of duplicates, Friis (1975) designated a neotype and isoneotypes.
- Examined material: Ethiopia: Bonga near R.C. Mission, alt. approx. 1800 m, W.J.J.O.de Wilde & B.E.E.de Wilde-Duyfjes 9440, 12.22.1965 (BR0000016273047, WAG.1322605); Kaffa prov. around Giren, alt. 2000 m, I.Friis, A.Getachew, F.Rasmussen & K.B.Vollesen 1556, 12.05.1972 (BR0000016273023).
- Nuriaea dender (Friis) Susanna, Calleja & Moreyra, comb. nov.
- ≡ Cirsium dender Friis in Norweg. J. Bot. 22(3): 203 (1975) [basionym].
- Holotype: Ethiopia, Kaffa Prov., Mt. Maigudo, ca. 40 km from Jimma-Addis road on Omo-Nadda track, 7°30′ N 37°23′ E, 2650 m a.s.l., Erica-Hagenia-Arundinara-Maesa-Agauria-Ilex-scrub, along the road, 12.03.1972, Danish–Ethiopian Botanical Expedition 1972-73 (I.Friis, G.Aweke, F.Rasmussen & K.Vollesen) 1444 (C10000338! [photo]). Isotypes: BR0000008872876! and BR0000008872227! (photo), ETH000000080! (photo), K000418444!, WAG 000541! and WAG 0000542!.
- Examined material: Ethiopia: Illubator Region, 55 km north of Tepi, along the new road to Gore, between Gecha and Macha, alt. 2200 m, I.Friis, G S.Bidgood, P.Host, D.Desissa & S.Kebede 7161, 11.15.1995 (BR0000016273009); Gamu-Gofa, in the mountains above Arba Minch near Chencha, alt. 2300 m, I.Friis, G.S.Bidgood, M.Wondafrash & G.Gibre-Hiwot 8788, 12.25.1997 (WAG.1322547); Kefa, Mt. Maigudo, ca. 40 km from Jimma-Addis road on Omo-Nadda track, alt. 2650 m, 12.03.1972, I.Friis, G.Aweke, F.Rasmussen & K.B.Vollesen 1444 (WAG0000541/42).
- 1
- Large plants more than 3 m high; globose capitula to 4–7 cm diam………………………2
- 1
- Smaller plants, capitula usually less than 4 cm…………………………………………4
- 2
- Stems to 5 m; leaves green above, white tomentose beneath; involucral bracts with flat apex; achenes linear–lanceloate; pappus plumose to 4 cm long…………………Nuriaea
- 2
- Stems to 3 m; leaves uniformly glabrous or pilose; achenes obpyramidal or globose……………………………………………………………………………………………………………………3
- 3
- Leaves white–variegated, conspicuously white–marbled along the veins above, with long–spiny, recurvate involucral bracts to 5 (7), the basalmost ones erect or reflexed; achenes oblong–ovoid; pappus barbellate……………………………………………..Silybum
- 3
- Leaves not white–variegated or conspicuously white–marbled; spines of the invlucral bracts usually straight; achenes obpyramidal; pappus plumose………………..Cynara
- 4
- Annual plants with pinnatisect leaves; lobes of the leaves linear, spiny; peripheral florets sterile and showy……………….……………………….…………………….Galactites
- 4
- Annual or perennial plants without showy sterile peripheral florets………………….5
- 5
- Pappus barbellate or scabrid; stamen filaments laterally acrescent…………………..6
- 5
- Pappus plumose; stamen filaments free…………………………………………………8
- 6
- Pappus bristles s-shaped; capitula less than 3 cm diam. on long leafless peduncles……………….……………………………….……………………….……………..Tyrimnus
- 6
- Pappus bristles straight; peduncles foliose………………….…………………….…….7
- 7
- Achenes with 10–15 longitudinal grooves; papillary hairs of the staminal filaments to 0.75 mm; epidermal cells on the dorsal corolla lobes undulate……………………Carduus
- 7
- Achenes with four lines; papillary hairs of the staminal filaments to 0.13 mm; epidermal cells on the dorsal corolla lobes straight……………………………………Afrocarduus
- 8
- Leaves white–veined; involucral bract apex with entire spine; corolla more deeply split on the abaxial side; apical elaiosome absent……………….………………….Notobasis
- 8
- White–veined leaves absent; corolla regularly split; apical elaiosome present……..9
- 9
- Involucral bracts pectinated on the margin and the appendix, rarely toothed…….10
- 9
- Involucral bracts entire or dentate…………………….………………………………….11
- 10
- Divaricately branched plants; achenes linear; capitula concealed by densely araneous hairs………………….….………………………………….…………….……………..Picnomon
- 10
- Unbranched plants; capitula not concealed by bracts……………………….……………………….…………….Afrocirsium
- 11
- Achenes broadly ovoid, not compressed, without nectary………………..Ptilostemon
- 11
- Achenes oblong, compressed, with apical nectary……………………….………………12
- 12
- Leaves green above, white–lanuginose beneath; elaiosome cylindrical…Lamyropsis
- 12
- Leaves glabrous or uniformly pilose; elaiosome globose……………………….Cirsium
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hooper, D.; Adair, E.; Cardinale, B.; Byrnes, J.E.K.; Hungate, B.A.; Matulich, K.L.; Gonzalez, A.; Duffy, J.E.; Gamfeldt, L.; O’Connor, M.I. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 2012, 486, 105–108. [Google Scholar] [CrossRef]
- IPBES. Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; Brondizio, E.S., Settele, J., Díaz, S., Ngo, H.T., Eds.; IPBES Secretariat: Bonn, Germany, 2019; pp. 1–1148. [Google Scholar] [CrossRef]
- Vane-Wright, R.I. Systematics and the Conservation of Biological Diversity. Ann. Mo. Bot. Gard. 1996, 83, 47–57. [Google Scholar] [CrossRef]
- Rudbeck, A.V.; Sun, M.; Tietje, M.; Gallagher, R.V.; Govaerts, R.; Smith, S.A.; Svenning, J.C.; Eiserhardt, W.L. The Darwinian shortfall in plants: Phylogenetic knowledge is driven by range size. Ecography 2022, 2022, e06142. [Google Scholar] [CrossRef]
- Mittermeier, R.A.; Turner, W.R.; Larsen, F.W.; Brooks, T.M.; Gascon, C. Global Biodiversity Conservation: The Critical Role of Hotspots. In Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas; Zachos, F.E., Habel, J.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–22. [Google Scholar]
- Vamosi, J.C.; Vamosi, S.M. Extinction Risk Escalates in the Tropics. PLoS ONE 2008, 3, e3886. [Google Scholar] [CrossRef] [PubMed]
- Laurance, W.F.; Sayer, J.; Cassman, K.G. Agricultural expansion and its impacts on tropical nature. Trends Ecol. Evol. 2014, 29, 107–116. [Google Scholar] [CrossRef]
- Linder, H.P. On the relationship between the vegetation and floras of the Afromontane and the Cape regions of Africa. Mitt. Inst. Allg. Bot. Hambg. 1990, 23b, 777–790. [Google Scholar]
- Galley, C.; Bytebier, B.; Bellstedt, D.U.; Linder, H.P. The Cape element in the Afrotemperate flora: From Cape to Cairo? Proc. R. Soc. B 2007, 274, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Walter, H. Vegetation of the Earth and Ecological Systems of the Geo-Biosphere, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 1985. [Google Scholar]
- White, F. The vegetation map of Africa: The history of a completed project. Boissiera 1976, 24, 659–666. [Google Scholar]
- Gehrke, B.; Linder, H.P. Species richness, endemism and species composition in the tropical Afroalpine flora. Alp. Bot. 2014, 124, 165–177. [Google Scholar] [CrossRef]
- Brochmann, C.; Gizaw, A.; Chala, D.; Kandziora, M.; Eilu, G.; Popp, M.; Pirie, M.D.; Gehrke, B. History and evolution of the afroalpine flora: In the footsteps of Olov Hedberg. Alp. Bot. 2021, 1, 65–87. [Google Scholar] [CrossRef]
- Susanna, A.; Garcia-Jacas, N. Tribe Cardueae. In The Families and Genera of Vascular Plants; Jeffrey, C., Kadereit, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 8, pp. 123–146. [Google Scholar]
- Häffner, E.; Hellwig, F. Phylogeny of the tribe Cardueae (Compositae) with emphasis on the subtribe Carduinae: An analysis based on ITS sequence data. Willdenowia 1999, 29, 27–39. [Google Scholar] [CrossRef]
- Garcia-Jacas, N.; Garnatje, T.; Susanna, A.; Vilatersana, R. Tribal and subtribal delimitation and phylogeny of the Cardueae (Asteraceae): A combined nuclear and chloroplast DNA analysis. Mol. Phylogenet. Evol. 2002, 22, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Häffner, E. On the phylogeny of the subtribe Carduinae (tribe Cardueae, Compositae). Englera 2000, 21, 3–208. [Google Scholar] [CrossRef]
- Susanna, A.; Garcia-Jacas, N. Cardueae (Carduoideae). In Systematics, Evolution, and Biogeography of Compositae; Funk, V.A., Susanna, A., Stuessy, T.F., Bayer, R.J., Eds.; IAPT: Vienna, Austria, 2009; pp. 293–313. [Google Scholar]
- Ackerfield, J.; Susanna, A.; Funk, V.; Kelch, D.; Park, D.S.; Thornhill, A.H.; Yildiz, B.; Arabaci, T.; Dirmenci, T. A Prickly Puzzle: Generic delimitations in the Carduus-Cirsium group (Compositae: Cardueae: Carduinae). Taxon 2020, 69, 715–738. [Google Scholar] [CrossRef]
- Del Guacchio, E.; Bureš, P.; Iamonico, D.; Carucci, F.; De Luca, D.; Zedek, F.; Caputo, P. Towards a monophyletic classification of Cardueae: Restoration of the genus Lophiolepis (= Cirsium pp) and new circumscription of Epitrachys. Plant Biosyst. 2022, 56, 1269–1290. [Google Scholar] [CrossRef]
- Kelch, D.G.; Baldwin, B.G. Phylogeny and ecological radiation of New World thistles 113 (Cirsium, Cardueae—Compositae) based on ITS and ETS rDNA sequence data. Mol. Ecol. 2003, 12, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Susanna, A.; Garcia-Jacas, N.; Hidalgo, O.; Vilatersana, R.; Garnatje, T. The Cardueae (Compositae) revisited: Insights from ITS, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Ann. Mo. Bot. Gard. 2006, 93, 150–171. [Google Scholar] [CrossRef]
- Carucci, F. Filogenesi Molecolare del Genere Cirsium Mill. Sect. Eriolepis (Cass.) Dumort. Ph.D. Thesis, University of Naples “Federico II”, Napoli, Italy, 2011. [Google Scholar]
- Slotta, T.A.B.; Horvath, D.P.; Foley, M.E. Phylogeny of Cirsium spp. in North America: Host specificity does not follow phylogeny. Plants 2012, 1, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Barres, L.; Sanmartín, I.; Anderson, C.L.; Susanna, A.; Buerki, S.; Galbany-Casals, M.; Vilatersana, R. Reconstructing the evolution and biogeographic history of tribe Cardueae (Compositae). Am. J. Bot. 2013, 100, 867–882. [Google Scholar] [CrossRef]
- Rundell, R.J.; Price, T.D. Adaptive radiation, nonadaptive radiation, ecological speciation and nonecological speciation. Trends Ecol. Evol. 2009, 24, 394–399. [Google Scholar] [CrossRef]
- Glor, R.E. Phylogenetic insights on adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 251–270. [Google Scholar] [CrossRef]
- López-Vinyallonga, S.; Mehregan, I.; Garcia-Jacas, N.; Tscherneva, O.; Susanna, A.; Kadereit, J.W. Phylogeny and evolution of the Arctium-Cousinia complex (Compositae, Cardueae-Carduinae). Taxon 2009, 58, 153–171. [Google Scholar] [CrossRef]
- Kazmi, S.M.A. Revision der Gattung Carduus (Compositae) I. Mitt. Bot. Staatssamml. München 1963, 5, 139–198. [Google Scholar]
- Fries, R.E. Revision der tropisch-afrikanischen Carduus-Arten. Acta Horti. Berg. 1925, 25, 11–38. [Google Scholar]
- Jeffrey, C. The Cynareae of East Tropical Africa. Kew Bull. 1968, 22, 107–140. [Google Scholar] [CrossRef]
- Herrando-Moraira, S.; Calleja, J.A.; Galbany-Casals, M.; Garcia-Jacas, N.; Liu, J.-Q.; López-Alvarado, J.; López-Pujol, J.; Mandel, J.R.; Massó, S.; Montes-Moreno, N.; et al. Nuclear and plastid DNA phylogeny of tribe Cardueae (Compositae) with Hyb-Seq data: A new subtribal classification and a temporal diversification framework. Mol. Phylogenet. Evol. 2019, 137, 313–332. [Google Scholar] [CrossRef]
- Firetti, F.; Zuntini, A.R.; Gaiarsa, J.W.; Oliveira, R.S.; Lohmann, L.G.; Van Sluys, M.A. Complete chloroplast genome sequences contribute to plant species delimitation: A case study of the Anemopaegma species complex. Am. J. Bot. 2017, 104, 1493–1509. [Google Scholar] [CrossRef] [PubMed]
- Draper, I.; Villaverde, T.; Garilleti, R.; Burleigh, J.G.; McDaniel, S.F.; Mazimpaka, V.; Calleja, J.A.; Lara, F. An NGS-Based Phylogeny of Orthotricheae (Orthotrichaceae, Bryophyta) with the Proposal of the New Genus Rehubryum from Zealandia. Front. Plant Sci. 2022, 13, 882960. [Google Scholar] [CrossRef] [PubMed]
- Siniscalchi, C.M.; Ackerfield, J.R.; Folk, R.A. Diversification and biogeography of North American thistles (Cirsium: Carduoideae: Compositae): Drivers of a rapid continent-wide radiation. Int. J. Plant Sci. 2023, 184, 5. [Google Scholar] [CrossRef]
- Mandel, J.R.; Dikow, R.B.; Funk, V.A.; Masalia, R.R.; Staton, S.E.; Kozik, A.; Michelmore, R.W.; Rieseberg, L.H.; Burke, J.M. A target enrichment method for gathering phylogenetic information from hundreds of loci: An example from the Compositae. J. Appl. Polym. Sci. 2014, 2, 1300085. [Google Scholar] [CrossRef]
- Fér, T.; Schmickl, R.E. HybPhyloMaker: Target Enrichment data analysis from raw reads to species trees. Evol. Bioinform. 2018, 14, 1–9. [Google Scholar] [CrossRef]
- Ufimov, R.; Gorospe, J.M.; Fér, T.; Kandziora, M.; Salomon, L.; Van Loo, M.; Schmickl, R. Utilizing paralogues for phylogenetic reconstruction has the potential to increase species tree support and reduce gene tree discordance in target enrichment data. Mol. Ecol. Resour. 2022, 22, 3018–3034. [Google Scholar] [CrossRef] [PubMed]
- Kandziora, M.; Sklenář, P.; Kolář, F.; Schmickl, R. How to tackle phylogenetic discordance in recent and rapidly radiating groups? Developing a workflow using Loricaria (Asteraceae) as an example. Front. Plant Sci. 2022, 12, 765719. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Johnson, M.G.; Gardner, E.M.; Liu, Y.; Medina, R.; Goffinet, B.; Shaw, A.J.; Zerega, N.J.C.; Wickett, N.J. HybPiper: Extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci. 2016, 4, 1600016. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Lemoine, F.; Domelevo Entfellner, J.B.; Wilkinson, E.; Correia, D.; Dávila Felipe, M.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.P.; Moret, B.M.E.; Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Stamatakis, A. Using RAxML-NG in Practice; Scornavacca, C., Delsuc, F., Galtier, N., Eds.; Phylogenetics in the Genomic Era, HAL-open science: Lyon, France, No commercial publisher, Authors open access book; 2020; pp. 1–25. Available online: https://hal.science/hal-02535311 (accessed on 27 June 2023).
- Simon, C. An evolving view of phylogenetic support. Syst. Biol. 2022, 71, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 2018, 19, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Sayyari, E.; Mirarab, S. Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 2016, 33, 1654–1668. [Google Scholar] [CrossRef]
- Sayyari, E.; Mirarab, S. Testing for polytomies in phylogenetic species trees using quartet frequencies. Genes 2018, 9, 132. [Google Scholar] [CrossRef]
- Xi, Z.; Rest, J.S.; Davis, C.C. Phylogenomics and coalescent analyses resolve extant seed plant relationships. PLoS ONE 2013, 8, e80870. [Google Scholar] [CrossRef]
- Xi, Z.; Liu, L.; Rest, J.S.; Davis, C.C. Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst. Biol. 2014, 63, 919–932. [Google Scholar] [CrossRef]
- Straub, S.C.K.; Moore, M.J.; Soltis, P.S.; Soltis, D.E.; Liston, A.; Livshultz, T. Phylogenetic signal detection from an ancient rapid radiation: Effects of noise reduction, long-branch attraction, and model selection in crown clade Apocynaceae. Mol. Phylogenet. Evol. 2014, 80, 169–185. [Google Scholar] [CrossRef]
- Bagley, J.C.; Uribe-Convers, S.; Carlsen, M.M.; Muchhala, N. Utility of targeted sequence capture for phylogenomics in rapid, recent angiosperm radiations: Neotropical Burmeistera bellflowers as a case study. Mol. Phylogenet. Evol. 2020, 152, 106769. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, K.; Cornetti, L.; Fields, P.D.; Ebert, D. Whole-Genome Phylogenetic Reconstruction as a Powerful Tool to Reveal Homoplasy and Ancient Rapid Radiation in Waterflea Evolution. Syst. Biol. 2022, 71, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Novaković, J.; Janaćković, P.; Susanna, A.; Lazarević, M.; Boršić, I.; Milanovici, S.; Lakušić, D.; Zlatković, B.; Marin, P.D.; Garcia-Jacas, N. Molecular Insights into the Centaurea calocephala Complex (Compositae) from the Balkans—Does Phylogeny Match Systematics? Diversity 2022, 14, 394. [Google Scholar] [CrossRef]
- Friis, I. The giant species of Cirsium (Asteraceae) in southern Ethiopia. Norweg. J. Bot. 1975, 22, 201–207. [Google Scholar]
- Wiklund, A. The genus Cynara L. (Asteraceae-Cardueae). Bot. J. Linn. 1992, 109, 75–123. [Google Scholar] [CrossRef]
- Bureš, P.; Ozcan, M.; Šmerda, J.; Michálková, E.; Horová, L.; Plačková, K.; Šmarda, P.; Elliott, T.L.; Veselý, P.; Ćato, S.; et al. Evolution of genome size and GC content in the tribe Carduinae (Asteraceae): Rare descending dysploidy and polyploidy, limited environmental control and strong phylogenetic signal. Preslia 2023, 95, 185–213. [Google Scholar] [CrossRef]
- Arnold, M.L. Natural hybridization as an evolutionary process. Annu. Rev. Ecol. Evol. Syst. 1992, 23, 237–261. [Google Scholar] [CrossRef]



| Previous Classification | n° of Accepted Species | New Classification Proposed Here | n° of Species |
|---|---|---|---|
| Carduus L. | ca. 100 | Afrocarduus (Kazmi) Garcia-Jacas, Moreyra & Susanna | 10 |
| Carduus L. | ca. 90 | ||
| Cirsium Mill. | ca. 450 | Afrocirsium Calleja, Garcia-Jacas, Moreyra & Susanna | 3 |
| Cirsium Mill. | ca. 450 | ||
| Nuriaea Susanna, Calleja & Moreyra (out of Carduus-Cirsium group) | 2 | ||
| Notobasis (Cass.) Cass. | 1 | Notobasis (Cass.) Cass. | 1 |
| Picnomon Adans. | 1 | Picnomon Adans. | 1 |
| Silybum Adans. | 2 | Silybum Adans. | 2 |
| Tyrimnus Cass. | 1 | Tyrimnus Cass. | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreyra, L.D.; Garcia-Jacas, N.; Roquet, C.; Ackerfield, J.R.; Arabacı, T.; Blanco-Gavaldà, C.; Brochmann, C.; Calleja, J.A.; Dirmenci, T.; Fujikawa, K.; et al. African Mountain Thistles: Three New Genera in the Carduus-Cirsium Group. Plants 2023, 12, 3083. https://doi.org/10.3390/plants12173083
Moreyra LD, Garcia-Jacas N, Roquet C, Ackerfield JR, Arabacı T, Blanco-Gavaldà C, Brochmann C, Calleja JA, Dirmenci T, Fujikawa K, et al. African Mountain Thistles: Three New Genera in the Carduus-Cirsium Group. Plants. 2023; 12(17):3083. https://doi.org/10.3390/plants12173083
Chicago/Turabian StyleMoreyra, Lucía D., Núria Garcia-Jacas, Cristina Roquet, Jennifer R. Ackerfield, Turan Arabacı, Carme Blanco-Gavaldà, Christian Brochmann, Juan Antonio Calleja, Tuncay Dirmenci, Kazumi Fujikawa, and et al. 2023. "African Mountain Thistles: Three New Genera in the Carduus-Cirsium Group" Plants 12, no. 17: 3083. https://doi.org/10.3390/plants12173083
APA StyleMoreyra, L. D., Garcia-Jacas, N., Roquet, C., Ackerfield, J. R., Arabacı, T., Blanco-Gavaldà, C., Brochmann, C., Calleja, J. A., Dirmenci, T., Fujikawa, K., Galbany-Casals, M., Gao, T., Gizaw, A., López-Alvarado, J., Mehregan, I., Vilatersana, R., Yıldız, B., Leliaert, F., Seregin, A. P., & Susanna, A. (2023). African Mountain Thistles: Three New Genera in the Carduus-Cirsium Group. Plants, 12(17), 3083. https://doi.org/10.3390/plants12173083











