Abstract
Arsenic (As) is one of the toxic heavy metal pollutants found in the environment. An excess of As poses serious threats to plants and diminishes their growth and productivity. NAC transcription factors revealed a pivotal role in enhancing crops tolerance to different environmental stresses. The present study investigated, for the first time, the functional role of SNAC3 in boosting As stress tolerance and grain productivity in rice (Oryza sativa L.). Two SNAC3-overexpressing (SNAC3-OX) and two SNAC3-RNAi transgenic lines were created and validated. The wild-type and transgenic rice plants were exposed to different As stress levels (0, 25, and 50 µM). The results revealed that SNAC3 overexpression significantly improved rice tolerance to As stress and boosted grain yield traits. Under both levels of As stress (25 and 50 µM), SNAC3-OX rice lines exhibited significantly lower levels of oxidative stress biomarkers and OsCRY1b (cryptochrome 1b) expression, but they revealed increased levels of gas exchange characters, chlorophyll, osmolytes (soluble sugars, proteins, proline, phenols, and flavonoids), antioxidant enzymes (SOD, CAT, APX, and POD), and stress-tolerant genes expression (OsSOD-Cu/Zn, OsCATA, OsCATB, OsAPX2, OsLEA3, OsDREB2B, OsDREB2A, OsSNAC2, and OsSNAC1) in comparison to wild-type plants. By contrast, SNAC3 suppression (RNAi) reduced grain yield components and reversed the aforementioned measured physio-biochemical and molecular traits. Taken together, this study is the first to demonstrate that SNAC3 plays a vital role in boosting As stress resistance and grain productivity in rice through modulating antioxidants, photosynthesis, osmolyte accumulation, and stress-related genes expression, and may be a useful candidate for further genetic enhancement of stress resistance in many crops.
1. Introduction
Plants often encounter various environmental stresses throughout their lifetime, such as heavy metals toxicity, salinity, and drought [1,2]. Among the extremely toxic heavy metal environmental pollutants, arsenic (As) is present in water and agricultural soils due to different anthropogenic activities [2,3]. An excess of As causes critical threats to plant, animal, and human health [2]. As toxicity drastically diminishes crop growth, development, metabolism, and yield through influencing the physiological and biochemical pathways and inducing generation of toxic reactive oxygen species (ROS) [4,5]. To combat such stress-induced adverse effects, plants employ a variety of complex processes at different levels, including accumulation of osmolytes, binding metallothionein, sequestration of metals into vacuoles, and induction of several antioxidant enzymes and stress-responsive genes [5,6,7].
The genetic engineering approach has also proved efficient for developing plant varieties with enhanced tolerance to environmental stresses. Over the last years, various transcription factor family members such as NAC (NAM, ATAF1/2, and CUC2), bHLH, AP2/ERF, WRKY, MYB, CAMTA, bZIP, DREB, and NF-Y have revealed remarkable functions in mediating plant resistance to adverse stresses [1,8,9,10,11,12,13,14,15,16]. The name “NAC” is derived from the initials of the three founding members of this transcription factor family: NAM, ATAF1/2 (Arabidopsis Transcription Activation Factor 1/2), and CUC2. The NAC transcription factor family regulates diverse developmental processes and stress responses in plants. NAC proteins are distinguished by remarkably maintained DNA-binding domains, recognized as NAC domains, and have demonstrated a fundamental role in the regulation of plant cell division, organ development, senescence, iron homeostasis, and environmental stress responsiveness [9,17,18,19,20]. Overexpression of ANAC072, ANAC055, or ANAC019 stimulated by ABA, salt, and drought stresses enhances transgenic Arabidopsis tolerance to drought stress [21]. Additionally, ANAC013 is a membrane-linked NAC protein that boosted transgenic Arabidopsis resistance to oxidative stress [20,22]. Another NAC factor, AtJUB1, mediated plant growth through suppressing the GA3ox1 and DWF4 genes, encoding brassinosteroids and gibberellic acid biosynthesis enzymes, as well as via inducing DELLA genes [1,23,24]. Moreover, Arabidopsis overexpressing AtJUB1 or banana overexpressing MusaNAC042 (AtJUB1 homologue) displayed increased resistance to multiple environmental stresses including excessive salinity and drought, while Arabidopsis AtJUB1 knockdown or tomato SlJUB1 silencing mitigated resistance to oxidative and drought stresses [25,26,27,28]. In the regulation of plant responsiveness to abiotic stress conditions, the expression of SlJUB1 and AtJUB1 can be promptly linked to SlDREB1, SlDREB2, and AtDREB2A promoters, respectively, which encode essential abiotic stress regulators, including AP2-type TFs [1,25,29]. Moreover, AtJUB1 could function downstream of AtHB13, which positively regulates drought stress tolerance [1,26]. Chen et al. [12] stated that ZmNAC2, a transcription factor from maize, could augment osmotic stress resistance in transgenic Arabidopsis. IPA1 also enhanced drought stress tolerance through inducing SNAC1 in rice [13]. Moreover, AhMYB30 boosted freezing and salt resistance in Arabidopsis [14]. AtMYB40 transcription factor also displayed a functional role in As tolerance in Arabidopsis [30]. Furthermore, OsARM1 overexpression enhanced rice sensitivity to As stress [31].
Rice (Oryza sativa L.) is amongst the foremost economically crucial cereal crops globally and supplies core food crops for inhabitants worldwide [20]. Environmental stresses negatively influence rice yield and grain quality. Moreover, rice retains the maximum level of As amongst all other crops because of the existence of aquaglyceroporins and phosphate transporters [2]. More importantly, due to the severe impacts of climate change and growing global population, there is an urgent requirement to develop more stress-resistant rice cultivars for use in breeding programs to boost crop yields to satisfy global food demands. Over the past decades, different rice varieties have been genetically engineered for enhanced stress tolerance via overexpressing several stress-related transcription factors, including NACs. For example, transgenic rice lines overexpressing SNAC2 displayed considerably increased resistance to salt and cold stresses [32], while the overexpression of the root-specific OsNAC9, OsNAC6, or OsNAC10 significantly augmented tolerance to drought in rice [33,34]. Overexpression of ONAC066 improved drought and oxidative stress resistance in rice [1]. ONAC095 plays different functional roles in cold- and drought stress responsiveness [35]. It negatively regulates drought responses but positively regulates cold responses in rice plants [35]. More importantly, overexpression of some stress-responsive NAC transcription factors significantly augmented rice resistance to severe environmental stress conditions without causing adverse impacts on crop yields [1,33,36,37], indicating the importance of the use of those NAC genes for further enhancement of stress tolerance and crop yield [38,39]. For example, transgenic rice plants overexpressing OsNAC5, OsNAC10, and SNAC1 and subjected to drought stress conditions displayed increased root diameter, drought tolerance, and grain yield [20,36,40,41].
Another rice NAC transcription factor, SNAC3, has been previously documented as a significant regulator of drought, heat, and oxidative stress responsiveness in rice plants via modulating abscisic acid–independent pathways and ROS [20]. However, the function of SNAC3 in mediating As stress tolerance and rice grain yield attributes has not been studied and validated yet. Therefore, the present study aimed to investigate whether SNAC3 has a vital role in augmenting rice tolerance to As stress and improving grain yield. SNAC3-overexpressing and SNAC3-RNAi transgenic rice lines were created utilizing an Agrobacterium-mediated transformation method. Various morpho-physiological, biochemical and molecular parameters as well as grain yield traits have been analyzed and recorded to reveal the significant differences between transgenics and wild-type rice plants in the current investigation.
2. Results and Discussion
2.1. Generation and Molecular Characterization of Transgenic Rice Lines
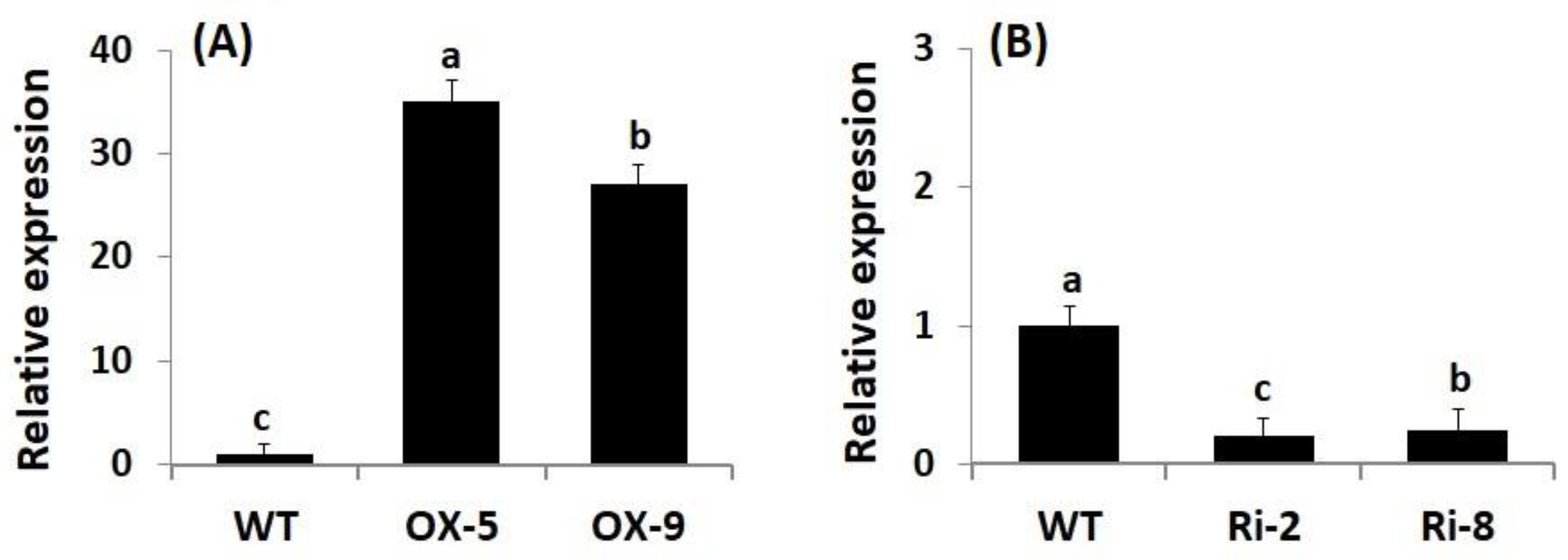
Genetic engineering has emerged as a promising approach for boosting crop stress resistance and accelerating plant breeding programs. To evaluate whether SNAC3 contributes significantly to enhancing rice resistance to As stress and boosting grain yield, SNAC3-OX and SNAC3-RNAi T3 transgenic rice lines were developed utilizing the Agrobacterium-mediated transformation method in the current study. qRT-PCR analysis indicated that the expression level of SNAC3 in SNAC3-OX lines, OX-5 and OX-9, were 34.9- and 27.1-fold higher, respectively, than that in wild-type (WT) plants (Figure 1A), whereas the expression level in SNAC3-RNAi lines, Ri-2 and Ri-8, were calculated to be 20 and 25% of the level in WT plants (Figure 1B), respectively. These transgenic lines were selected for use in the As stress experiment and for subsequent morpho-physio-biochemical and transcriptional analyses of the present study.
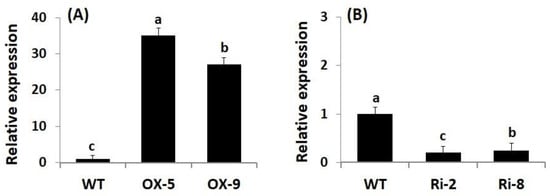
Figure 1.
Characterization of transgenic rice lines. (A) Expression of SNAC3 in SNAC3-overexpressing lines; and (B) expression of SNAC3 in SNAC3-RNAi lines. Data are means ± SE (n = 5). Different letters above bars (a, b, c) indicate a significant difference among rice lines (p ≤ 0.05).
2.2. SNAC3 Overexpression Enhances While SNAC3 Suppression Reduces Survival Rate, Leaf Relative Water Content, and Growth Characteristics of Rice under Arsenic Stress
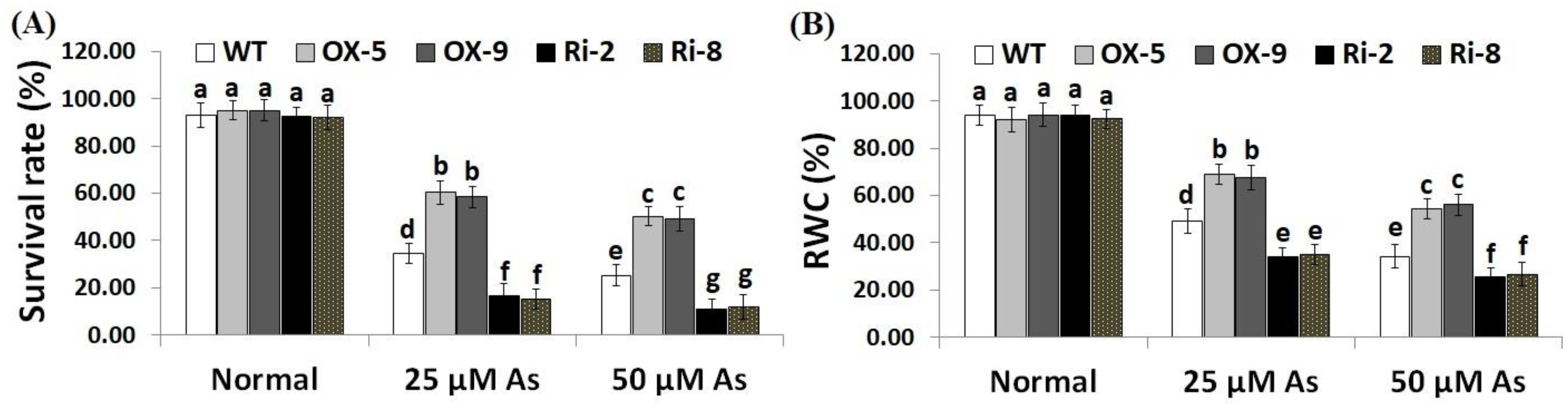
To explore whether SNAC3 could contribute to As stress resistance, a 10-day recovery period was applied after As stress treatment to record plants’ survival rate. The results showed that under both doses of As stress (25 and 50 µM), the survival rate of SNAC3-OX rice lines (OX-5 and OX-9) was remarkably higher than that of the wild-type plants, while SNAC3-RNAi lines (Ri-2 and Ri-8) exhibited significantly lower survival rates comparing to non-transformed plants (Figure 2A). In addition, leaf relative water content (RWC) represents one of the main indicators of water level balancing in plants. RWC was therefore estimated in the leafy tissues of the non-transgenic, SNAC3-OX, and SNAC3-RNAi plants. The findings indicate that both levels of As stress (25 and 50 µM) significantly reduced the RWC of non-transgenic and transgenic rice plants comparing to those grown under optimal growth conditions (Figure 2B). Moreover, RWC of As stress-treated SNAC3-OX plants was noticeably higher than that of As stress-treated non-transgenic plants, while As stress-treated SNAC3-RNAi plants had significantly lower RWC, comparing to As stress-treated non-transgenic plants. These results suggest that SNAC3 overexpression enhances As stress resistance in transgenic rice plants via maintaining the water status of plants. Our results agree with those demonstrated by Fang et al. [20], who revealed the enhanced survival rate of SNAC3-overexpressing rice plants under drought circumstances. Similarly, Yuan et al. [1] reported enhanced survival rate and RWC of ONAC066-overexpressing rice plants cultivated under drought circumstances.
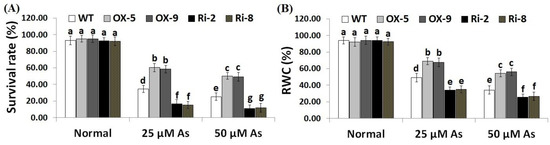
Figure 2.
Survival rate (A) and RWC (B) of the wild type (WT) and transformed lines subjected to varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
To further validate the function of SNAC3 in As stress tolerance, plant growth traits were measured. The findings demonstrated that both of As stress concentrations (25 and 50 µM) significantly restricted plant height, fresh weights of root and shoot, and dry weights of root and shoot of the non-transgenic and transgenic rice plants, in comparison to those grown under normal circumstances (Table 1). However, no significant differences in these growth parameters were detected between the non-transgenic and transgenic plants grown under optimal (0 As) treatment. On the other hand, under both levels of As stress (25 and 50 µM), all growth parameters of SNAC3-OX rice lines (OX-5 and OX-9) were remarkably higher than that of the wild-type plants, while SNAC3-RNAi lines (Ri-2 and Ri-8) exhibited significantly reduced growth parameters, comparing to the non-transgenic plants (Table 1). Such results further indicate that SNAC3 has a crucial function in rice As stress resistance through improving plant growth characteristics. Our results agree with that of Fang et al. [20], who recorded enhanced plant height of SNAC3-overexpressing transgenic rice plants under drought circumstances. Additionally, Yuan et al. [1] reported augmented performance of ONAC066-overexpressing rice plants grown under drought circumstances.

Table 1.
Morphological parameters of the wild type (WT) and transformed plants under varying doses of arsenic stress for 12 days.
2.3. Overexpression of SNAC3 Reduces While Suppression of SNAC3 Increases Arsentic Uptake and Oxidative Stress Biomarkers in Rice under Arsenic Stress
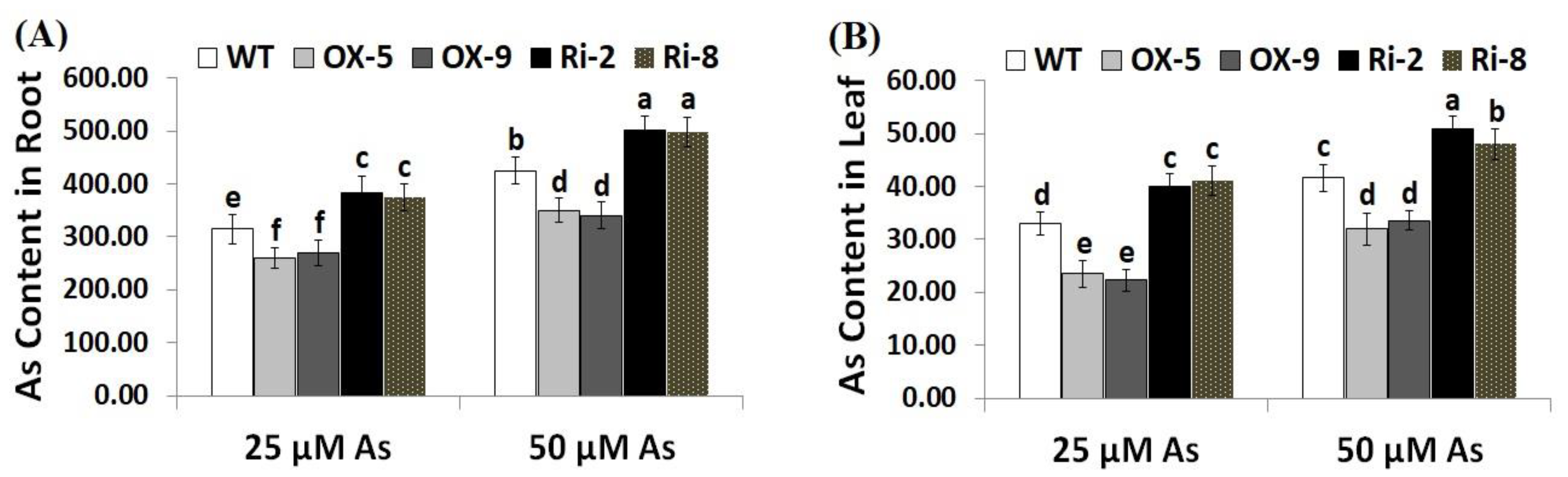
Arsenic concentration was assessed in the roots and leaves of the wild-type, SNAC3-OX, and SNAC3-RNAi plants. The results revealed that As concentration was higher in the plant root than shoot (Figure 3A,B). Moreover, As uptake increased in the roots and shoots of rice plants with increasing As stress concentration. On the other hand, the roots of SNAC3-OX rice lines, OX-5 and OX-9, exhibited significant reductions in As content by 17.5 and 14.6% at 25 µM As, respectively, and by 17.6 and 20% at 50 µM As, respectively, while the roots of SNAC3-RNAi lines, Ri-2 and Ri-8, had significant increases in As by 21.6 and 18.7% at 25 µM As, respectively, and by 18.1 and 17.2% at 50 µM As, respectively, as compared to As stress-treated wild-type plants (Figure 3A). Moreover, the leaves of SNAC3-OX rice lines, OX-5 and OX-9, exhibited significant reductions in As content by 28.8 and 32.4% at 25 µM As, respectively, and by 23.1 and 19.5% at 50 µM As, respectively, while the leaves of SNAC3-RNAi lines, Ri-2 and Ri-8, exhibited significant increases in As by 21.2 and 24.2% at 25 µM As, respectively, and by 22.6 and 15.4% at 50 µM As, respectively, as compared to As stress-treated wild-type plants (Figure 3B). The results indicate that the SNAC3 gene contributes to mitigating As toxicity in rice plants grown under As stress circumstances via reducing As uptake. Similarly, WRKY6 transcription factor conferred As stress tolerance in Arabidopsis via restricting As uptake [42].
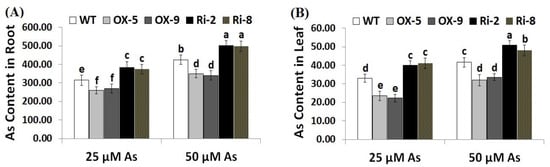
Figure 3.
Arsenic concentration (µg g−1 DW) in the roots (A) and leaves (B) of the wild type (WT) and transformed lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
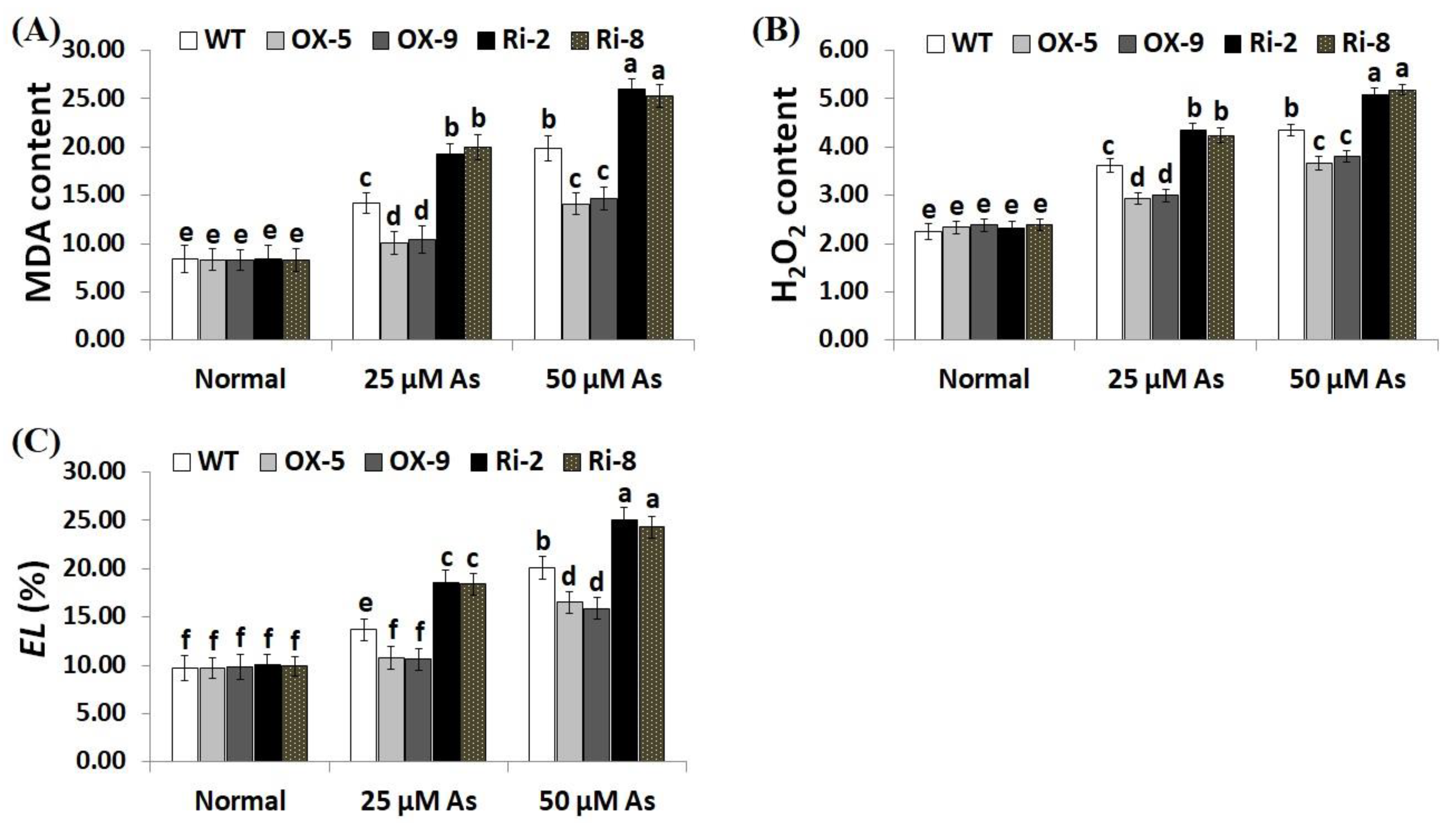
Overproduction of ROS causes toxic effects and destruction to plant cells. Electrolyte leakage (EL) represents a main indicator for the damage of the cell membrane [43]. Moreover, MDA is a key indicator for the free radicals-induced damage of lipid peroxidation end products [44]. Consequently, developing more stress-resistant crop cultivars is urgently required. To explore whether SNAC3-OX lines scavenge ROS, levels of oxidative stress markers (MDA, H2O2, and EL) were measured in the leaves of transformed and non-transformed plants under optimal and As treatments. As stress concentrations (25 and 50 µM) significantly enhanced the level of oxidative stress markers of the transgenic and non-transgenic rice plants, in comparison with those grown under optimal growth circumstances (Figure 4A–C). However, no considerable variances were recorded in MDA, H2O2, and EL between the transgenic and non-transgenic plants under standard conditions. In contrast, under both concentrations of As stress, levels of oxidative stress markers of the two SNAC3-OX lines were considerably lower than those of the non-transgenic plants, while the two SNAC3-RNAi lines demonstrated considerably higher level of MDA, H2O2, and EL, comparing to non-transgenic plants (Figure 4A–C). These findings indicate that SNAC3 overexpression in rice plants counteracts the toxic impacts of ROS accumulation, mitigates the oxidative damage, maintains a lower degree of cell membrane lipid peroxidation, and confers enhanced tolerance to As stress. Our results agree with that demonstrated by Fang et al. [20], who demonstrated reduced H2O2 levels in SNAC3-overexpressing transgenic rice plants under heat stress circumstances. Yuan et al. [1] also recorded reduced H2O2 content in ONAC066-overexpressing rice plants grown under drought stress conditions. WRKY6 transcription factor also reduced the oxidative stress level and conferred As stress tolerance in Arabidopsis [42].
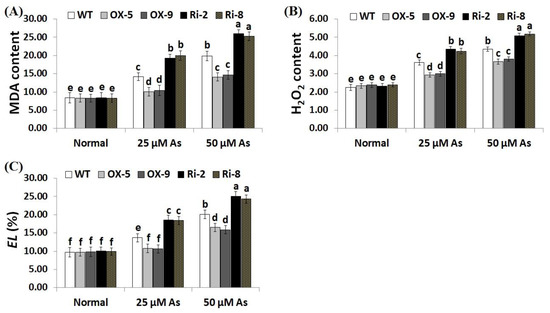
Figure 4.
Levels of lipid peroxidation (MDA, µM g−1 FW) (A), H2O2 (µM g−1 FW) (B), and electrolyte leakage (EL) (C) of the non-transformed (WT) and transformed rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
2.4. Overexpression of SNAC3 Enhances While Suppression of SNAC3 Reduces Chlorophyll Content, Gas-Exchange Parameters, and Osmolyte Accumulation in Rice under Arsenic Stress
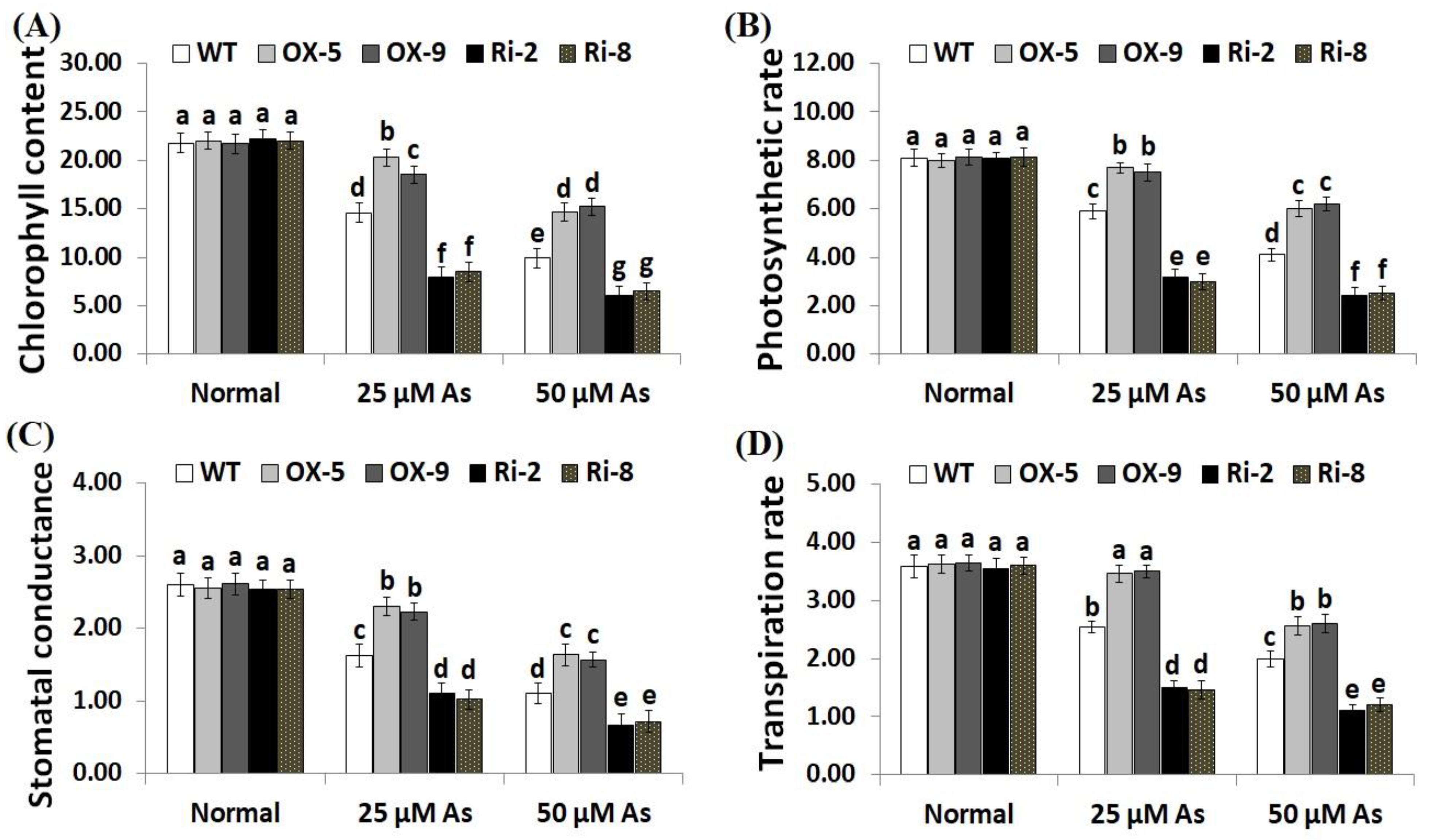
Environmental factors have adverse impacts on chlorophyll and gas exchange attributes in plants. To investigate whether SNAC3 could improve chlorophyll concentration and gas-exchange traits in rice plants grown under As stress, we measured the chlorophyll content, photosynthesis rate (Pn), stomatal conductance (gs) and transpiration rate (E) in the non-transformed, SNAC3-OX, and SNAC3-RNAi plants grown under normal conditions and both levels of As stress (25 and 50 µM). The findings indicate that both concentrations of As stress significantly decreased the rates of chlorophyll and gas-exchange parameters of the non-transformed and transformed rice plants in comparison to those grown under standard growth circumstances (Figure 5A–D). Conversely, under standard circumstances, no significant variations were detected in chlorophyll content and gas-exchange parameters between the non-transformed and transformed lines. In contrast, under both concentrations of As stress, levels of chlorophyll, Pn, gs, and E of the two SNAC3-OX lines were noticeably greater than those of the non-transgenic plants, while the two SNAC3-RNAi lines showed markedly lower levels of chlorophyll and gas-exchange parameters, comparing to the non-transformed plants (Figure 5A–D). These findings indicate that SNAC3 overexpression enhances As stress resistance through boosting photosynthesis and gas-exchange processes in rice leaves. Similarly, Yuan et al. [1] reported improved chlorophyll concentration in ONAC066-overexpressing rice plants cultivated under stress circumstances.
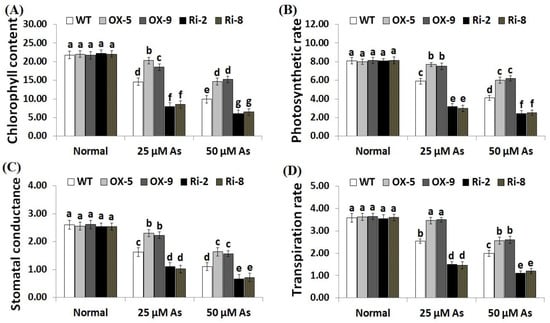
Figure 5.
Chlorophyll content (mg g−1 FW) (A), photosynthesis rate (Pn, μmol m2s−1) (B), stomatal conductance (gs, mol m2s−1) (C), and transpiration rate (E, mmol m2s−1) (D) of the non-transformed (WT) and transformed rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
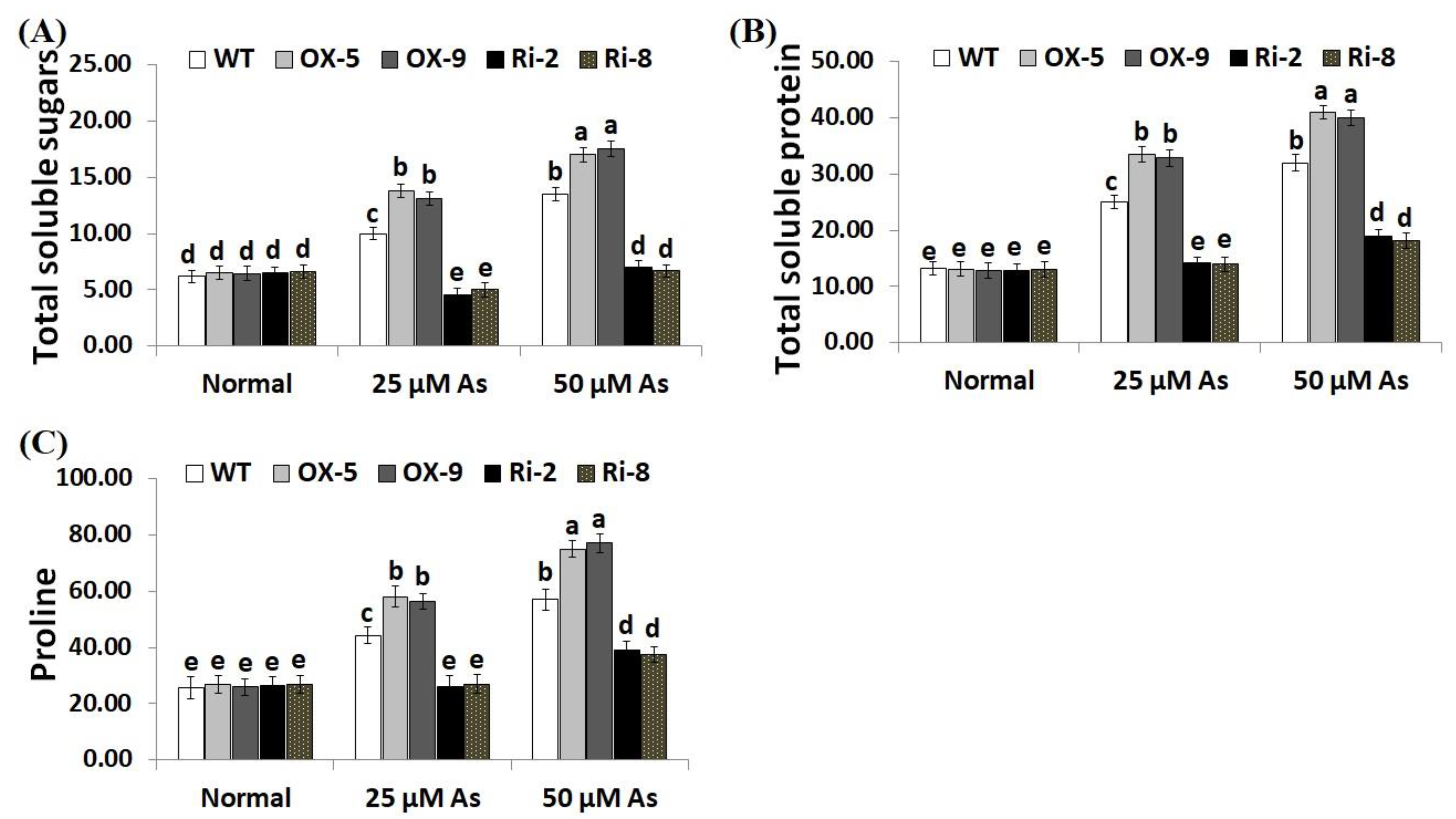
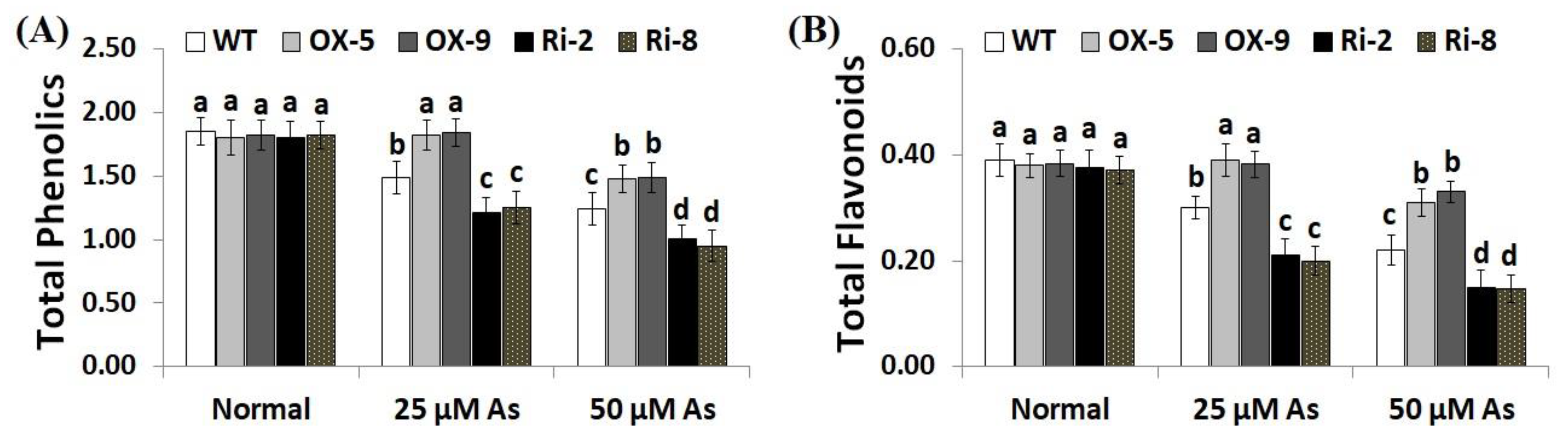
Soluble sugars, proline, proteins, phenolics, flavonoids, and other compatible solutes are useful osmoprotectants for plants under stress circumstances [45,46]. Soluble sugars and proteins also induce the efficiency of plant cells to resist dehydration and maintain biological molecules’ functions [47]. To dissect the mechanistic roles of SNAC3 in As stress tolerance and osmoregulation process, we estimated the contents of soluble proteins, sugars, proline, total phenolics, and total flavonoids in the non-transformed and transformed rice lines grown under As treatments. The results revealed that no noticeable variations were recorded in the level of sugars, soluble proteins, proline, total phenolics, and total flavonoids between the transformed and non-transformed plants grown under optimal circumstances (Figure 6A–C; Figure 7A,B). However, under both concentrations of As stress, level of soluble sugars, proteins, proline, total phenolics, and total flavonoids of the two SNAC3-OX lines were considerably higher than those of the non-transformed plants, while the two SNAC3-RNAi lines exhibited significantly lower levels of soluble sugars, soluble proteins, proline, phenolics, and flavonoids, comparing to the non-transgenic plants (Figure 6A–C; Figure 7A,B). These findings revealed that SNAC3 overexpression enhanced As stress tolerance in rice plants through increasing osmolyte accumulation in plant cells. Similarly, Yuan et al. [1] revealed enhanced synthesis of proline and soluble sugars in ONAC066-overexpressing rice plants grown under drought circumstances.
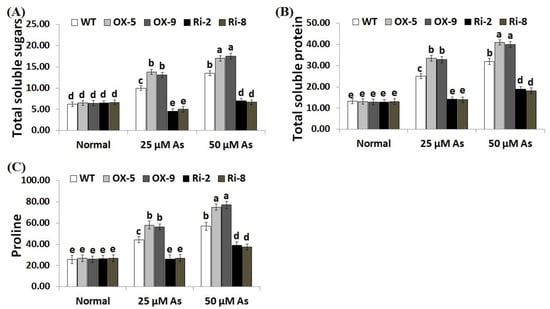
Figure 6.
Contents of total soluble sugars (mg g−1 FW) (A), total soluble proteins (mg g−1 FW) (B), and proline (µg g−1 FW) (C) of the wild-type (WT) and transformed rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
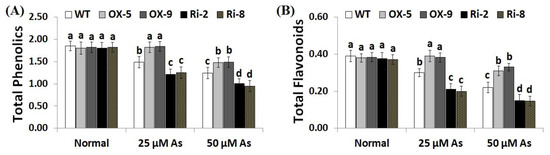
Figure 7.
The total phenolics (mg gallic acid/g extract) (A) and total flavonoids (mg catechin/g extract) (B) of the wild-type (WT) and transformed rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
2.5. Overexpression of SNAC3 Induces While Suppression of SNAC3 Reduces Antioxidant Enzyme Activities in Rice under Arsenic Stress
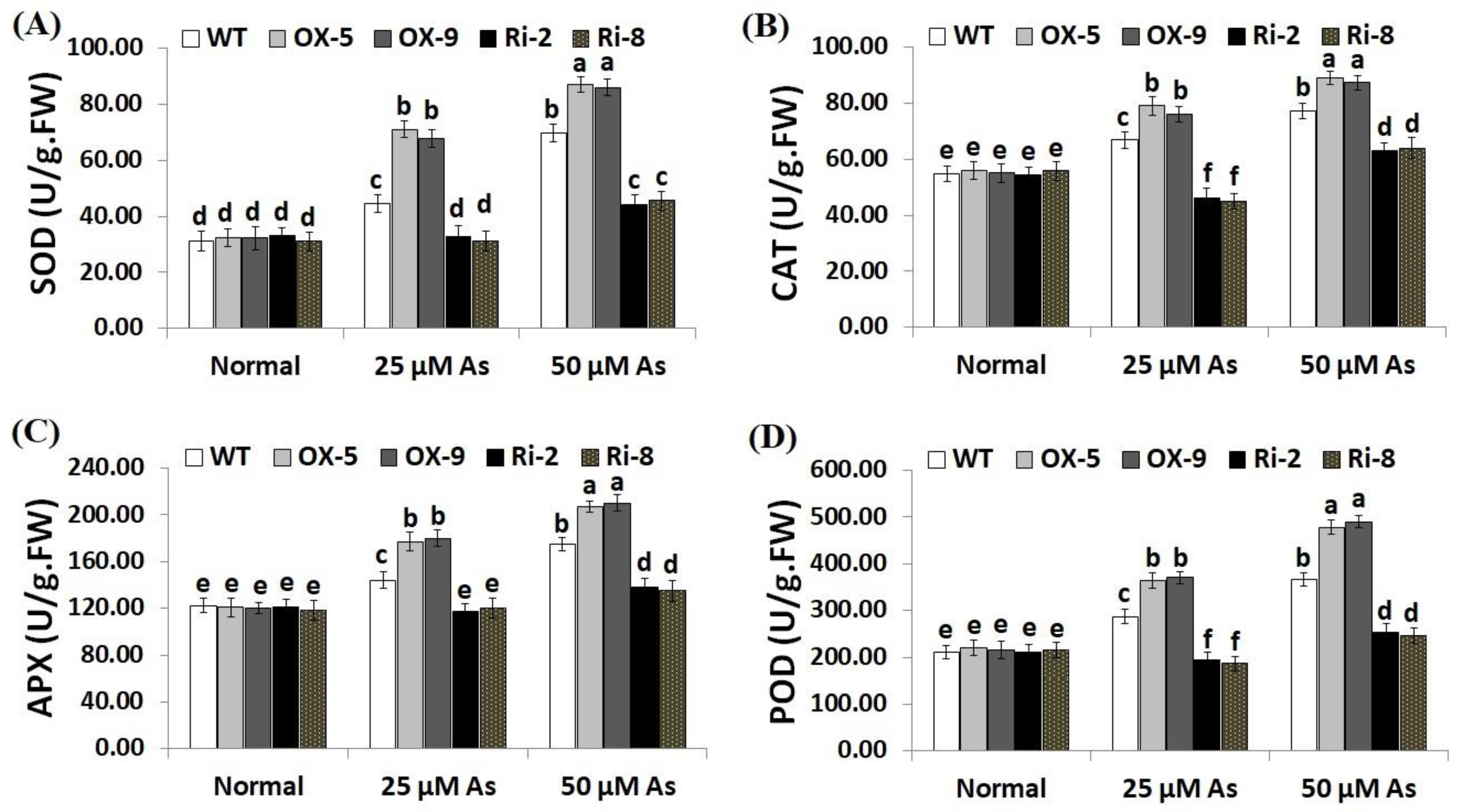
Enzymic antioxidants have a crucial role in ROS detoxification and thus boosting plant ability to tolerate stresses [48]. CAT assists in the detoxification of ROS under stress, while APX assists in the fine-tuning of ROS signals [20,49]. We therefore measured the activities of SOD, CAT, POD and APX in the non-transgenic and transgenic rice plants grown under As stress circumstances (Figure 8A–D). The results revealed no obvious differences in the enzymic antioxidant levels among the transgenic and non-transgenic lines grown under optimal circumstances (Figure 8A–D). Conversely, under both concentrations of As stress (25 and 50 µM), the levels of SOD, CAT, POD and APX of the two SNAC3-OX lines were considerably higher than those of the non-transformed plants, while the two SNAC3-RNAi lines showed significantly lower levels of SOD, CAT, APX, and POD in comparison with the non-transgenic plants (Figure 8A–D). These findings indicate that SNAC3 overexpression induces the enzymic antioxidant levels of transgenic rice lines which in turn scavenge ROS, causing decreased oxidative stress and enhanced As stress tolerance.
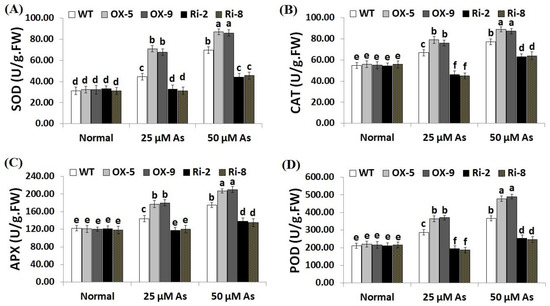
Figure 8.
Levels of enzymic antioxidants ((A): SOD, (B): CAT, (C): APX and (D): POD) of the wild-type (WT) and transformed rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
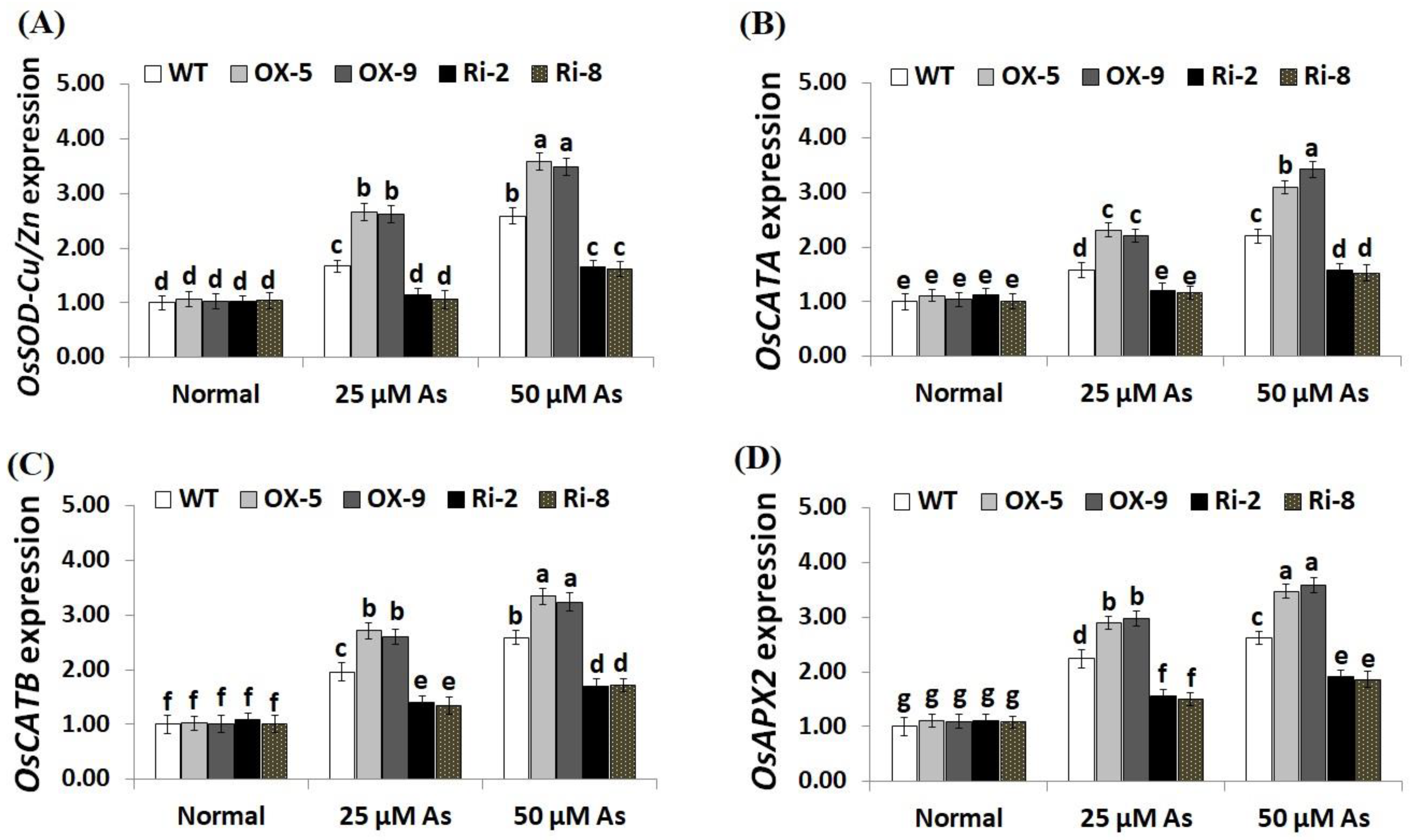
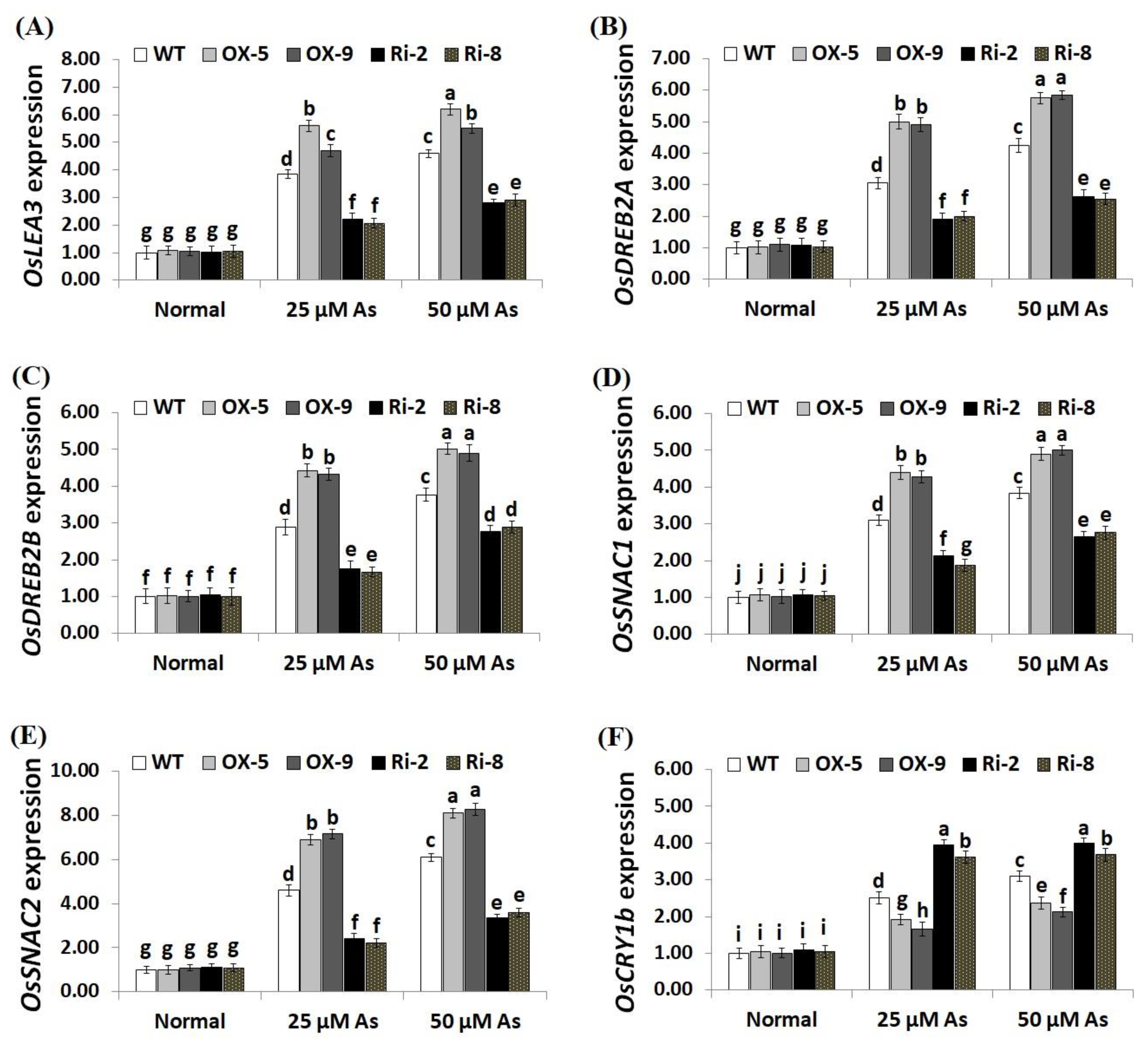
2.6. SNAC3 Overexpression and Suppression Modulate Abiotic Stress-Associated Genes Expression in Rice under Arsenic Stress
To unravel the downstream mechanisms and regulatory function of SNAC3 in As stress resistance, the expression levels of antioxidant genes (OsSOD-Cu/Zn, OsCATA, OsCATB, and OsAPX2) and stress-related genes (OsLEA3, OsDREB2B, OsDREB2A, OsSNAC2, OsSNAC1, and OsCRY1b) were assessed in the transgenic and non-transgenic rice lines grown under As stress circumstances utilizing qRT-PCR. The results revealed no significant variations in the expression levels of these genes among transgenic and non-transgenic plants grown under normal circumstances (Figure 9A–D; Figure 10A–F). By contrast, under both levels of As stress (25 and 50 µM), the expression levels of the antioxidant genes and five stress-related genes (OsLEA3, OsDREB2B, OsDREB2A, OsSNAC2, and OsSNAC1) of the two SNAC3-OX lines were higher than that of the non-transformed plants, while the two SNAC3-RNAi lines indicated remarkably lower levels, comparing to the non-transformed plants (Figure 9A–D; Figure 10A–E). More importantly, under both levels of As stress, the expression level of OsCRY1b in the two SNAC3-OX lines were considerably lower than those in the non-transgenic plants, while the two SNAC3-RNAi lines showed significantly enhanced expression levels of OsCRY1b, comparing to the non-transgenic plants (Figure 10F). These findings reveal that SNAC3 overexpression alleviates As toxicity and associated oxidative damage in rice through inducing genes that mediate free-radical-scavenging pathways and proteins involved in defense mechanisms. Moreover, SNAC3 overexpression suppressed the expression of OsCRY1b (rice cryptochrome 1b gene), thereby enhancing As resistance in rice plants. These results were in concordance with those of the enzymic antioxidants assayed in the present study. Our results were also in line with the findings of Yuan et al. [1] and Fang et al. [20], who recorded enhanced stress-responsive gene expression in ONAC066- and SNAC3-overexpressing lines, respectively, comparing to the non-transgenic stressed-plants. The findings were also in line with those recorded previously by El-Esawi et al. [50] and Cai et al. [51] who revealed that Rab7- and nNOS-overexpressing rice plants, respectively, showed higher expression levels of abiotic stress-related genes in comparison with the non-transgenic plants grown under stress conditions. Furthermore, our findings agree with the outcomes of Yu et al. [52], who revealed that OsEm1-overexpressing rice plants showed increased expression of the late embryogenesis abundant protein (OsLEA3) mediating stress resistance, comparing to the non-transgenic plants grown under stress conditions.
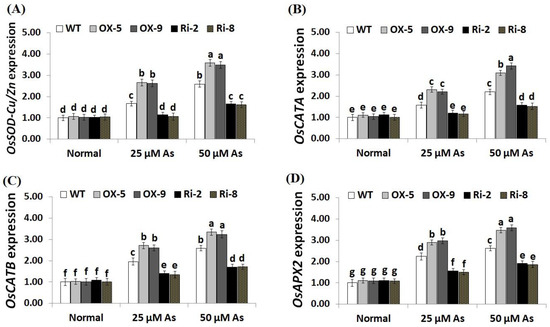
Figure 9.
Expression of OsSOD-Cu/Zn (A), OsCATA (B), OsCATB (C), and OsAPX2 (D) genes of the wild-type (WT) and transgenic rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
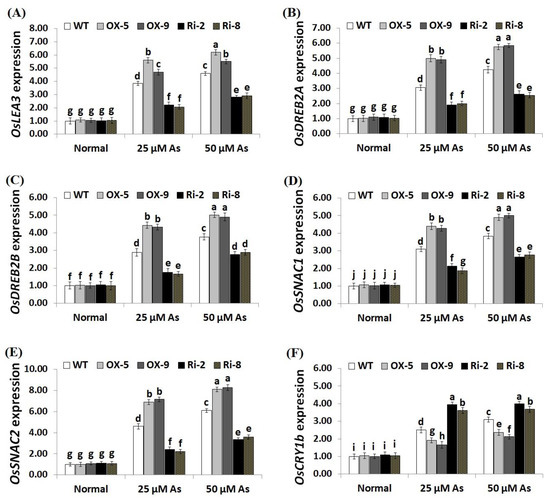
Figure 10.
Expression of OsLEA3 (A), OsDREB2A (B), OsDREB2B (C), OsSNAC1 (D), OsSNAC2 (E), and OsCRY1b (F) genes of the wild-type (WT) and transgenic rice lines under varying arsenic doses for 12 days. Values represent means ± SE (n = 5). Different letters above bars represent a significant difference among lines (p ≤ 0.05).
2.7. Overexpression of SNAC3 Improves While Suppression of SNAC3 Reduces Rice Grain Yield and Yield Components under Arsenic Stress
Environmental stresses influence crop grain yields. To assess the key role of SNAC3 gene in enhancing rice grain yields, several yield traits, including panicle length, number of spikelets per hill, number of spikelets per panicle, filling rate, number of filled grains per hill, and total grain weight were recorded for the transgenic and non-transgenic plants grown under optimal and As stress treatments. The results revealed no considerable variations in the measured yield parameters among the non-transgenic and transgenic lines grown under optimal circumstances (Table 2). However, under both levels of As stress (25 and 50 µM), all the yield traits of the two SNAC3-OX lines were considerably greater than those of the wild-type rice plants, while the two SNAC3-RNAi lines showed significantly lower rates of yield parameters, comparing to the non-transgenic plants (Table 2). For instance, the two SNAC3-OX lines had a greater filling rate than that of the non-transgenic plants grown under As stress circumstances, leading to significant increments in the total weight of grain. These outcomes demonstrate the key role of SNAC3 in boosting the grain yield of rice plants grown under As stress conditions. Our findings are in agreement with those of other reports that revealed enhanced grain yields in rice plants overexpressing OsRab7 [50], TIFY [53], AP37 [54], OsNRT2.1 [55], and OsNAC2 [56].

Table 2.
Yield traits of the wild-type (WT) and transgenic rice lines subjected to normal and arsenic stress circumstances for 12 days.
3. Materials and Methods
3.1. Plant Material and Growth Circumstances
Oryza sativa subsp. japonica cultivar Giza 177, received from the Egyptian Agricultural Research Center, was employed in all the experiments of the current study. After surface-sterilizing in sodium hypochlorite (0.5%) and washing with H2O five times, plant seeds were sown and grown on wet papers for 6 days. Healthy uniform plantlets were chosen, transplanted into plastic containers comprising perlite, peat, and sand (1:1:1, v/v/v), and then left to grow in a growth room at 70% humidity, 300–400 µmol m−2 s−1 light intensity, 16/8 h light/dark and 27/21 °C. A daily irrigation with a Hoagland nutrient solution [57] was applied to the plants.
3.2. Construction of Plasmids and Transformation of Rice Plants
SNAC3-overexpression (SNAC3-OX) and SNAC3-RNAi (SNAC3-Ri) constructs were created as stated by Fang et al. [20]. Briefly, to generate the SNAC3-OX construct, RNA was taken out from 3-week-old rice cultivar Giza 177 plants utilizing an RNeasy Plant Mini kit (Qiagen, Hilden, Germany), and cDNA was then generated using a Reverse Transcription kit (Qiagen, Hilden, Germany). cDNA of SNAC3 was then amplified and introduced into the pU1301 vector under Ubiquitin promoter control. To create the SNAC3-RNAi construct, a SNAC3-specific fragment (334 bp) was obtained and inserted into the pDS1301 vector. Obtained constructs were transferred to Agrobacterium tumefaciens EHA105, which was then transformed into rice cultivar Giza 117 plants according to Agrobacterium-mediated transformation protocol reported by Lin and Zhang [58] with some modifications. Briefly, callus induction was performed on Murashige and Skoog (MS) medium comprising 500 mg/L glutamine, 3.0 mg/l 2,4-dichlorophenoxyacetic acid (2,4-D), 3% sucrose, 500 mg/L proline, and 0.25% phytagel at pH 5.8. Subculture was conducted on MS medium comprising 500 mg/L proline, 2.5 mg/l 2,4-D, 3% maltose, 500 mg/L glutamine, and 0.25% phytagel at pH 5.8. SNAC3-OX and SNAC3-Ri constructs were created using the primers reported by Fang et al. [20].
3.3. Molecular Characterization of Transformed Rice Lines
Transgenic lines of the T2 generation were screened for hygromycin-resistant phenotype segregation on 1/2 MS (Murashige and Skoog) medium supplemented with 50 mg/L hygromycin. Plants exhibiting 3:1 segregation for hygromycin-resistant phenotype were selected as single-copy transformed lines. T3 transformed lines exhibiting 100% hygromycin-resistant phenotype on selective medium were chosen as homozygous lines. Homozygous single-copy T3 transgenic lines were utilized for all subsequent analyses. Leaves of three-week-old wild-type, SNAC3-OX, and SNAC3-RNAi plants were harvested for analyzing SNAC3 expression level utilizing quantitative real-time PCR (qRT-PCR). In brief, total RNA and cDNA were prepared from those collected leaf samples as mentioned above. qRT-PCR analysis was then performed in 5 replications utilizing Qiagen QuantiTect SYBR Green PCR kit in order to analyze SNAC3 expression level. PCR conditions and SNAC3-specific primers were utilized as described by Fang et al. [20]. The housekeeping gene, Ubiquitin, was assayed. Relative expression level of SNAC3 was then calculated as reported by Livak and Schmittgen [59].
3.4. Arsenic Stress Treatments
Wild-type, two T3 homozygous SNAC3-OX (OX-5 and OX-9), and two T3 homozygous SNAC3-RNAi (Ri-2 and Ri-8) rice lines were used for the As stress treatment experiment. Seeds of these 5 lines were disinfected in sodium hypochlorite (0.5%), washed with H2O, and left to grow on wet papers for 6 days. Healthy uniform seedlings were moved into containers comprising peat, sand, and perlite (1:1:1, v/v/v). Containers were arranged in a complete randomized design in a growth room at 70% humidity, 27/21 °C, 16/8 h (light/dark), and a light intensity of 250–300 µmoL m−2 s−1. The plants received daily irrigation with a Hoagland nutrient solution for 3 weeks. The 27-day-old plants were assigned to groups representing different treatments as follows: (i) control plants irrigated with a Hoagland nutrient solution; (ii) As-stressed plants irrigated with a Hoagland solution containing 25 µM As; and (iii) As-stressed plants irrigated with a Hoagland solution containing 50 µM As. As concentrations were prepared using sodium arsenite (NaAsO2) and selected based on our preliminary standardization experiments and previous rice studies [60,61]. After 12 days of As stress treatments, some plant samples were harvested for use in physio-biochemical and molecular assays. Following a 10-day recovery period, the survival rate of plants was estimated.
To estimate the plant yield traits under As stress circumstances, the wild-type and T3 transformed rice plants were moved into larger containers filled with natural paddy soil. Containers were placed in a complete randomized design in growth rooms at 27/21 °C. Plants were fed daily with a Hoagland solution. Approximately 12–14 days before panicle heading stage, plants were assigned to different treatment groups and irrigated with Hoagland nutrient solutions containing 0 (control), 25 µM As, or 50 µM As. All these treatments continued for 24 days, then the plants were grown at standard growth conditions till harvesting. At maturity stage, the yield traits such as total number of spikelets per hill, panicle length (cm), number of spikelets per panicle, total grain weight (g), filling rate (%), and number of filled grains per hill were estimated.
3.5. Estimating the Morphological Characteristics and Relative Water Content
The plant samples were collected and washed using distilled water. A measuring scale was utilized for determining the plant height. The root and shoot samples were then separated. Fresh weights of shoots and roots were estimated. Dry weights of shoots and roots were also estimated after exposure to 70 °C for 48 h. Moreover, the methodology of Yamasaki and Dillenburg [62] was adopted to calculate the leaf relative water content (RWC).
3.6. Estimating Arsenic Content in Plant Roots and Leaves
As concentration were assessed in plant roots and leaves as previously stated by Mousavi et al. [61] with slight adjustments. Briefly, root and leaf tissue samples were cleaned with water and then oven-dried at 70 °C for 72 h. Dried tissue samples were mixed with a solution of HNO3:H2O2 (1:4 ratio). As concentration was then measured using ICP–mass spectrometry (Optima 7900DV, PerkinElmer, MA, USA).
3.7. Measurement of Oxidative Stress Biomarkers
Estimation of the leaf content of hydrogen peroxide (H2O2) was carried out using the methodology indicated by Velikova et al. [63] with slight adjustments. In brief, 0.5 g of fresh leafy sample was dissolved in 5 mL of trichloroacetic acid (0.1 %) and then centrifuged at 10,000× g for 20 min. A mixture of supernatant (0.5 mL), 10 mM potassium phosphate buffer (0.5 mL) and 1 M KI (1 mL) was prepared and then vortexed before measuring the absorbance at 390 nm. H2O2 concentration was calculated utilizing H2O2 standard curve.
Production of leaf malondialdehyde (MDA) was estimated using the methodology of Heath and Packer [64] with slight modifications. Leafy samples (1 g) were homogenized in TCA (20 mL, 0.1%), followed by a 7-min centrifugation at 14,000× g. A mixture of supernatant (1.5 mL) and thiobarbituric acid (6.0 mL, 0.5%) in TCA (20%) was prepared and then agitated at 95 °C for 25 min. This was followed by chilling on ice and a 10-min centrifugation at the highest speed. Absorbance of supernatants was monitored at 532 and 660 nm. Moreover, the methodology of Dionisio-Sese and Tobita [65] was adopted to estimate leaf electrolyte leakage (EL).
3.8. Estimation of Total Chlorophyll, Gas-Exchange Parameters, Soluble Protein, Sugars, Proline, Phenolics, and Flavonoids
Estimation of total leaf chlorophyll was done as indicated by Lichtenthaler [66]. In brief, maceration of fresh leaves (of 0.2 g) was performed in acetone (50 mL, 80%). The extract was centrifuged at 14,000× g for 8 min. Supernatant absorbance was measured at 663 and 645 nm. Moreover, estimation of net photosynthesis rate (Pn), stomatal conductance (gs), and transpiration rate (E) was performed on the leaf midrib in the early morning following the protocol reported by Holá et al. [67].
To determine the total protein content of leaves, samples of fresh leaves were macerated in Tris buffer (100 mM, pH 8.0) using a cold mortar. The extracts were exposed to a 15-min centrifugation at 20,000× g. Bradford assay [68] was then utilized to determine total protein content. Total soluble sugar of leaves was also spectrophotometrically estimated at 485 nm following the reported methodology of Dey [69].
Determination of proline level was performed as indicated by Bates et al. [70] with slight adjustments. Briefly, 0.5 g of leaf sample was macerated in 3 mL of 5% (w/v) sulfosalicylic acid, followed by centrifugation at 9000× g for 8 min. Supernatant (0.5 mL) was diluted with sterile H2O (1 mL) and mixed with 2% ninhydrin (2 volumes) by a vortex mixer. This was followed by a 35-min incubation at 95 °C and then cooling. An equivalent amount of toluene was added into the solution, and the absorbance was measured at 520 nm for the upper aqueous layer using a spectrophotometer. The L-proline calibration curve was utilized as a standard for calculation.
The concentration of total phenolics in plant leaves was estimated through the homogenization of leafy samples (2 g) in 80% of methanol (10 mL), followed by heating up at 70 °C for 20 min. A volume of extract (2 mL) was diluted with H2O (10 mL) containing 1 N Folin–Ciocalteau reagent (500 μL), followed by incubation at 30 °C. To estimate the contents of phenols, the mixture absorbance was read at 725 nm [71]. Gallic acid was utilized as a standard.
The content of total flavonoids in plant leaves was also estimated according to Zhishen et al. [72]. Dried leaf powder (1 g) was dissolved in sterilized H2O (100 mL), followed by filtration of the extract and mixing with AlCl3, distilled H2O, and NaNO2 solution. Then, NaOH solution was transferred to the mixture, followed by dilution with distilled H2O. The absorbance was monitored at 510 nm. The catechin standard curve was utilized.
3.9. Antioxidant Enzyme Assays
Approximately 1 gm of leaf sample was macerated in 100 mM Tris-HCl (pH 7.5) combined with 10 mM MgCl2, PVP-40 (1.5%), 5 mM Dithiothreitol, 1 mM EDTA, 1 mM phenylmethanesulfonyl fluoride, 5 mM magnesium acetate, and 1 μg.mL−1 aproptinin. The suspension was refined, followed by a 10-min centrifugation at 11,000 rpm. Enzyme activities were monitored using the supernatant. For APX activity analysis, 2 mM AsA was utilized in the homogenization of leaf sample.
Ascorbate peroxidase (APX) level in the plant leaf was estimated following the methodology of Nakano and Asada [73], and the optical density was monitored at 265 nm using a spectrophotometer. The activity of APX was expressed in enzyme unit per gram fresh weight (U/g.FW). Superoxide dismutase (SOD) activity was recorded following the methodology of Kono [74] and according to nitroblue tetrazolium photoreduction. A spectrophotometer was used to record the absorbance at 540 nm. Moreover, determination of catalase (CAT) activity was performed following the reported methodology of Aebi [75]. Absorbance was monitored at 240 nm. Peroxidase (POD) enzyme level was also assessed according to Putter and Becker [76]. Production rate of oxidized guaiacol was read at 436 nm. The activity of POD, CAT and SOD was also expressed as U/g.FW.
3.10. Analysis of Stress-Tolerant Genes Expression
Expression of antioxidant genes (OsSOD-Cu/Zn, OsCATB, OsCATA, and OsAPX2) and 6 abiotic stress-associated genes (OsLEA3, OsDREB2B, OsDREB2A, OsSNAC2, OsSNAC1, and OsCRY1b) were measured in the transformed and non-transformed plants under As stress conditions. In brief, total RNA and cDNA were isolated from the harvested leaf samples as mentioned above. qRT-PCR analysis was then performed utilizing the Qiagen QuantiTect SYBR Green PCR kit. PCR circumstances were set up as outlined by Vighi et al. [77] and Cai et al. [51]. A set of genes-specific primers [51,52,77,78] were applied for amplification. An internal reference gene, UBQ10, was used [77]. The relative expression levels of genes were calculated as reported by Livak and Schmittgen [59].
3.11. Statistical Analysis
One-way analysis of variance and Duncan’s multiple range test were applied to analyze the obtained data. Values indicate means ± standard error (SE), and are significantly different at p ≤ 0.05.
4. Conclusions
Arsenic toxicity adversely restricts rice growth and yield. The crucial function of SNAC3 in boosting As stress resistance and grain yield in rice has been investigated. Two SNAC3-OX lines and two SNAC3-RNAi lines were created and subjected to As stress treatments. SNAC3 overexpression significantly augmented rice tolerance to As stress and boosted grain yield, while SNAC3 suppression increased rice sensitivity to As and reduced grain yield. SNAC3 enhanced As stress resistance and grain productivity in rice through modulating antioxidant machinery, osmolyte synthesis, and stress-responsive genes, and could be a useful prospect for further improvement of crop stress resistance and grain productivity. Further analysis might be carried out to unravel the regulatory network of SNAC3 in greater depth and to further dissect the molecular mechanisms underlying stress resistance in rice.
Author Contributions
Conceptualization, M.A.E.-E. and M.A.; methodology, E.M.E.-B., M.P. and M.A.E.-E.; software, E.M.E.-B., M.A.E.-E., H.M.A., N.J., M.A. and C.H.; validation, M.P., E.M.E.-B., M.A.E.-E. and H.M.A.; formal analysis, E.M.E.-B., M.P., N.J., C.H., M.A. and M.A.E.-E.; investigation and visualization, M.P., E.M.E.-B., H.M.A., N.J., C.H., M.A. and M.A.E.-E.; writing—original draft preparation, E.M.E.-B., M.P. and M.A.E.-E.; writing—review and editing, E.M.E.-B., M.P., M.A.E.-E., H.M.A., N.J., M.A. and C.H.; supervision, E.M.E.-B., M.A. and M.A.E.-E. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Researchers Supporting Project number (RSP2023R123), King Saud University, Riyadh, Saudi Arabia.
Data Availability Statement
All the data of the present study are included in this published article.
Acknowledgments
Authors would like to extend their sincere appreciation to the Researchers Supporting Project number (RSP2023R123), King Saud University, Riyadh, Saudi Arabia. Authors would also like to acknowledge the support provided by Sorbonne University, Tanta University and Embassy of France in Egypt.
Conflicts of Interest
The authors declare no competing interests.
References
- Yuan, X.; Wang, H.; Cai, J.; Bi, Y.; Li, D.; Song, F. Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 2019, 19, 278. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Thounaojam, T.C.; Upadhyaya, H. Arsenic stress in Rice (Oryza sativa) and its amelioration approaches. Plant Stress 2022, 4, 100076. [Google Scholar] [CrossRef]
- Singh, V.; Singh, N.; Rai, S.N.; Kumar, A.; Singh, A.K.; Singh, M.P.; Sahoo, A.; Shekhar, S.; Vamanu, E.; Mishra, V. Heavy Metal Contamination in the Aquatic Ecosystem: Toxicity and Its Remediation Using Eco-Friendly Approaches. Toxics 2023, 11, 147. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Weyens, N.; Vangronsveld, J. Phytoremediation: State-of-the-art and a key role for the plant microbiome in future trends and research prospects. Int. J. Phytoremediat. 2017, 19, 23–38. [Google Scholar] [CrossRef]
- Noor, I.; Sohail, H.; Sun, J.; Nawaz, M.A.; Li, G.; Hasanuzzaman, M.; Liu, J. Heavy metal and metalloid toxicity in horticultural plants: Tolerance mechanism and remediation strategies. Chemosphere 2022, 303, 135196. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Alayafi, A.A.; Witczak, J.; Ahmad, M. Analysis of Genetic Variation and Enhancement of Salt Tolerance in French Pea (Pisum Sativum L.). Int. J. Mol. Sci. 2018, 19, 2433. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, U.; Ayub, A.; Hussain, S.; Waraich, E.; El-Esawi, M.; Ishfaq, M.; Ahmad, M.; Ali, N.; Maqsood, M. Cadmium Toxicity in Plants: Recent Progress on Morpho-physiological Effects and Remediation Strategies. J. Soil Sci. Plant Nutr. 2022, 22, 212–269. [Google Scholar] [CrossRef]
- Castilhos, G.; Lazzarotto, F.; Spagnolo-Fonini, L.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci. 2014, 223, 1–7. [Google Scholar] [CrossRef]
- Shao, H.B.; Wang, H.Y.; Tang, X.L. NAC transcription factors in plant multiple abiotic stress responses: Progress and prospects. Front Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Ahmad, M. Overexpression of AtWRKY30 Transcription Factor Enhances Heat and Drought Stress Tolerance in Wheat (Triticum aestivum L.). Genes 2019, 10, 163. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Alayafi, A.A. Overexpression of StDREB2 Transcription Factor Enhances Drought Stress Tolerance in Cotton (Gossypium barbadense L.). Genes 2019, 10, 142. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, X.; Xie, X.; Liu, L.; Fu, J.; Wang, Q. Maize transcription factor ZmNAC2 enhances osmotic stress tolerance in transgenic Arabidopsis. J. Plant Physiol. 2023, 282, 153948. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhang, H.; Li, H.; Lian, L.; Wei, Y.; Lin, Y.; Wang, L.; He, W.; Cai, Q.; Xie, H.; et al. IPA1 improves drought tolerance by activating SNAC1 in rice. BMC Plant Biol. 2023, 23, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Pan, L.; Yang, Z.; Su, M.; Xu, J.; Jiang, X.; Yin, X.; Wang, T.; Wan, F.; Chi, X. A MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic Arabidopsis through both DREB/CBF and ABA-signaling pathways. Front. Plant Sci. 2023, 14, 1136626. [Google Scholar] [CrossRef]
- Oliveira, P.N.d.; Matias, F.; Martínez-Andújar, C.; Martinez-Melgarejo, P.A.; Prudencio, Á.S.; Galeano, E.; Pérez-Alfocea, F.; Carrer, H. Overexpression of TgERF1, a Transcription Factor from Tectona grandis, Increases Tolerance to Drought and Salt Stress in Tobacco. Int. J. Mol. Sci. 2023, 24, 4149. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, C.; Wang, L.; Zhong, H.; Xu, X.; Cheng, Y.; Nian, H.; Liu, W.; Chen, P.; Zhang, A.; et al. GmABR1 encoding an ERF transcription factor enhances the tolerance to aluminum stress in Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1125245. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J.A. NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Satoh, K.; Moumeni, A.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Attia, K.; Kikuchi, S. Comprehensive gene expression analysis of the NAC gene family under normal growth conditions, hormone treatment, and drought stress conditions in rice using near-isogenic lines (NILs) generated from crossing Aday selection (drought tolerant) and IR64. Mol. Gen. Genom. 2012, 287, 389–410. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, I.; Vermeirssen, V.; Van Aken, O.; Vandepoele, K.; Murcha, M.W.; Law, S.R.; Inzé, A.; Ng, S.; Ivanova, A.; Rombaut, D.; et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013, 25, 3472–3490. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Nobmann, B.; Devi Allu, A.; Balazadeh, S. JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav. 2016, 11, e1181245. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2016, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Ebrahimian-Motlagh, S.; Ribone, P.A.; Thirumalaikumar, V.P.; Allu, A.D.; Chan, R.L.; Mueller-Roeber, B.; Balazadeh, S. JUNGBRUNNEN1 confers drought tolerance downstream of the HD-zip I transcription factor AtHB13. Front Plant Sci. 2017, 8, 2118. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 2017, 254, 803–816. [Google Scholar] [CrossRef]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, H.-Y.; Chen, Y.-F. The transcription factor MYB40 is a central regulator in arsenic resistance in Arabidopsis. Plant Comm. 2021, 2, 100234. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Z.; Chen, M.X.; Yu, L.J.; Xie, L.J.; Yuan, L.B.; Qi, H.; Xiao, M.; Guo, W.; Chen, Z.; Yi, K.; et al. OsARM1, an R2R3 MYB transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 2017, 8, 1868. [Google Scholar] [CrossRef]
- Hu, H.H.; You, J.; Fang, Y.J.; Zhu, X.Y.; Qi, Z.Y.; Xiong, L.Z. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Redillas, M.C.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Huang, L.; Hong, Y.; Zhang, H.; Li, D.; Song, F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016, 16, 203. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Lv, B.; Li, J.; Luo, L.; Lu, S.; Zhang, X.; Ma, H.; Ming, F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014, 55, 604–619. [Google Scholar] [CrossRef]
- Tran, L.S.; Nishiyama, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 2010, 1, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Revi. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, K.; Schauer, N.; Larson, T.R.; Graham, I.A.; Fernie, A.R.; Leaver, C.J. The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 2006, 47, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, G.; Sánchez-Bermejo, E.; de Lorenzo, L.; Crevillén, P.; Fraile-Escanciano, A.; Tc, M.; Mouriz, A.; Catarecha, P.; Sobrino-Plata, J.; Olsson, S.; et al. WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 2013, 25, 2944–2957. [Google Scholar] [CrossRef]
- Bajji, M.; Kinet, J.; Lutts, S. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 2002, 36, 61–70. [Google Scholar] [CrossRef]
- Ganguli, L.J. Lipid peroxidation-DNA damage by malondialdehyde. Mutat. Res. Fundam. Mol. Mech. 1999, 424, 83–95. [Google Scholar]
- Ben, K.R.; Abdelly, C.; Savouré, A. Proline, a multifunctional amino-acid involved in plant adaptation to environmental constraints. Biol. Aujourdhui 2012, 206, 291. [Google Scholar]
- Chaleff, R.S. Further characterization of picloram tolerant mutance of Nicotinana tabacum. Theor. Appl. Genet. 1980, 58, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, P.; Zhu, J. Effect of magnesium (Mg) on contents of free proline, soluble sugar and protein in soybean leaves. J. Henan Agric. Sci. 2004, 6, 35–38. [Google Scholar]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Alzahrani, S.M.; Ali, H.M.; Alayafi, A.A.; Ahmad, M. Serratia liquefaciens KM4 Improves Salt Stress Tolerance in Maize by Regulating Redox Potential, Ion Homeostasis, Leaf Gas Exchange and Stress-Related Gene Expression. Int. J. Mol. Sci. 2018, 19, 3310. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Alayafi, A.A. Overexpression of Rice Rab7 Gene Improves Drought and Heat Tolerance and Increases Grain Yield in Rice (Oryza sativa L.). Genes 2019, 10, 56. [Google Scholar] [CrossRef]
- Cai, W.; Liu, W.; Wang, W.S.; Fu, Z.W.; Han, T.T.; Lu, Y.T. Overexpression of rat neurons nitric oxide synthase in rice enhances drought and salt tolerance. PLoS ONE 2015, 10, e0131599. [Google Scholar] [CrossRef]
- Yu, J.; Lai, Y.; Wu, X.; Wu, G.; Guo, C. Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem. Biophys. Res. Commun. 2016, 478, 703–709. [Google Scholar] [CrossRef]
- Hakata, M.; Kuroda, M.; Ohsumi, A.; Hirose, T.; Nakamura, H.; Muramatsu, M.; Ichikawa, H.; Yamakawa, H. Overexpression of a rice TIFY gene increases grain size through enhanced accumulation of carbohydrates in the stem. Biosci. Biotechnol. Biochem. 2012, 76, 2129–2134. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.J.; Kim, Y.S.; Kwon, C.W.; Park, H.K.; Jeong, J.S.; Kim, J.K. Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 2009, 150, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Luo, B.; Chen, J.; Zhu, L.; Liu, S.; Li, B.; Lu, H.; Ye, G.; Xu, G.; Fan, X. Overexpression of a high-affinity nitrate transporter OsNRT2.1 increases yield and manganese accumulation in rice under alternating wet and dry condition. Front. Plant Sci. 2018, 9, 1192. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Chen, W.; Dong, J.; Li, J.; Yang, F.; Wu, Z.; Zhou, H. Overexpression of OsmiR164b-resistant OsNAC2 improves plant architecture and grain yield in rice. J. Exp. Bot. 2018, 69, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.; Arnon, D. Physiological aspects of availability of nutrients for plant growth. Soil Sci. 1941, 51, 431–444. [Google Scholar] [CrossRef]
- Lin, Y.J.; Zhang, Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005, 23, 540–547. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Singh, A.P.; Dixit, G.; Mishra, S.; Dwivedi, S.; Tiwari, M.; Mallick, S.; Pandey, V.; Trivedi, P.K.; Chakrabarty, D.; Tripathi, R.D. Salicylic acid modulates arsenic toxicity by reducing its root to shoot translocation in rice (Oryza sativa L.). Front. Plant Sci. 2015, 6, 340. [Google Scholar] [CrossRef]
- Mousavi, S.R.; Niknejad, Y.; Fallah, H.; Tari, D.B. Methyl jasmonate alleviates arsenic toxicity in rice. Plant Cell Rep. 2020, 39, 1041–1060. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Dillenburg, L.C. Measurements of Leaf Relative Water Content in Araucaria angustifolia. R. Bras. Fisiol. Veg. 1999, 11, 69–75. [Google Scholar]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant responses of rice seedlings to salinity stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Meth. Enzymol. 1987, 148, 350–382. [Google Scholar]
- Holá, D.; Benešová, M.; Honnerová, J.; Hnilička, F.; Rothová, O.; Kočová, M.; Hniličková, H. The evaluation of photosynthetic parameters in maize inbred lines subjected to water deficiency: Can these parameters be used for the prediction of performance of hybrid progeny? Photosynthetica 2010, 48, 545–558. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Dey, P.M. Oligosaccharides. In Methods in Plant Biochemistry, Carbohydrates; Dey, P.M., Ed.; Academic Press: London, UK, 1990; Volume 2, pp. 189–218. [Google Scholar]
- Bates, L.; Waldren, P.P.; Teare, J.D. Rapid determination of free proline of water stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Zieslin, N.; Ben-Zaken, R. Peroxidase activity and presence of phenolic substances in peduncles of rose flowers. Plant Physiol. Biochem. 1993, 31, 333–339. [Google Scholar]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Kono, Y. Generation of superoxide radical during autoxidation of hydroxylamine and an assay for superoxide dismutase. Arch. Biochem. Biophys. 1978, 186, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Aebi, H. Catalase in vitro. Meth. Enzymol. 1984, 105, 121–126. [Google Scholar] [CrossRef]
- Putter, J.; Becker, R. Methods of Enzymatic Analysis; Academic Press: New York, NY, USA, 1974; p. 685. [Google Scholar]
- Vighi, I.L.; Benitez, L.C.; Amaral, M.N.; Moraes, G.P.; Auler, P.A.; Rodrigues, G.S.; Deuner, S.; Maia, L.C.; Braga, E.J.B. Functional characterization of the antioxidant enzymes in rice plants exposed to salinity stress. Biol. Plant. 2017, 61, 1–11. [Google Scholar] [CrossRef]
- Hwang, O.J.; Back, K. Suppression of Rice Cryptochrome 1b Decreases Both Melatonin and Expression of Brassinosteroid Biosynthetic Genes Resulting in Salt Tolerance. Molecules 2021, 26, 1075. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).