Evaluation of Inbred Maize (Zea mays L.) for Tolerance to Low Phosphorus at the Seedling Stage
Abstract
1. Introduction
2. Results
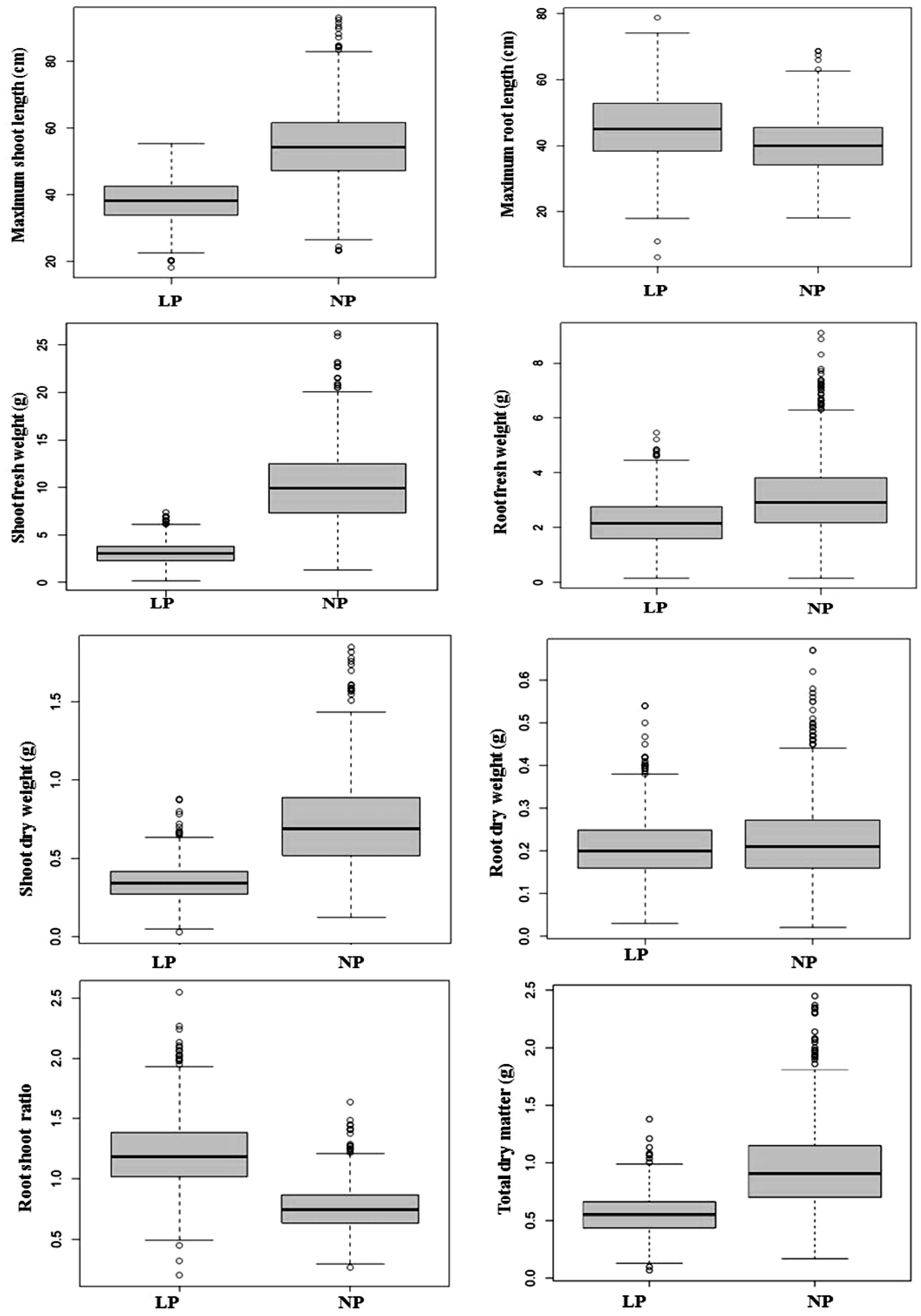
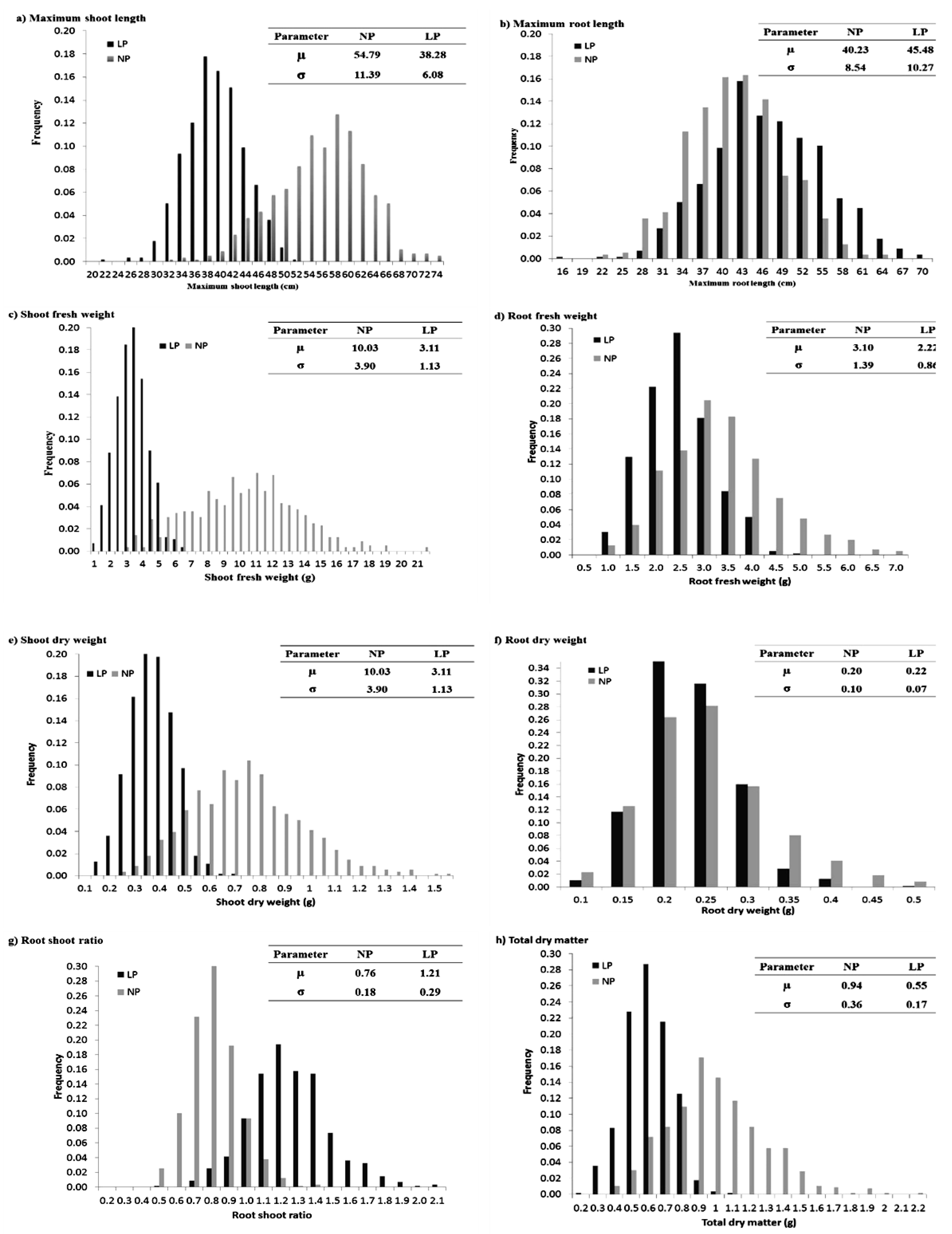
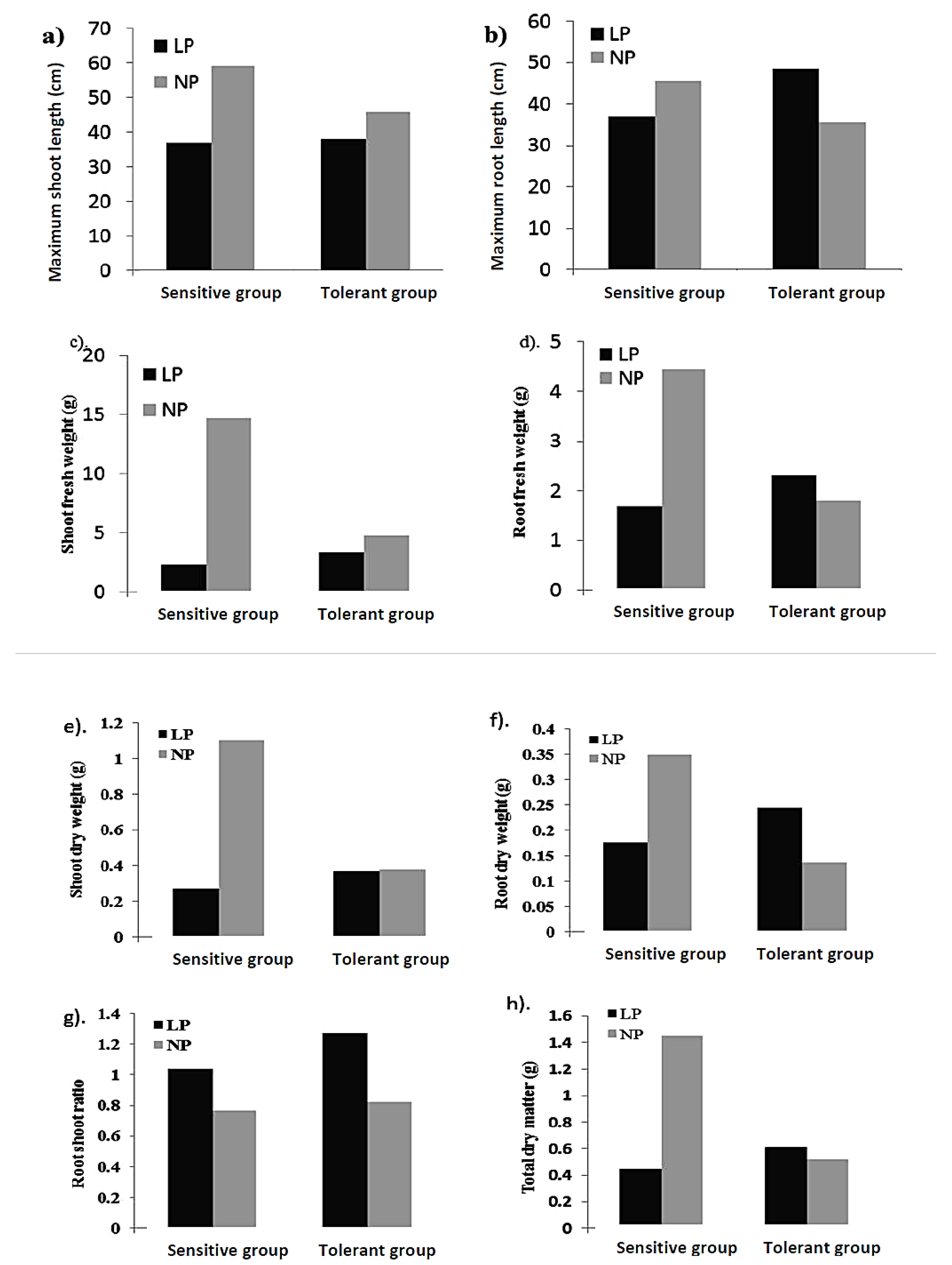
2.1. Analysis of Variance (ANOVA) and Frequency Distribution of Traits
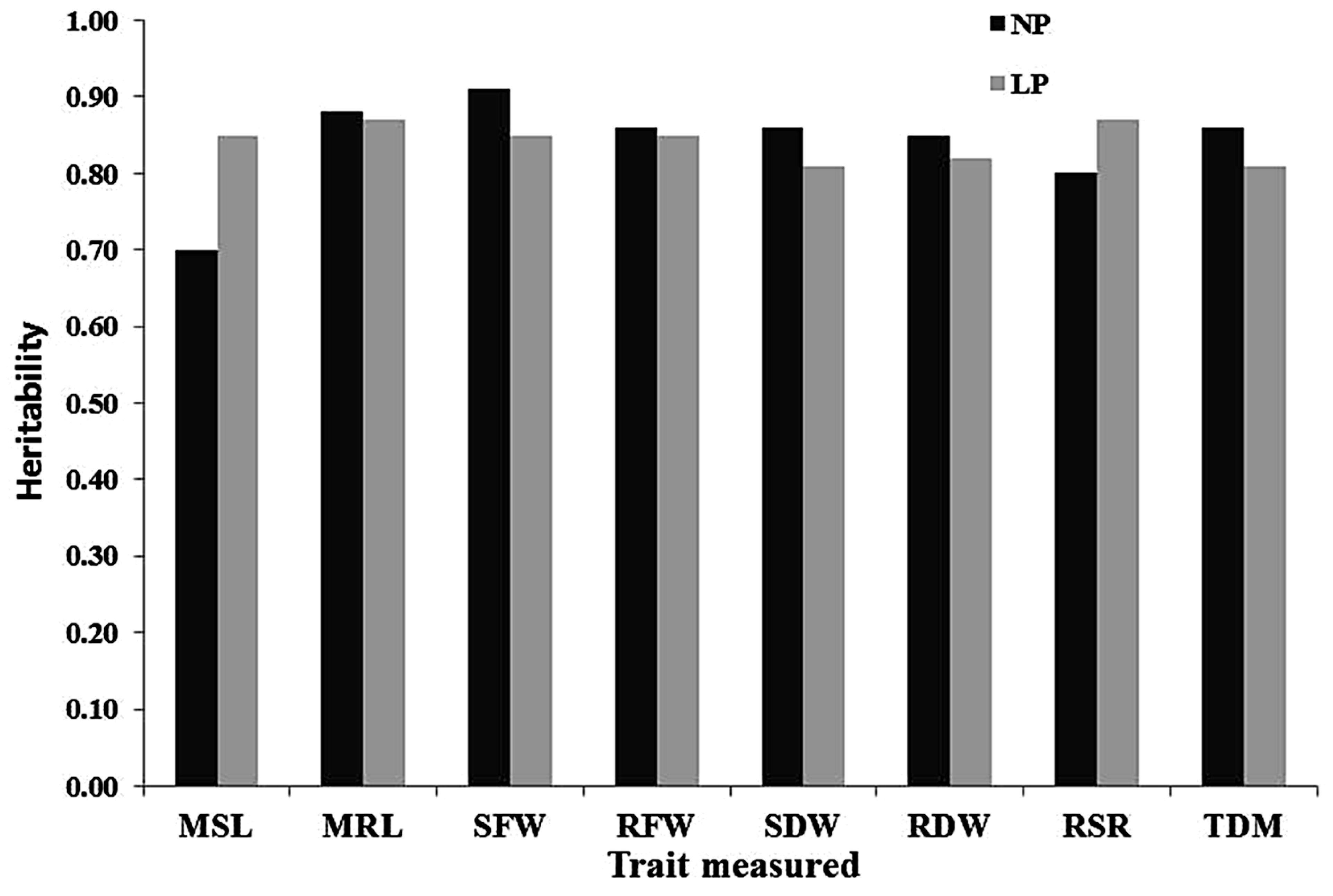
2.2. Estimated Heritabilities
2.3. Phenotypic Correlation Coefficients under Low-Phosphorus (LP) Conditions
2.4. Phenotypic Correlation Coefficients under NP Conditions
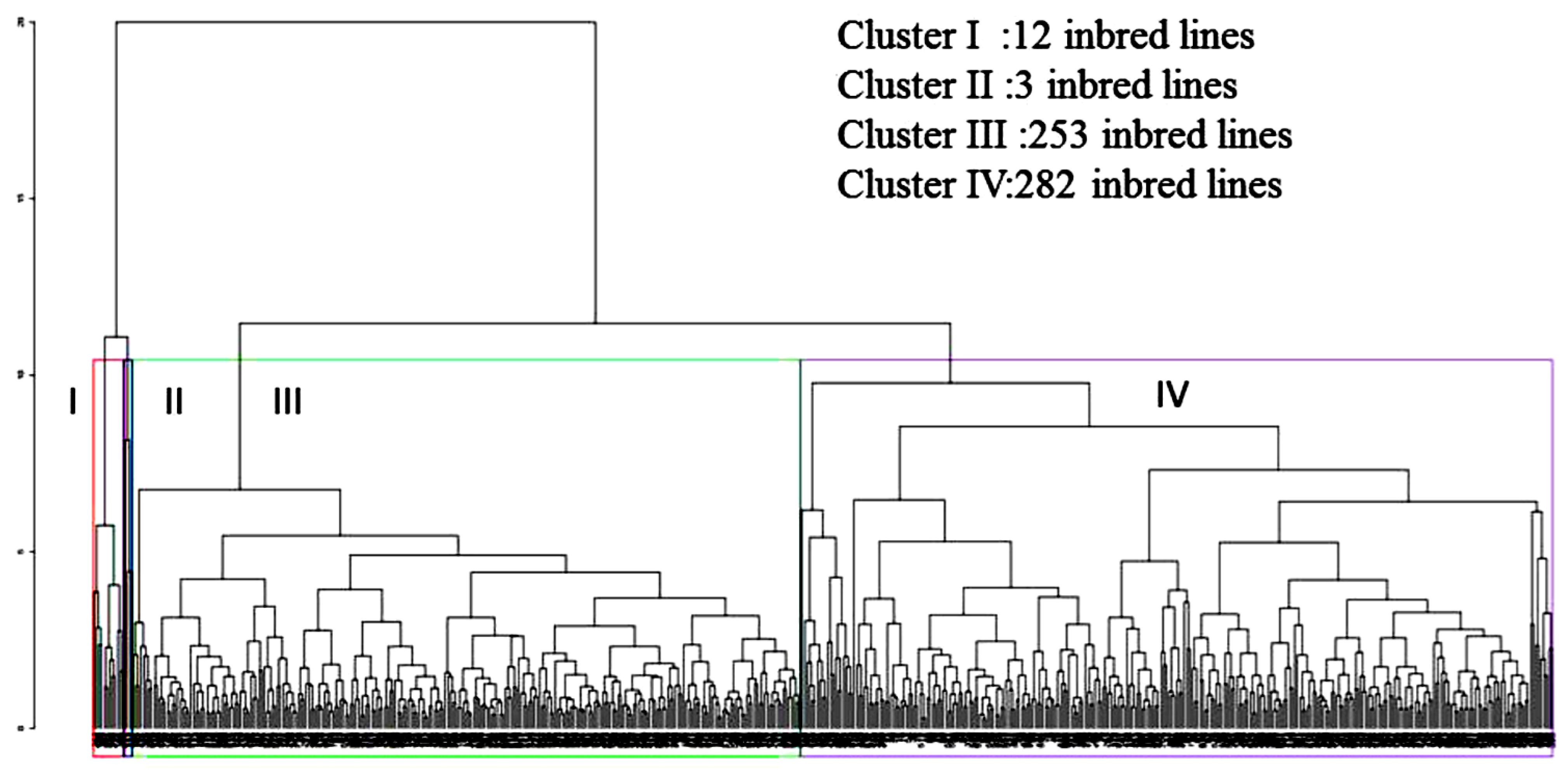
2.5. Multivariate Analysis
2.6. Selection of Genotypes with a Multi-Trait Index
2.7. LP Tolerance and P Responsiveness
3. Discussion
4. Materials and Methods
4.1. Experimental Site
4.2. Plant Materials
4.3. Plant Growth Conditions
4.4. Statistical Analysis
4.4.1. Multivariate Analysis
4.4.2. Calculation of the Multi-Trait Selection Index
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muitire, C.; Kamutando, C.; Moyo, M. Building Stress Resilience of Cereals under Future Climatic Scenarios: ‘The Case of Maize, Wheat, Rice and Sorghum’. In Cereal Grains; IntechOpen: London, UK, 2021. [Google Scholar]
- Flint-Garcia, S.A. Kernel Evolution: From Teosinte to Maize. In The Book Maize Kernel Development; University of Nebraska-Lincoln: Lincoln, NE, USA, 2017; pp. 1–16. [Google Scholar]
- Palacios-Rojas, N.; McCulley, L.; Kaeppler, M.; Titcomb, T.J.; Gunaratna, N.S.; Lopez-Ridaura, S.; Tanumihardjo, S.A. Mining maize diversity and improving its nutritional aspects within agro-food systems. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1809–1834. [Google Scholar] [CrossRef] [PubMed]
- Scheben, A.; Yuan, Y.; Edwards, D. Advances in genomics for adapting crops to climate change. Curr. Plant Biol. 2016, 6, 2–10. [Google Scholar] [CrossRef]
- Lynch, J.P.; Brown, K.M. Root strategies for phosphorus acquisition. In The Ecophysiology of Plant-Phosphorus Interactions; Springer: Berlin/Heidelberg, Germany, 2008; pp. 83–116. [Google Scholar]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef] [PubMed]
- Childers, D.; Corman, J.; Edwards, M.; Elser, J. Sustainability Challenges of Phosphorus and Food: Solutions from Closing the Human Phosphorus Cycle. Bioscience 2011, 61, 117–124. [Google Scholar] [CrossRef]
- Tirado, R.; Allsopp, M. Phosphorus in agriculture: Problems and solutions. In Greenpeace Research Laboratories Technical Report (Review); Greenpeace International Ottho Heldringstraat: Amsterdam, The Netherlands, 2012; p. 2. [Google Scholar]
- Gamuyao, R.; Chin, J.H.; Pariasca-Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Slamet-Loedin, I.; Tecson-Mendoza, E.M.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef]
- Elser, J.J. Phosphorus: A limiting nutrient for humanity? Curr. Opin. Biotechnol. 2012, 23, 833–838. [Google Scholar] [CrossRef]
- Song, C.; Wang, W.; Gan, Y.; Wang, L.; Chang, X.; Wang, Y.; Yang, W. Growth promotion ability of phosphate-solubilizing bacteria from the soybean rhizosphere under maize–soybean intercropping systems. J. Sci. Food Agric. 2021, 102, 1430–1442. [Google Scholar] [CrossRef]
- Jia, X.; Liu, P.; Lynch, J.P. Greater lateral root branching density in maize improves phosphorus acquisition from low phosphorus soil. J. Exp. Bot. 2018, 69, 4961–4970. [Google Scholar] [CrossRef]
- Zhang, L.; Yan, C.; Guo, Q.; Zhang, J.; Ruiz-Menjivar, J. The impact of agricultural chemical inputs on environment: Global evidence from informetrics analysis and visualization. Int. J. Low-Carbon Technol. 2018, 13, 338–352. [Google Scholar] [CrossRef]
- Galindo-Castañeda, T.; Brown, K.M.; Lynch, J.P. Reduced root cortical burden improves growth and grain yield under low phosphorus availability in maize. Plant Cell Environ. 2018, 41, 1579–1592. [Google Scholar] [CrossRef]
- Li, D.; Chen, Z.; Wang, M.; Leiser, W.L.; Weiß, T.M.; Zhao, Z.; Cheng, S.; Chen, S.; Chen, F.; Yuan, L.; et al. Dissecting the phenotypic response of maize to low phosphorus soils by field screening of a large diversity panel. Euphytica 2021, 217, 1–12. [Google Scholar] [CrossRef]
- Wissuwa, M.; Gamat, G.; Ismail, A.M. Is root growth under phosphorus deficiency affected by source or sink limitations? J. Exp. Bot. 2005, 56, 1943–1950. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.M.; Klein, S.P.; Hanlon, M.T.; Nord, E.A.; Kaeppler, S.; Brown, K.M.; Warry, A.; Bhosale, R.; Lynch, J.P. Genetic control of root architectural plasticity in maize. J. Exp. Bot. 2020, 71, 3185–3197. [Google Scholar] [CrossRef]
- Tucker, S.L.; Dohleman, F.G.; Grapov, D.; Flagel, L.; Yang, S.; Wegener, K.M.; Kosola, K.; Swarup, S.; Rapp, R.A.; Bedair, M.; et al. Evaluating maize phenotypic variance, heritability, and yield relationships at multiple biological scales across agronomically relevant environments. Plant Cell Environ. 2019, 43, 880–902. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.G.; Sarker, U.; Uddin, M.S. Screening maize (Zea mays L.) genotypes for phosphorus deficiency at the seedling stage. Turk. J. Agric. For. 2022, 46, 802–821. [Google Scholar] [CrossRef]
- Li, D.; Wang, H.; Wang, M.; Li, G.; Chen, Z.; Leiser, W.L.; Weiß, T.M.; Lu, X.; Wang, M.; Chen, S.; et al. Genetic Dissection of Phosphorus Use Efficiency in a Maize Association Population under Two P Levels in the Field. Int. J. Mol. Sci. 2021, 22, 9311. [Google Scholar] [CrossRef]
- Mäkelä, P.S.A.; Wasonga, D.O.; Hernandez, A.S.; Santanen, A. Seedling Growth and Phosphorus Uptake in Response to Different Phosphorus Sources. Agronomy 2020, 10, 1089. [Google Scholar] [CrossRef]
- Reddy, V.R.P.; Aski, M.S.; Mishra, G.P.; Dikshit, H.K.; Singh, A.; Pandey, R.; Singh, M.P.; Ramtekey, V.; Rai, N.; Nair, R.M. Genetic variation for root architectural traits in response to phosphorus deficiency in mungbean at the seedling stage. PLoS ONE 2020, 15, e0221008. [Google Scholar] [CrossRef]
- Schillaci, M.; Arsova, B.; Walker, R.; Smith, P.M.C.; Nagel, K.A.; Roessner, U.; Watt, M. Time-resolution of the shoot and root growth of the model cereal Brachypodium in response to inoculation with Azospirillum bacteria at low phosphorus and temperature. Plant Growth Regul. 2020, 93, 149–162. [Google Scholar] [CrossRef]
- Zhang, L.-T.; Li, J.; Rong, T.-Z.; Gao, S.-B.; Wu, F.-K.; Xu, J.; Li, M.-L.; Cao, M.-J.; Wang, J.; Hu, E.-L.; et al. Large-scale screening maize germplasm for low-phosphorus tolerance using multiple selection criteria. Euphytica 2014, 197, 435–446. [Google Scholar] [CrossRef]
- Linkohr, B.I.; Williamson, L.C.; Fitter, A.H.; Leyser, H.O. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002, 29, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Woll, K.; Borsuk, L.A.; Stransky, H.; Nettleton, D.; Schnable, P.S.; Hochholdinger, F. Isolation, Characterization, and Pericycle-Specific Transcriptome Analyses of the Novel Maize Lateral and Seminal Root Initiation Mutant rum1. Plant Physiol. 2005, 139, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Arrones, A.; Vilanova, S.; Plazas, M.; Mangino, G.; Pascual, L.; Díez, M.J.; Prohens, J.; Gramazio, P. The Dawn of the Age of Multi-Parent MAGIC Populations in Plant Breeding: Novel Powerful Next-Generation Resources for Genetic Analysis and Selection of Recombinant Elite Material. Biology 2020, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Benavente, E.; Giménez, E. Modern Approaches for the Genetic Improvement of Rice, Wheat and Maize for Abiotic Constraints-Related Traits: A Comparative Overview. Agronomy 2021, 11, 376. [Google Scholar] [CrossRef]
- Cockerton, H.M.; Li, B.; Stavridou, E.; Johnson, A.; Karlström, A.; Armitage, A.D.; Martinez-Crucis, A.; Galiano-Arjona, L.; Harrison, N.; Barber-Pérez, N. Genetic and phenotypic associations between root architecture, arbuscular mycorrhizal fungi colonisation and low phosphate tolerance in strawberry (Fragaria × ananassa). BMC Plant Biol. 2020, 20, 154 . [Google Scholar] [CrossRef]
- Singhal, T.; Satyavathi, C.T.; Singh, S.P.; Kumar, A.; Sankar, S.M.; Bhardwaj, C.; Mallik, M.; Bhat, J.; Anuradha, N.; Singh, N. Multi-Environment Quantitative Trait Loci Mapping for Grain Iron and Zinc Content Using Bi-parental Recombinant Inbred Line Mapping Population in Pearl Millet. Front. Plant Sci. 2021, 12, 659789. [Google Scholar] [CrossRef]
- Scheiner, S.M.; Levis, N.A. The loss of phenotypic plasticity via natural selection: Genetic assimilation. In Phenotypic Plasticity & Evolution; CRC Press: Boca Raton, FL, USA, 2021; pp. 161–181. [Google Scholar]
- Tuberosa, R.; Salvi, S.; Giuliani, S.; Sanguineti, M.; Frascaroli, E.; Conti, S.; Landi, P. Genomics of root architecture and functions in maize. In Root Genomics; Costa de Oliveira, A., Varshney, R.K., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 179–204. [Google Scholar] [CrossRef]
- Tuberosa, R.; Salvi, S.; Sanguineti, M.C.; Maccaferri, M.; Giuliani, S.; Landi, P. Searching for quantitative trait loci controlling root traits in maize: A critical appraisal. Plant Soil 2003, 255, 35–54. [Google Scholar] [CrossRef]
- Oliveira, G.H.; Amaral, C.B.; Silva, F.A.; Dutra, S.M.; Marconato, M.B.; Môro, G.V. Mixed models and multivariate analysis for selection of superior maize genotypes. Chil. J. Agric. Res. 2016, 76, 427–431. [Google Scholar] [CrossRef]
- Dos Santos, W.F.; Maciel, L.C.; Júnior, B.L.D.; Peluzio, J.M.; Sodré, L.F.; Júnior, O.J.F.; De Oliveira, M.; Da Silva, R.M.; Barbosa, A.S. Genetic Divergence in Corn Genotypes for High and Low Phosphorus in Pará, Brazil. Annu. Res. Rev. Biol. 2020, 35, 82–90. [Google Scholar] [CrossRef]
- Ganie, A.H.; Ahmad, A.; Pandey, R.; Aref, I.M.; Yousuf, P.Y.; Ahmad, S.; Iqbal, M. Metabolite Profiling of Low-P Tolerant and Low-P Sensitive Maize Genotypes under Phosphorus Starvation and Restoration Conditions. PLoS ONE 2015, 10, e0129520. [Google Scholar] [CrossRef]
- Torres, L.G.; Caixeta, D.G.; Rezende, W.M.; Schuster, A.; Azevedo, C.F.; e Silva, F.F.; DeLima, R.O. Genotypic variation and relationships among traits for root morphology in a panel of tropical maize inbred lines under contrasting nitrogen levels. Euphytica 2019, 215, 51. [Google Scholar] [CrossRef]
- Uddin, S.; Azam, G.; Billah, M.; Bagum, S.A.; Biswas, P.L.; Khaldun, A.B.M.; Hossain, N.; Gaber, A.; Althobaiti, Y.S.; Abdelhadi, A.A.; et al. High-Throughput Root Network System Analysis for Low Phosphorus Tolerance in Maize at Seedling Stage. Agronomy 2021, 11, 2230. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Jiang, Z. Using the linear mixed-effect model framework to estimate generalizability variance components in R: A lme4 package application. Methodology. Eur. J. Res. Methods Behav. Soc. Sci. 2018, 14, 133–142. [Google Scholar] [CrossRef]
- STAR (Statistical Tool for Agricultural Research); International Rice Research Institute (IRRI): Manila, Philippines, 2014.
- SAS: Statistical Analysis Software. Users’ Guide Statistics Version 9.4; SAS Institute Inc., SAS Campus Drive: Cary, NC, USA, 2013. [Google Scholar]
- Bayuelo-Jiménez, J.S.; Gallardo-Valdéz, M.; Pérez-Decelis, V.A.; Magdaleno-Armas, L.; Ochoa, I.; Lynch, J.P. Genotypic variation for root traits of maize (Zea mays L.) from the Purhepecha Plateau under contrasting phosphorus availability. Field Crop. Res. 2011, 121, 350–362. [Google Scholar] [CrossRef]


| Source of Variation | Mean Square | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Df | MSL | MRL | SFWP | RFW | SDW | RDW | RSR | TDM | |
| Replication | 1 | 3185.98 | 18.17 | 4.57 | 0.79 | 0.002 | 0.001 | 0.52 | 0.002 |
| Treatment | 1 | 151,909.92 | 15,335.49 | 26,685.54 | 430.54 | 78.381 | 0.148 | 113.40 | 85.344 |
| Genotype | 549 | 176.87 | 222.85 | 18.14 | 3.13 | 0.094 | 0.015 | 0.13 | 0.168 |
| Genotype × Treatment | 549 | 77.25 | 94.15 | 11.87 | 1.52 | 0.064 | 0.009 | 0.07 | 0.107 |
| Error | 1098 | 33.67 | 20.14 | 1.46 | 0.34 | 0.011 | 0.002 | 0.01 | 0.020 |
| Significance level | *** | *** | *** | *** | *** | *** | *** | *** | |
| MSL | MRL | SFW | RFW | SDW | RDW | RSR | TDM | |
|---|---|---|---|---|---|---|---|---|
| MSL | 0.26 * | 0.74 ** | 0.49 ** | 0.65 ** | 0.51 ** | −0.42 ** | 0.64 ** | |
| MRL | 0.27 * | 0.41 ** | 0.59 ** | 0.43 ** | 0.44 ** | 0.76 ** | 0.47 ** | |
| SFW | 0.71 ** | 0.40 ** | 0.73 ** | 0.87 ** | 0.66 ** | −0.11 | 0.85 ** | |
| RFW | 0.71 ** | 0.57 ** | 0.74 ** | 0.76 ** | 0.73 ** | 0.23 | 0.81 ** | |
| SDW | 0.76 ** | 0.41 ** | 0.85 ** | 0.74 ** | 0.71 ** | −0.02 | 0.96 ** | |
| RDW | 0.73 ** | 0.43 ** | 0.66 ** | 0.73 ** | 0.71 ** | 0.07 | 0.88 ** | |
| RSR | −0.41 ** | 0.75 ** | −0.11 | 0.20 | −0.04 | 0.06 | 0.01 | |
| TDM | 0.64 ** | 0.45 ** | 0.84 ** | 0.79 ** | 0.96 ** | 0.89 ** | 0.01 |
| MSL | MRL | SFW | RFW | SDW | RDW | RSR | TDM | |
|---|---|---|---|---|---|---|---|---|
| MSL | 0.41 ** | 0.81 ** | 0.63 ** | 0.78 ** | 0.57 ** | −0.46 ** | 0.76 ** | |
| MRL | 0.36 * | 0.47 ** | 0.41 ** | 0.39 * | 0.33 * | 0.60 ** | 0.39 * | |
| SFW | 0.70 ** | 0.45 ** | 0.76 ** | 0.87 ** | 0.74 ** | −0.23 | 0.88 ** | |
| RFW | 0.55 ** | 0.41 ** | 0.75 ** | 0.73 ** | 0.81 ** | −0.13 | 0.78 ** | |
| SDW | 0.67 ** | 0.38 * | 0.87 ** | 0.72 ** | 0.79 ** | −0.28 * | 0.99 ** | |
| RDW | 0.49 ** | 0.32 * | 0.74 ** | 0.80 ** | 0.79 ** | −0.17 | 0.88 ** | |
| RSR | −0.51 ** | 0.58 ** | −0.21 | −0.11 | −0.25 | −0.15 | −0.27 * | |
| TDM | 0.65 ** | 0.38 * | 0.87 ** | 0.77 ** | 0.99 ** | 0.88 ** | −0.24 * |
| Parameter | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| Maximum shoot length (cm) | 0.27 | −0.40 | −0.72 | 0.12 |
| Maximum root length (cm) | 0.26 | 0.51 | −0.53 | 0.15 |
| Shoot fresh weight (g) | 0.42 | −0.09 | 0.00 | −0.25 |
| Root fresh weight (g) | 0.39 | 0.06 | 0.05 | −0.82 |
| Shoot dry weight (g) | 0.42 | −0.04 | 0.20 | 0.26 |
| Root dry weight (g) | 0.39 | −0.02 | 0.33 | 0.28 |
| Root shoot ratio | 0.04 | 0.75 | 0.01 | 0.02 |
| Total dry matter (g) | 0.44 | −0.03 | 0.24 | 0.29 |
| Cumulative% of the total variance | 59.35 | 81.27 | 89.78 | 94.23 |
| Tolerance Group | C5 | C6 | Tropical | Temperate | Total | |
|---|---|---|---|---|---|---|
| LPTI all | Sensitive | 0 | 29 | 1 | 0 | 30 |
| Tolerant | 9 | 8 | 12 | 1 | 30 | |
| TR | 1 | 2 | 1 | 1 | 5 | |
| TNR | 1 | 1 | 2 | 0 | 4 | |
| SNR | 0 | 6 | 0 | 0 | 6 | |
| SR | 0 | 3 | 0 | 0 | 3 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, M.S.; Akter, F.; Azam, M.G.; Bagum, S.A.; Hossain, N.; Billah, M.; Biswas, P.L.; Hasibuzzaman, A.S.M.; Khaldun, A.B.M.; Alsuhaibani, A.M.; et al. Evaluation of Inbred Maize (Zea mays L.) for Tolerance to Low Phosphorus at the Seedling Stage. Plants 2023, 12, 2520. https://doi.org/10.3390/plants12132520
Uddin MS, Akter F, Azam MG, Bagum SA, Hossain N, Billah M, Biswas PL, Hasibuzzaman ASM, Khaldun ABM, Alsuhaibani AM, et al. Evaluation of Inbred Maize (Zea mays L.) for Tolerance to Low Phosphorus at the Seedling Stage. Plants. 2023; 12(13):2520. https://doi.org/10.3390/plants12132520
Chicago/Turabian StyleUddin, Md. Shalim, Farzana Akter, Md. Golam Azam, Shamim Ara Bagum, Neelima Hossain, Masum Billah, Priya Lal Biswas, Abu Sayeed Md. Hasibuzzaman, Abul Bashar Mohammad Khaldun, Amnah Mohammed Alsuhaibani, and et al. 2023. "Evaluation of Inbred Maize (Zea mays L.) for Tolerance to Low Phosphorus at the Seedling Stage" Plants 12, no. 13: 2520. https://doi.org/10.3390/plants12132520
APA StyleUddin, M. S., Akter, F., Azam, M. G., Bagum, S. A., Hossain, N., Billah, M., Biswas, P. L., Hasibuzzaman, A. S. M., Khaldun, A. B. M., Alsuhaibani, A. M., Gaber, A., & Hossain, A. (2023). Evaluation of Inbred Maize (Zea mays L.) for Tolerance to Low Phosphorus at the Seedling Stage. Plants, 12(13), 2520. https://doi.org/10.3390/plants12132520









