Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan
Abstract
:1. Introduction
2. Results
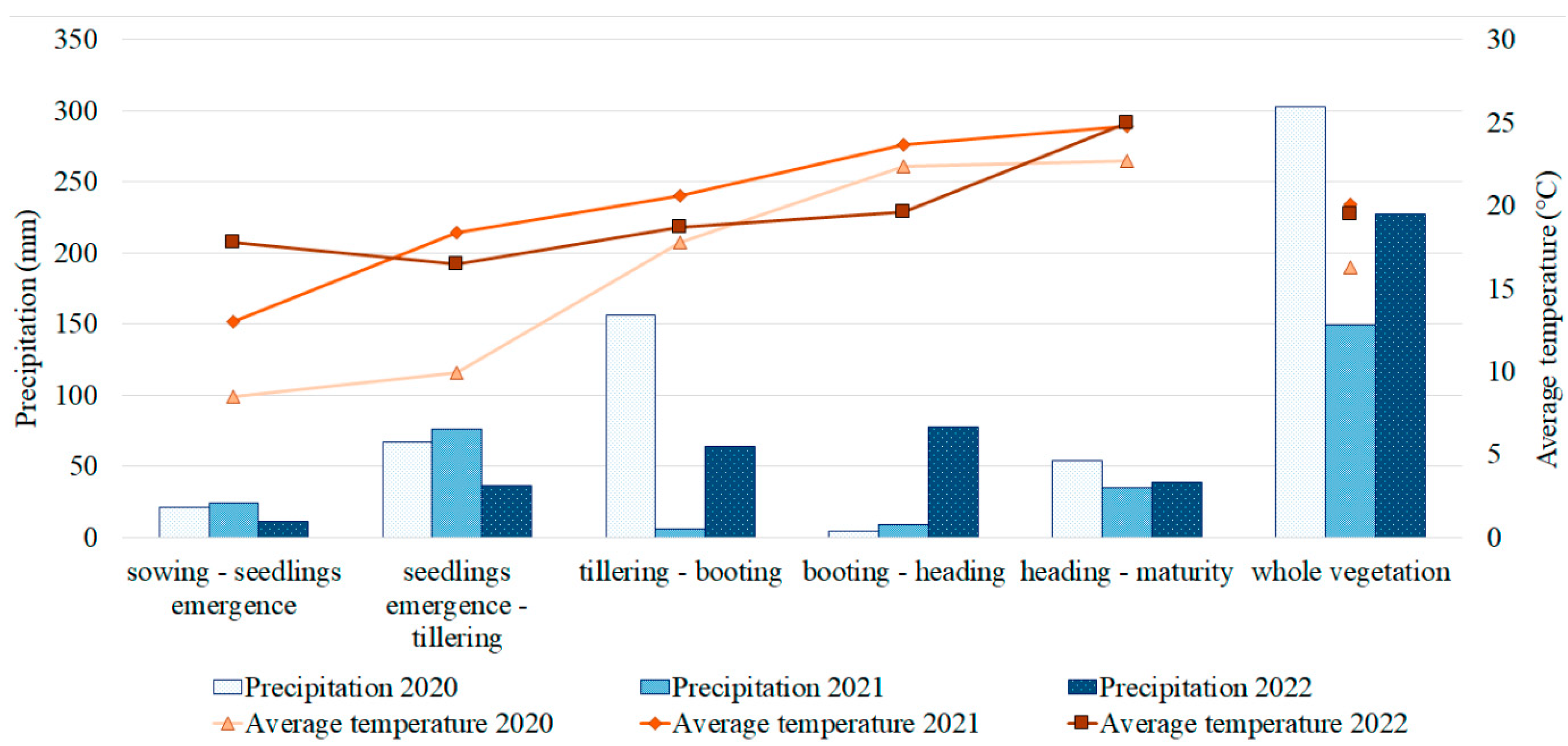
2.1. PM Resistance and Its Correlation with Agronomic Traits in the Studied Barley Collection
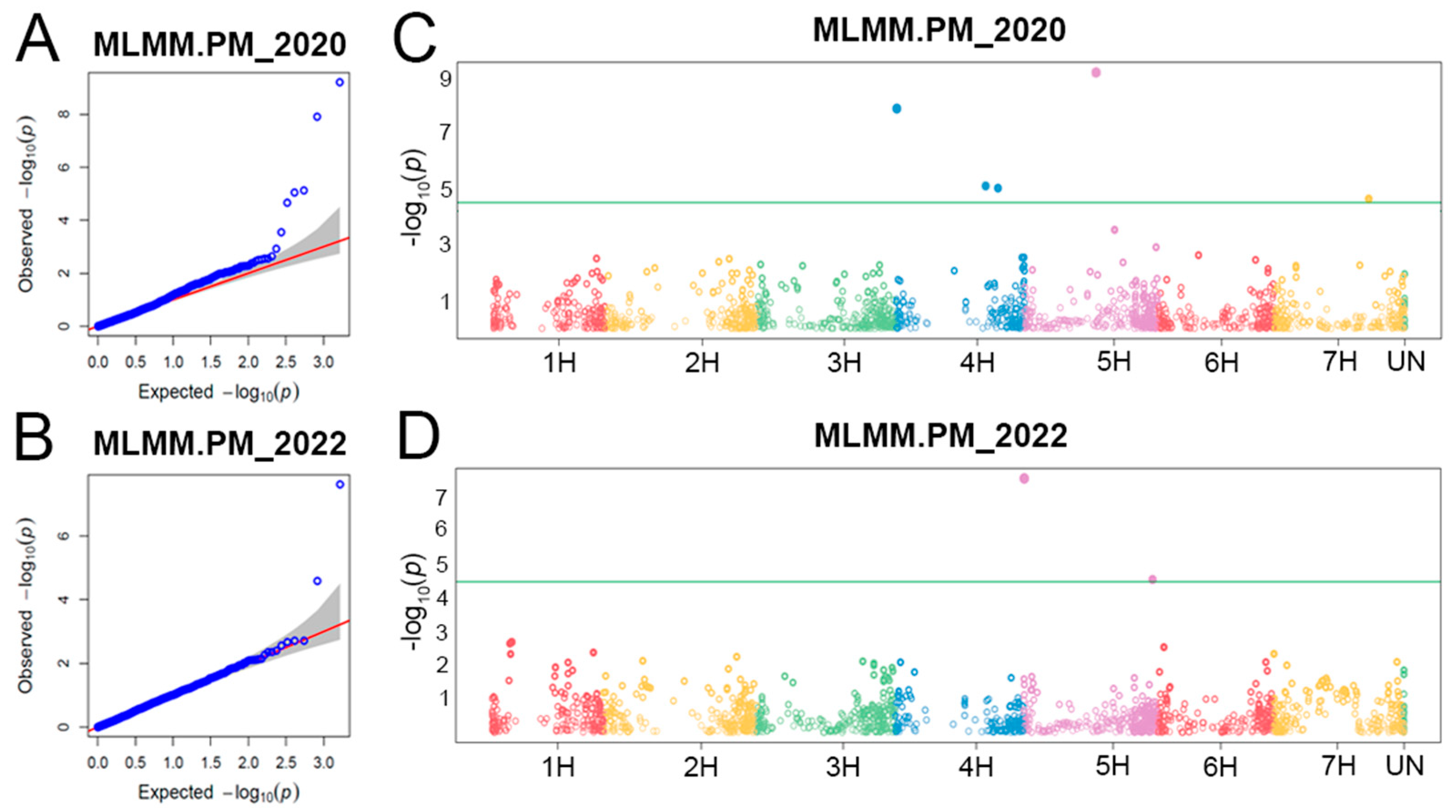
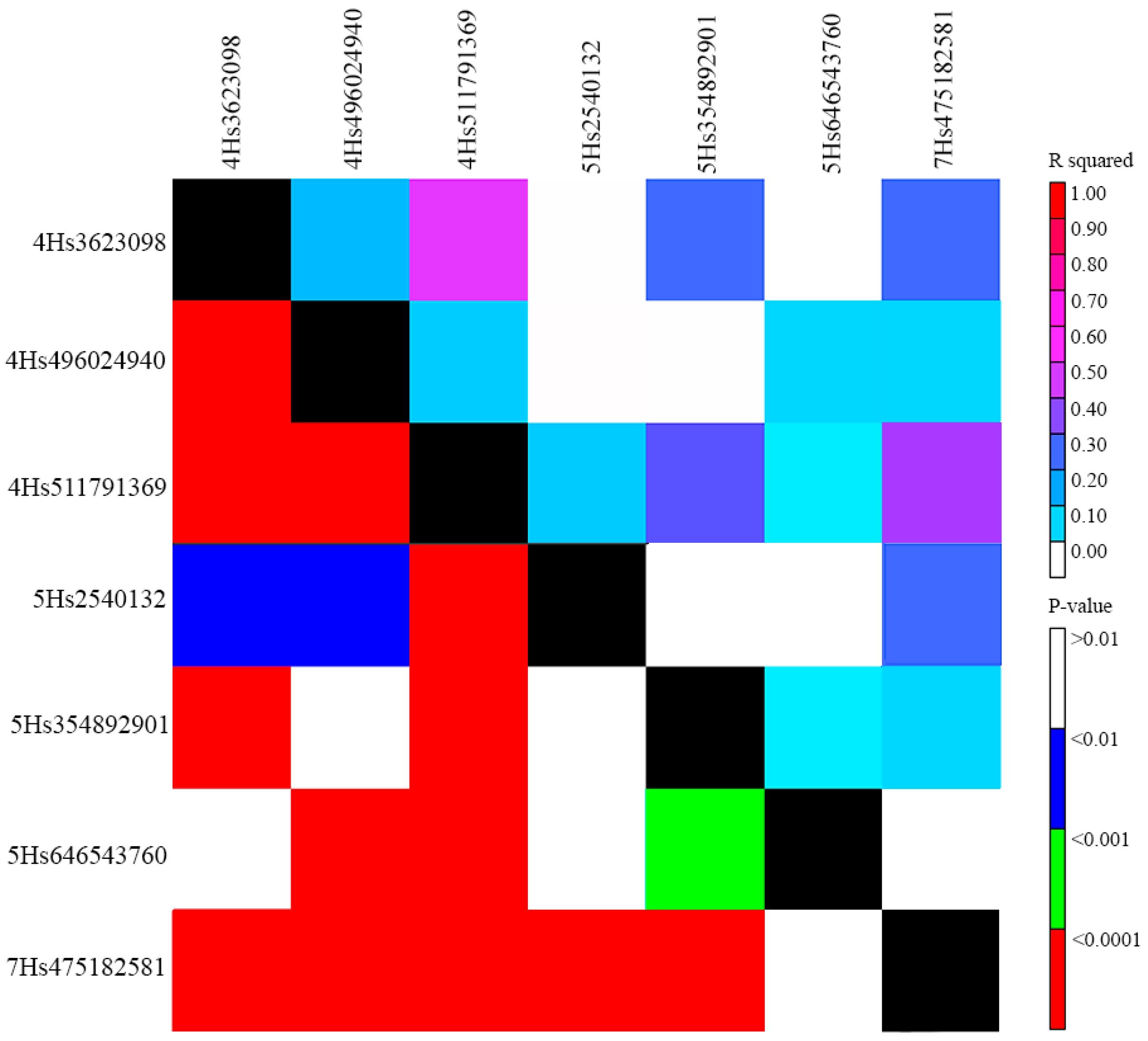
2.2. Populaton Structure, QTLs for Powdery Mildew PM Resistance, and Linkage Disequilibrium (LD) among Them
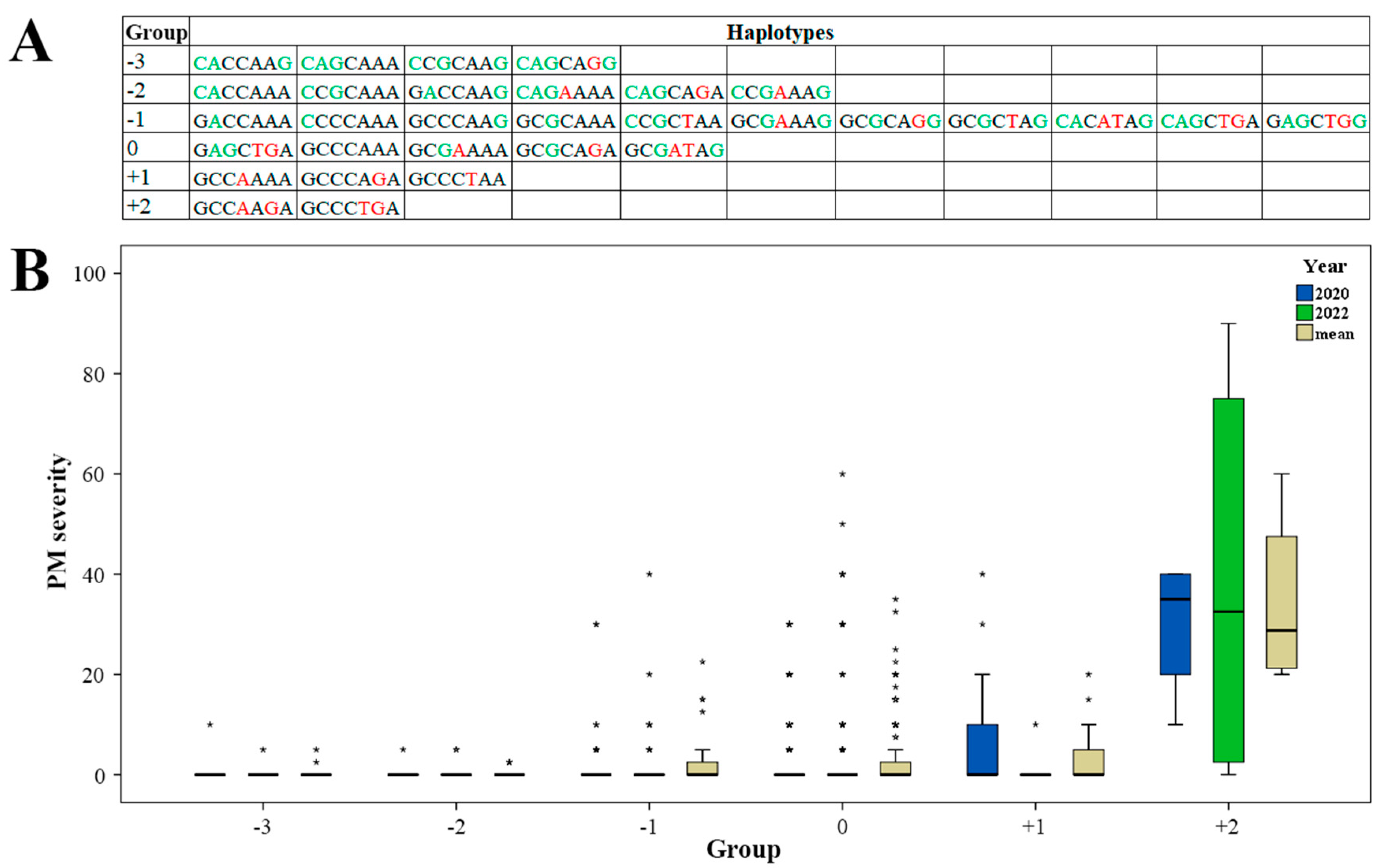
2.3. Effect of Haplotypes
3. Discussion
3.1. PM Resistance in the Field and Its Relations with Yield Components
3.2. QTLs for PM Resistance
3.3. Promising Haplotypes for PM Resistance
4. Materials and Methods
4.1. Barley Germplasm Panel and SNP Genotyping
4.2. Field Trials and PM Resistance Evaluation
4.3. Genome-Wide Association Study (GWAS), Linkage Disequilibrium (LD), Population Structure, and Haplotype Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Statista Database. Available online: https://www.statista.com/ (accessed on 20 January 2023).
- Agency for Strategic Planning and Reforms of the Republic of Kazakhstan, Bureau of National Statistics. Available online: https://stat.gov.kz/ (accessed on 20 January 2023).
- U.S. Department of Agriculture. Foreign Agricultural Service. Available online: https://www.fas.usda.gov/ (accessed on 21 January 2023).
- Glawe, D.A. The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef]
- Chaure, P.; Gurr, S.J.; Spanu, P. Stable transformation of Erysiphe graminis an obligate biotrophic pathogen of barley. Nat. Biotechnol. 2000, 18, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Czembor, J.H. Resistance to powdery mildew in selections from Moroccan barley landraces. Euphytica 2002, 125, 397–409. [Google Scholar] [CrossRef]
- Jørgensen, J.H.; Wolfe, M. Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 1994, 13, 97–119. [Google Scholar] [CrossRef]
- Koishibayev, M. Diseases of Cereals. Kazakhstan, 1st ed.; Publishing House “Bastau”: Almaty, Kazakhstan, 2002. [Google Scholar]
- Rsaliyev, A.S.; Chudinov, V.A.; Amirkhanova, N.T. Resistance of selection materials of barley at Karabalyk agricultural experimental station to net blotch and powdery mildew. Rep. Natl. Acad. Sci. RK 2016, 4, 79–87. [Google Scholar]
- Patel, N.; Desai, P.; Patel, N.; Jha, A.; Gautam, H.K. Agronanotechnology for plant fungal disease management: A review. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 71–84. [Google Scholar]
- Burdon, J.J.; Barrett, L.G.; Rebetzke, G.; Thrall, P.H. Guiding deployment of resistance in cereals using evolutionary principles. Evol. Appl. 2014, 7, 609–624. [Google Scholar] [CrossRef]
- Jørgensen, I.H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Kusch, S.; Panstruga, R. mlo-based resistance: An apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant-Microbe Interact. 2017, 30, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Lyngkjær, M.; Newton, A.; Atzema, J.; Baker, S. The barley mlo-gene: An important powdery mildew resistance source. Agronomie 2000, 20, 745–756. [Google Scholar] [CrossRef] [Green Version]
- Reinstädler, A.; Müller, J.; Czembor, J.H.; Piffanelli, P.; Panstruga, R. Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant Biol. 2010, 10, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.L.; Jones, A.T.; Duncan, G.H.; Roberts, I.M.; Reddy, D.V.R. Characterisation of a novel mite-transmitted virus associated with pigeonpea sterility mosaic disease. Phytopathology 2001, 91, 51. [Google Scholar]
- McGrann, G.R.D.; Stavrinides, A.; Russell, J.; Corbitt, M.M.; Booth, A.; Chartrain, L.; Thomas, W.T.B.; Brown, J.K.M. A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. J. Exp. Bot. 2014, 65, 1025–1037. [Google Scholar] [CrossRef] [Green Version]
- Leng, Y.; Zhao, M.; Wang, R.; Steffenson, B.J.; Brueggeman, R.S.; Zhong, S. The gene conferring susceptibility to spot blotch caused by Cochliobolus sativus is located at the Mla locus in barley cultivar Bowman. Theor. Appl. Genet. 2018, 131, 1531–1539. [Google Scholar] [CrossRef]
- Halterman, D.A.; Wise, R.P. A single-amino acid substitution in the sixth leucine-rich repeat of barley MLA6 and MLA13 alleviates dependence on RAR1 for disease resistance signaling. Plant J. 2014, 38, 215–226. [Google Scholar] [CrossRef]
- Schüller, C.; Backes, G.; Fischbeck, G.; Jahoor, A. RFLP markers to identify the alleles on the Mla locus conferring powdery mildew resistance in barley. Theor. Appl. Genet. 1992, 84, 330–338. [Google Scholar] [CrossRef]
- Giese, H.; Holm-Jensen, A.G.; Jensen, H.P.; Jensen, J. Localization of the Laevigatum powdery mildew resistance gene to barley chromosome 2 by the use of RFLP markers. Theor. Appl. Genet. 1993, 85, 897–900. [Google Scholar] [CrossRef]
- Piechota, U.; Czembor, P.C.; Słowacki, P.; Czembor, J.H. Identifying a novel powdery mildew resistance gene in a barley landrace from Morocco. J. Appl. Genet. 2019, 60, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Görg, R.; Hollricher, K.; Schulze-Lefert, P. Functional analysis and RFLP-mediated mapping of the Mlg resistance locus in barley. Plant J. 1993, 3, 857–866. [Google Scholar] [CrossRef]
- Schönfeld, M.; Ragni, A.; Fischbeck, G.; Jahoor, A. RFLP mapping of three new loci for resistance genes to powdery mildew (Erysiphe graminis f. sp. hordei) in barley. Theor. Appl. Genet. 1996, 93, 48–56. [Google Scholar] [CrossRef]
- Dreiseitl, A. Specific resistance of barley to powdery mildew, its use and beyond: A concise critical review. Genes 2020, 11, 971. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.A.; Jayasena, K.; Ellwood, S.R.; Oliver, R.P. Pathotype variation of barley powdery mildew in Western Australia. Australas. Plant Pathol. 2013, 42, 617–623. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, G.; Wang, F.; Lang, X.; Zhao, X.; Zhu, J.; Hu, C.; Hu, J.; Zhang, Y.; Yao, X.; et al. Virulence Variability and Genetic Diversity in Blumeria graminis f. sp. hordei in Southeastern and Southwestern China. Plant Dis. 2023, 107, 809–819. [Google Scholar]
- Rsaliyev, A.; Pahratdinova, Z.; Rsaliyev, S. Characterizing the pathotype structure of barley powdery mildew and effectiveness of resistance genes to this pathogen in Kazakhstan. BMC Plant Biol. 2017, 17, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhunusova, A.; Sarbaev, A. Resistance of barley varieties to major fungal diseases. Biosaf. Biotechnol. 2022, 10, 31–43. (In Kazakh) [Google Scholar] [CrossRef]
- Hinze, K.; Thompson, R.D.; Ritter, E.; Salamini, F.P.; Schulze-Lefert, P. Restriction fragment length polymorphism-mediated targeting of the mlo resistance locus in barley (Hordeum vulgare). Proc. Natl. Acad. Sci. USA 1991, 88, 3691–3695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Řepková, J.; Dreiseitl, A.; Lízal, P. New CAPS marker for selection of a barley powdery mildew resistance gene in the Mla locus. Cereal Res. Commun. 2009, 37, 93–99. [Google Scholar] [CrossRef] [Green Version]
- Ge, C.; Wentzel, E.; D’Souza, N.; Chen, K.; Oliver, R.P.; Ellwood, S.R. Adult resistance genes to barley powdery mildew confer basal penetration resistance associated with broad-spectrum resistance. Plant Genome 2011, 14, e20129. [Google Scholar] [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.T.; Zhou, G.; Hill, C.B. Genome-wide association study of salinity tolerance during germination in barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 118. [Google Scholar] [CrossRef] [Green Version]
- Thabet, S.G.; Moursi, Y.S.; Karam, M.A.; Graner, A.; Alqudah, A.M. Genetic basis of drought tolerance during seed germination in barley. PLoS ONE 2018, 13, e0206682. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Hernandez, J.; Herb, D.; Baenziger, P.S.; Bochard, A.M.; Capettini, F.; Casas, A.; Cuesta-Marcos, A.; Einfeldt, C.; Fisk, S.; et al. Perspectives on low temperature tolerance and vernalization sensitivity in barley: Prospects for facultative growth. Front. Plant Sci. 2020, 11, 585927. [Google Scholar] [CrossRef] [PubMed]
- Dang, V.H.; Hill, C.B.; Zhang, X.-Q.; Angessa, T.T.; McFawn, L.-A.; Li, C. Multi-locus genome-wide association studies reveal novel alleles for flowering time under vernalisation and extended photoperiod in a barley MAGIC population. Theor. Appl. Genet. 2022, 135, 3087–3102. [Google Scholar] [CrossRef] [PubMed]
- Alqudah, A.M.; Sharma, R.; Pasam, R.K.; Graner, A.; Kilian, B.; Schnurbusch, T. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS ONE 2014, 9, e113120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, T.; Hill, C.B.; Angessa, T.T.; Zhang, X.-Q.; Chen, K.; Moody, D.; Telfer, P.; Westcott, S.; Li, C. Gene-set association and epistatic analyses reveal complex gene interaction networks affecting flowering time in a worldwide barley collection. J. Exp. Bot. 2019, 70, 5603–5616. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Sharma, R.; Tondelli, A.; Russell, J.; Comadran, J.; Schnaithmann, F.; Pillen, K.; Kilian, B.; Cattivelli, L.; Thomas, W.T.; et al. Genome-wide association analysis of grain yield-associated traits in a pan-European barley cultivar collection. Plant Genome 2018, 11, 170073. [Google Scholar] [CrossRef]
- Sharma, R.; Draicchio, F.; Bull, H.; Herzig, P.; Maurer, A.; Pillen, K.; Thomas, W.T.B.; Flavell, A.J. Genome-wide association of yield traits in a nested association mapping population of barley reveals new gene diversity for future breeding. J. Exp. Bot. 2018, 69, 3811–3822. [Google Scholar] [CrossRef]
- Shu, X.; Rasmussen, S.K. Quantification of amylose, amylopectin, and β-glucan in search for genes controlling the three major quality traits in barley by genome-wide association studies. Front. Plant Sci. 2014, 5, 197. [Google Scholar] [CrossRef] [Green Version]
- Matthies, I.E.; Malosetti, M.; Roeder, M.S.; van Eeuwijk, F. Genome-wide association mapping for kernel and malting quality traits using historical European barley records. PLoS ONE 2014, 9, e110046. [Google Scholar] [CrossRef] [Green Version]
- Genievskaya, Y.; Almerekova, S.; Abugalieva, S.; Chudinov, V.; Blake, T.; Abugalieva, A.; Turuspekov, Y. Identification of SNP markers associated with grain quality traits in a barley collection (Hordeum vulgare L.) harvested in Kazakhstan. Agronomy 2022, 12, 2431. [Google Scholar] [CrossRef]
- Novakazi, F.; Afanasenko, O.; Lashina, N.; Platz, G.J.; Snowdon, R.; Loskutov, I.; Ordon, F. Genome-wide association studies in barley (Hordeum vulgare) diversity set reveal a limited number of loci for resistance to spot blotch (Bipolaris sorokiniana). Plant Breed. 2020, 139, 521–535. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, S.; Larkin, P.; Xu, R.; Hayden, M.; Forrest, K.; Meinke, H.; Hu, H.; Zhou, M.; Fan, Y. Genome wide association study reveals novel QTL for barley yellow dwarf virus resistance in wheat. BMC Genom. 2019, 20, 891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gyawali, S.; Mamidi, S.; Chao, S.; Bhardwaj, S.C.; Shekhawat, P.S.; Selvakumar, R.; Gangwar, O.P.; Verma, R.P.S. Genome-wide association studies revealed novel stripe rust resistance QTL in barley at seedling and adult-plant stages. Euphytica 2021, 217, 3. [Google Scholar] [CrossRef]
- Turuspekov, Y.; Ormanbekova, D.; Rsaliev, A.; Abugalieva, S. Genome-wide association study on stem rust resistance in Kazakh spring barley lines. BMC Plant Biol. 2016, 16, 6. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Bai, G.; Lin, M.; Xu, X.; Zheng, W. Genome-wide association analysis of powdery mildew resistance in US winter wheat. Sci. Rep. 2017, 7, 11743. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Xu, W.; Peng, C.; Li, C.; Zhang, Y.; Hu, L. Identification and validation of a novel locus, Qpm-3BL, for adult plant resistance to powdery mildew in wheat using multilocus GWAS. BMC Plant Biol. 2021, 21, 357. [Google Scholar] [CrossRef]
- Admassu-Yimer, B.; Klos, K.E.; Griffiths, I.; Cowan, A.; Howarth, C. Mapping of crown rust (Puccinia coronata f. sp. avenae) resistance Gene Pc54 and a novel quantitative trait locus effective against powdery mildew (Blumeria graminis f. sp. avenae) in the Oat (Avena sativa) Line Pc54. Phytopathology 2022, 112, 1316–1322. [Google Scholar] [CrossRef]
- Vendelbo, N.M.; Mahmood, K.; Sarup, P.; Kristensen, P.S.; Orabi, J.; Jahoor, A. Discovery of a novel powdery mildew (Blumeria graminis) resistance locus in rye (Secale cereale L.). Sci. Rep. 2021, 11, 23057. [Google Scholar] [CrossRef]
- Novakazi, F.; Krusell, L.; Jensen, J.D.; Orabi, J.; Jahoor, A.; Bengtsson, T. You had me at “MAGIC”!: Four barley MAGIC populations reveal novel resistance QTL for powdery mildew. Genes 2020, 11, 1512. [Google Scholar] [CrossRef]
- Bengtsson, T.; Åhman, I.; Manninen, O.; Reitan, L.; Christerson, T.; Jensen, J.D.; Krusell, L.; Jahoor, A.; Orabi, J. A novel QTL for powdery mildew resistance in nordic spring barley (Hordeum vulgare L. ssp. vulgare) revealed by genome-wide association study. Front. Plant Sci. 2017, 8, 1954. [Google Scholar] [CrossRef] [Green Version]
- Czembor, J.H.; Czembor, E.; Suchecki, R.; Watson-Haigh, N.S. Genome-wide association study for powdery mildew and rusts adult plant resistance in European spring barley from Polish gene bank. Agronomy 2021, 12, 7. [Google Scholar] [CrossRef]
- Ames, N.; Dreiseitl, A.; Steffenson, B.J.; Muehlbauer, G.J. Mining wild barley for powdery mildew resistance. Plant Pathol. 2015, 64, 1396–1406. [Google Scholar] [CrossRef] [Green Version]
- Genievskaya, Y.; Almerekova, S.; Abugalieva, S.; Abigalieva, A.; Sato, K.; Turuspekov, Y. Identification of SNPs associated with grain quality traits in spring barley collection grown in southeastern Kazakhstan. Agronomy 2023, 13, 1560. [Google Scholar] [CrossRef]
- Aust, H.J.; Hoyningen-Huene, J. Microclimate in relation to powdery mildew epidemics. Annu. Rev. Phytopathol. 1986, 24, 491–510. [Google Scholar] [CrossRef]
- Carver, T.L.W.; Griffiths, E. Relationship between powdery mildew infection, green leaf area and grain yield of barley. Ann. Appl. Biol. 1981, 99, 255–266. [Google Scholar] [CrossRef]
- Thomas, W.; Baird, E.; Fuller, J.; Lawrence, P.; Young, G.; Russell, J.; Ramsay, L.; Waugh, R.; Powell, W. Identification of a QTL decreasing yield in barley linked to Mlo powdery mildew resistance. Mol. Breed. 1998, 4, 381–393. [Google Scholar] [CrossRef]
- Ugarte, C.; Calderini, D.F.; Slafer, G.A. Grain weight and grain number responsiveness to pre-anthesis temperature in wheat, barley and triticale. Field Crops Res. 2007, 100, 240–248. [Google Scholar] [CrossRef]
- Ensembl Plants Database. Hordeum vulgare L. Available online: https://plants.ensembl.org/Hordeum_vulgare/Info/Index (accessed on 2 March 2023).
- Genievskaya, Y.; Almerekova, S.; Sariev, B.; Chudinov, V.; Tokhetova, L.; Sereda, G.; Ortaev, A.; Tsygankov, V.; Blake, T.; Chao, S.; et al. Marker-trait associations in two-rowed spring barley accessions from Kazakhstan and the USA. PLoS ONE 2018, 13, e0205421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waugh, R.; Jannink, J.L.; Muehlbauer, G.J.; Ramsay, L. The emergence of whole genome association scans in barley. Curr. Opin. Plant Biol. 2009, 12, 218–222. [Google Scholar] [CrossRef]
- Muñoz-Amatriaín, M.; Cuesta-Marcos, A.; Hayes, P.M.; Muehlbauer, G.J. Barley genetic variation: Implications for crop improvement. Brief Funct. Genom. 2014, 13, 341–350. [Google Scholar] [CrossRef] [Green Version]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; et al. Development and Evaluation of a Barley 50k iSelect SNP Array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Triticeae Toolbox (T3) Database. Barley. Available online: https://barley.triticeaetoolbox.org/ (accessed on 13 March 2023).
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 5 February 2023).
- IBM SPSS Statistics. Available online: https://www.ibm.com/products/spss-statistics (accessed on 25 February 2023).
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021, 19, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [Green Version]
| QTL | Marker | Chr. | Position (bp) | MAF | 2020 | 2022 | Candidate Loci | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-Value | FDR p-Value | PVE (%) | Alleles | Effect | p-Value | FDR p-Value | PVE (%) | Alleles | Effect | ||||||
| Qhv_PM-4H.1 | 12_20274 | 4H | 3,623,098 | 0.139 | 1.2 × 10−8 | 0.000010 | 85.3 | G/C | −6.3 | Bgh-qtl-4H-11_10319 [56] | |||||
| Qhv_PM-4H.2 | 11_10509 | 4H | 496,024,940 | 0.066 | 7.5 × 10−6 | 0.003696 | 0.0 | C/A | −4.4 | ||||||
| Qhv_PM-4H.3 | 11_10914 | 4H | 511,791,369 | 0.178 | 8.9 × 10−6 | 0.003696 | 0.0 | C/G | −4.1 | ||||||
| Qhv_PM-5H.1 | 12_30980 | 5H | 2,540,132 | 0.071 | 2.4 × 10−8 | 0.00004 | 20.6 | C/A | 5.4 | Qrbg_5H_1 [53] | |||||
| Qhv_PM-5H.2 | 11_11240 | 5H | 354,892,901 | 0.101 | 6.2 × 10−10 | 0.000001 | 0.0 | A/T | 5.0 | ||||||
| Qhv_PM-5H.3 | 12_10769 | 5H | 646,543,760 | 0.051 | 2.6 × 10−5 | 0.02137 | 12.8 | A/G | 4.2 | ||||||
| Qhv_PM-7H.1 | 12_31513 | 7H | 475,182,581 | 0.112 | 2.2 × 10−5 | 0.007203 | 0.4 | A/G | −4.9 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genievskaya, Y.; Zatybekov, A.; Abugalieva, S.; Turuspekov, Y. Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan. Plants 2023, 12, 2375. https://doi.org/10.3390/plants12122375
Genievskaya Y, Zatybekov A, Abugalieva S, Turuspekov Y. Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan. Plants. 2023; 12(12):2375. https://doi.org/10.3390/plants12122375
Chicago/Turabian StyleGenievskaya, Yuliya, Alibek Zatybekov, Saule Abugalieva, and Yerlan Turuspekov. 2023. "Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan" Plants 12, no. 12: 2375. https://doi.org/10.3390/plants12122375
APA StyleGenievskaya, Y., Zatybekov, A., Abugalieva, S., & Turuspekov, Y. (2023). Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Spring Barley under Conditions of Southeastern Kazakhstan. Plants, 12(12), 2375. https://doi.org/10.3390/plants12122375





