Haplotype-Based Genome-Wide Association Analysis Using Exome Capture Assay and Digital Phenotyping Identifies Genetic Loci Underlying Salt Tolerance Mechanisms in Wheat
Abstract
1. Introduction
2. Results
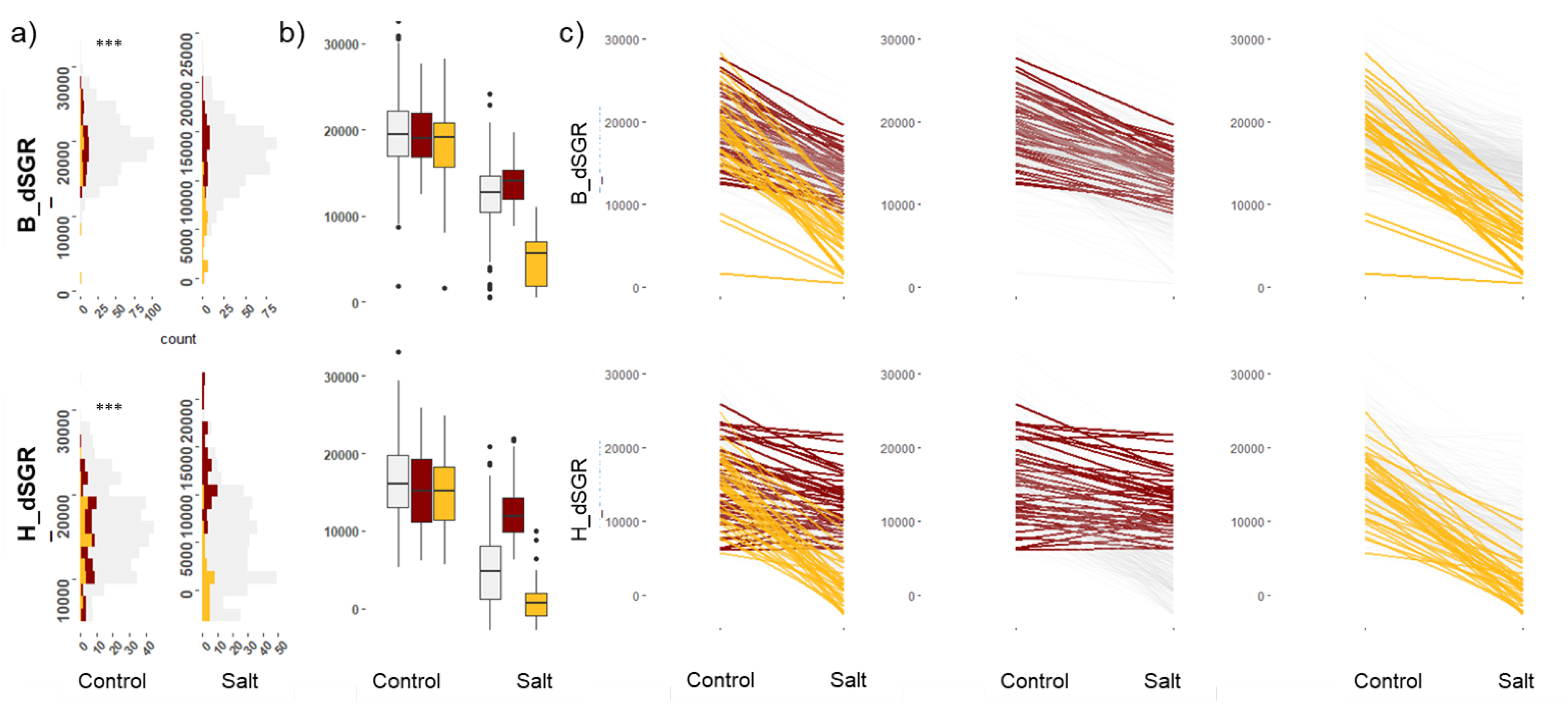
2.1. Estimation of Digital Shoot Growth Rate (dSGR) and Its Variation in Wheat
2.2. Salt Susceptibility Index (SSI) to Determine Tolerance
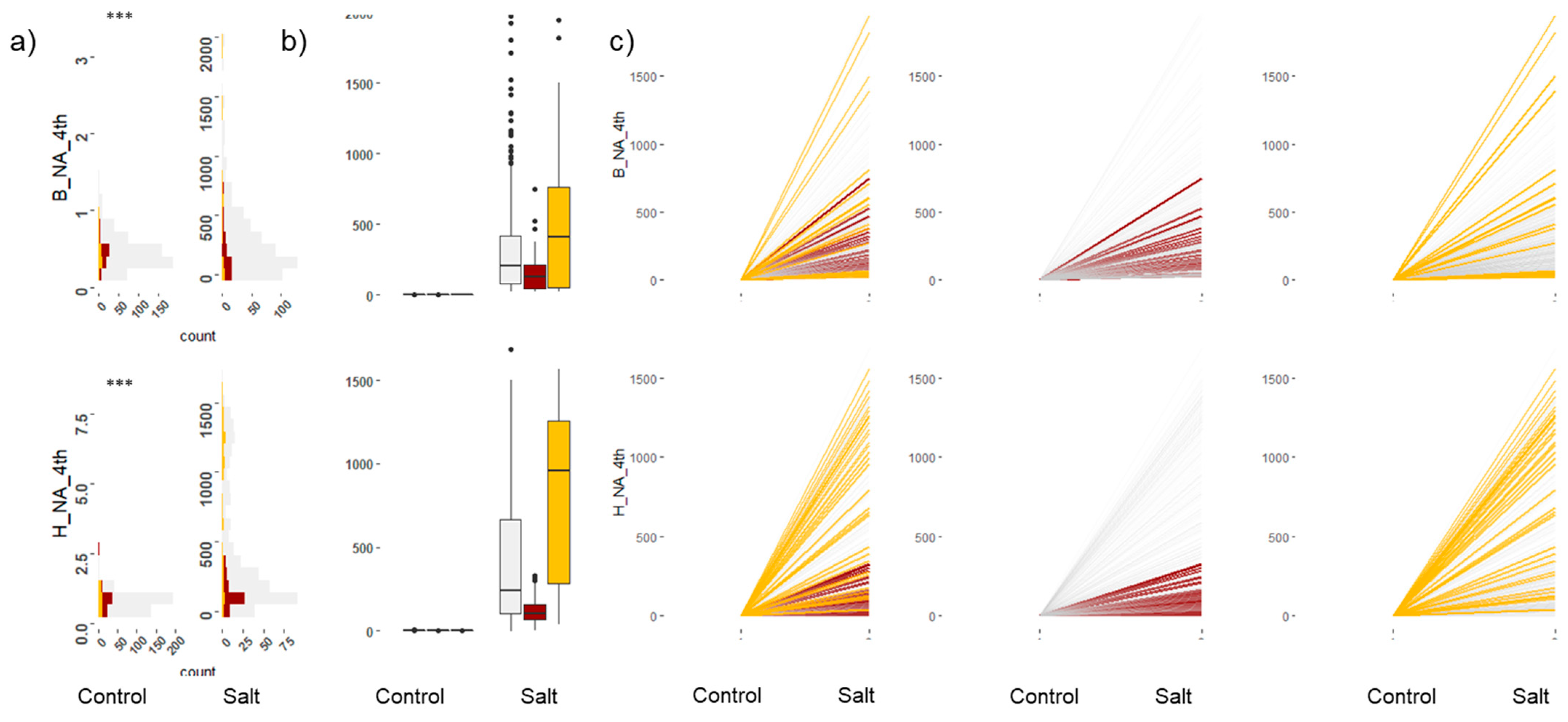
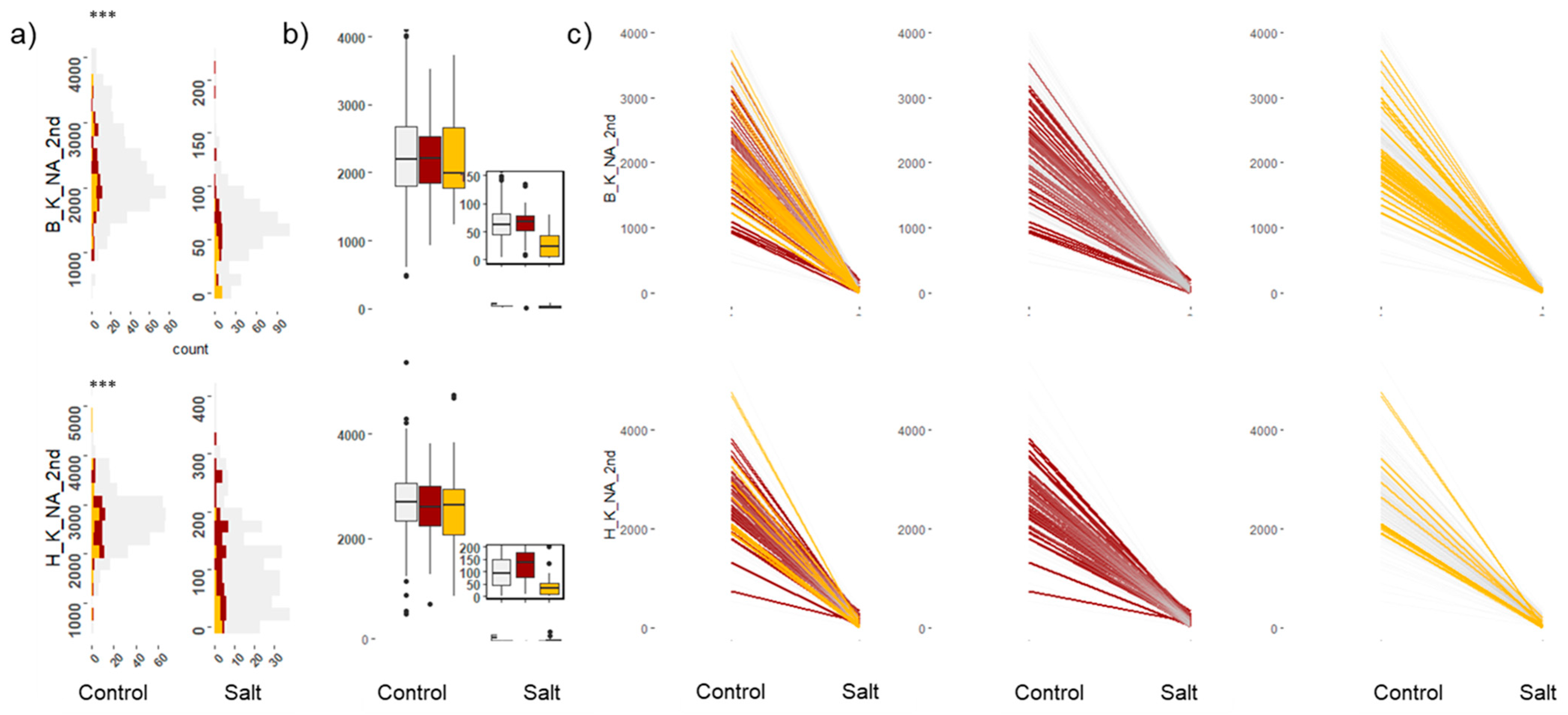
2.3. Leaf Elemental Analysis
2.4. Haplotype Analysis
2.5. Haplotype-Based Genome-Wide Association Analysis (HGWAS) and Genetic Correlations
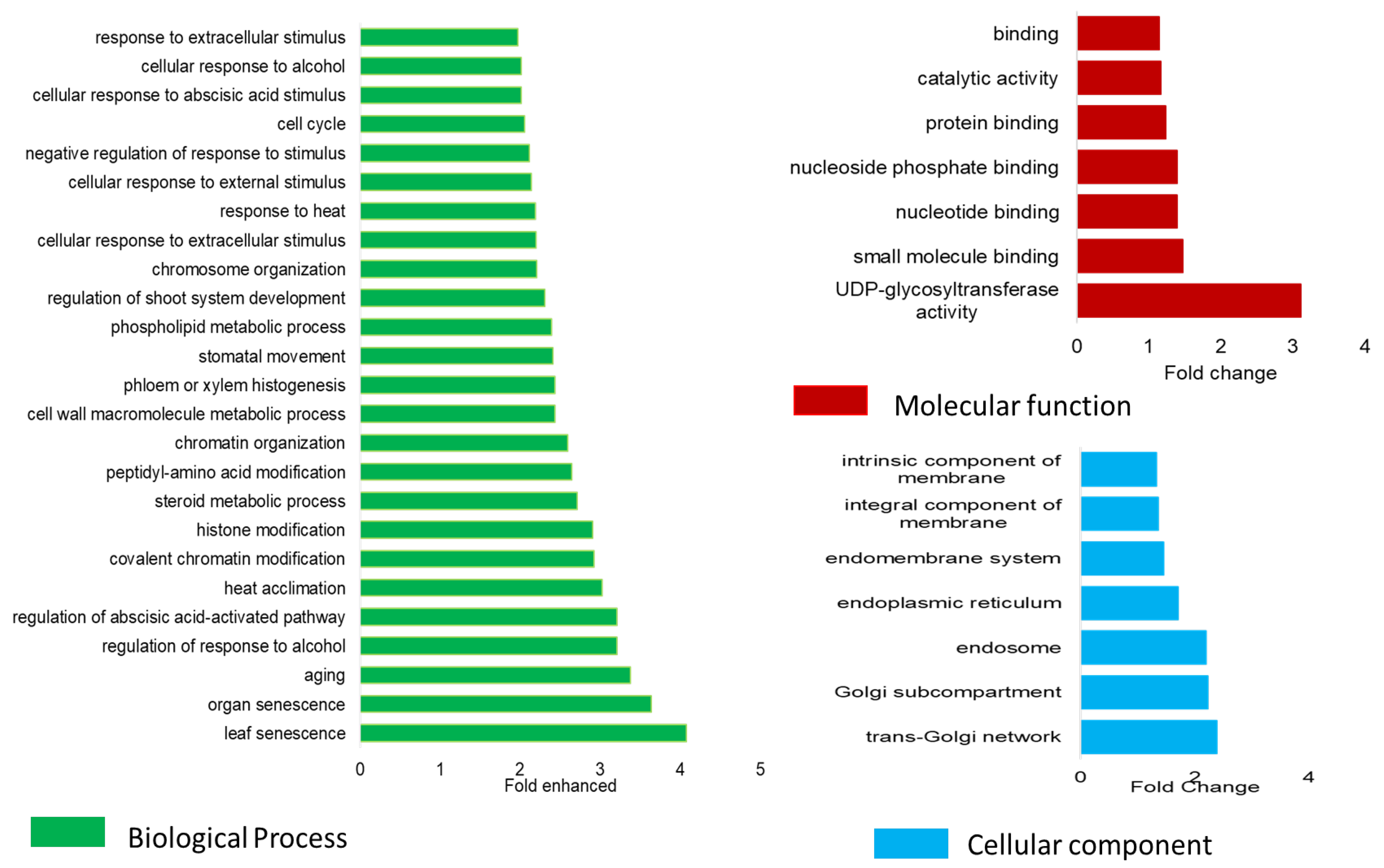
2.6. Gene-Based Meta-Analysis and Gene Ontology Terms Enrichment
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Growth Conditions and Phenotyping Trait Measures
4.3. Phenotype Data Analysis
4.4. Ion Analysis of Leaf
4.5. Genotype Data Analysis and Haplotype Blocks
4.6. Haplotype-Based GWAS (HGWAS)
4.7. Significant Marker Trait Associations and Cross-Validation of HGWAS
4.8. Gene-Based Meta-Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Butcher, K.; Wick, A.; DeSutter, T.; Chatterjee, A.; Harmon, J. Soil Salinity: A Threat to Global Food Security. Agron. J. 2016, 108, 2189–2200. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Rengasamy, P. World salinisation with emphasis on Australia. Comp. Biochem. Phys. A 2005, 141, 337–348. [Google Scholar]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New Insights on Plant Salt Tolerance Mechanisms and Their Potential Use for Breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Zelm Ev Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Ladeiro, B. Saline Agriculture in the 21st Century: Using Salt Contaminated Resources to Cope Food Requirements. J. Bot. 2012, 7, 310705. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Munns, R.; Rasheed, A.; Ogbonnaya, F.C.; Ali, N.; Hollington, P.; Dundas, I.; Saeed, N.; Wang, R.; Rengasamy, P.; et al. Breeding Strategies for Structuring Salinity Tolerance in Wheat. Adv. Agron. 2019, 155, 121–187. [Google Scholar]
- Asif, M.A.; Garcia, M.; Tilbrook, J.; Brien, C.; Dowling, K.; Berger, B.; Schilling, R.K.; Short, L.; Trittermann, C.; Gilliham, M.; et al. Identification of salt tolerance QTL in a wheat RIL mapping population using destructive and non-destructive phenotyping. Funct. Plant Biol. 2021, 48, 131–140. [Google Scholar] [CrossRef]
- Genc, Y.; Taylor, J.; Lyons, G.; Li, Y.; Cheong, J.; Appelbee, M.; Oldach, K.; Sutton, T. Bread Wheat With High Salinity and Sodicity Tolerance. Front. Plant Sci. 2019, 10, 1280. [Google Scholar] [CrossRef]
- Genc, Y.; Taylor, J.; Rongala, J.; Oldach, K. A Major Locus for Chloride Accumulation on Chromosome 5A in Bread Wheat. PLoS ONE 2014, 9, e98845. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zheng, Q.; Luo, Q.; Teng, W.; Li, H.; Li, B.; Li, Z. Genome-wide association study of yield and related traits in common wheat under salt-stress conditions. BMC Plant Biol. 2021, 21, 27. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Zhang, Q.; Fu, B.; Cai, J.; Wu, J.; Chen, Y. Genome-wide association analysis of quantitative trait loci for salinity-tolerance related morphological indices in bread wheat. Euphytica 2018, 214, 176. [Google Scholar] [CrossRef]
- Hasan, H.; Ali, M.; Javaid, A.; Liaqat, A.; Hussain, S.; Siddique, R.; Fayaz, T.; Gul, A. Chapter 4—Cellular mechanism of salinity tolerance in wheat. In Climate Change and Food Security with Emphasis on Wheat; Ozturk, M., Gul, A., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 55–76. [Google Scholar]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Cuin, T.A.; Bose, J.; Stefano, G.; Jha, D.; Tester, M.; Mancuso, S.; Shabala, S. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: In planta quantification methods. Plant Cell Environ. 2011, 34, 947–961. [Google Scholar] [CrossRef]
- Rajendran, K.; Tester, M.; Roy, S.J. Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 2009, 32, 237–249. [Google Scholar] [CrossRef]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1;5-Like Cation Transporters Linked to Na+ Exclusion Loci in Wheat Nax2 and Kna1. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Spielmeyer, W.; Lagudah, E.S.; James, R.A.; Platten, J.D.; Dennis, E.S.; Munns, R. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006, 142, 1718–1727. [Google Scholar] [CrossRef]
- Dubcovsky, J.; María, G.S.; Epstein, E.; Luo, M.C.; Dvořák, J. Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor. Appl. Genet. 1996, 92, 448–454. [Google Scholar] [CrossRef]
- Wu, H.; Shabala, L.; Liu, X.; Azzarello, E.; Zhou, M.; Pandolfi, C.; Chen, Z.-H.; Bose, J.; Mancuso, S.; Shabala, S. Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front. Plant Sci. 2015, 6, 71. [Google Scholar] [CrossRef]
- Xu, H.; Jiang, X.; Zhan, K.; Cheng, X.; Chen, X.; Pardo, J.M.; Cui, D. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 2008, 473, 8–15. [Google Scholar] [CrossRef]
- Frouin, J.; Languillaume, A.; Mas, J.; Mieulet, D.; Boisnard, A.; Labeyrie, A.; Bettembourg, M.; Bureau, C.; Lorenzini, E.; Portefaix, M.; et al. Tolerance to mild salinity stress in japonica rice: A genome-wide association mapping study highlights calcium signaling and metabolism genes. PLoS ONE 2018, 13, e0190964. [Google Scholar] [CrossRef]
- Zhou, Y.-B.; Liu, C.; Tang, D.-Y.; Yan, L.; Wang, D.; Yang, Y.-Z.; Gui, J.-S.; Zhao, X.-Y.; Li, L.-G.; Tang, X.-D.; et al. The Receptor-Like Cytoplasmic Kinase STRK1 Phosphorylates and Activates CatC, Thereby Regulating H2O2 Homeostasis and Improving Salt Tolerance in Rice. Plant Cell 2018, 30, 1100–1118. [Google Scholar] [CrossRef]
- Hairmansis, A.; Berger, B.; Tester, M.; Roy, S.J. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice 2014, 7, 16. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Yeo, A.R.; Yeo, M.E.; Flowers, S.A.; Flowers, T.J. Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 1990, 79, 377–384. [Google Scholar] [CrossRef]
- Meng, R.; Saade, S.; Kurtek, S.; Berger, B.; Brien, C.; Pillen, K.; Tester, M.; Sun, Y. Growth curve registration for evaluating salinity tolerance in barley. Plant Methods 2017, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Oyiga, B.C.; Sharma, R.C.; Shen, J.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Identification and Characterization of Salt Tolerance of Wheat Germplasm Using a Multivariable Screening Approach. J. Agron. Crop Sci. 2016, 202, 472–485. [Google Scholar] [CrossRef]
- Lee, U.; Chang, S.; Putra, G.A.; Kim, H.; Kim, D.H. An automated, high-throughput plant phenotyping system using machine learning-based plant segmentation and image analysis. PLoS ONE 2018, 13, e0196615. [Google Scholar] [CrossRef]
- Neumann, K.; Zhao, Y.; Chu, J.; Keilwagen, J.; Reif, J.C.; Kilian, B.; Graner, A. Genetic architecture and temporal patterns of biomass accumulation in spring barley revealed by image analysis. BMC Plant Biol. 2017, 17, 137. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Pasam, R.K.; Bansal, U.; Daetwyler, H.D.; Forrest, K.L.; Wong, D.; Petkowski, J.; Willey, N.; Randhawa, M.; Chhetri, M.; Miah, H.; et al. Detection and validation of genomic regions associated with resistance to rust diseases in a worldwide hexaploid wheat landrace collection using BayesR and mixed linear model approaches. Theor. Appl. Genet. 2017, 130, 777–793. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.-Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef]
- Wang, S.-B.; Feng, J.-Y.; Ren, W.-L.; Huang, B.; Zhou, L.; Wen, Y.-J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.-M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-J.; Zhang, H.; Ni, Y.-L.; Huang, B.; Zhang, J.; Feng, J.-Y.; Wang, S.-B.; Dunwell, J.M.; Zhang, Y.-M.; Wu, R. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 2017, 19, 700–712. [Google Scholar] [CrossRef]
- Abed, A.; Belzile, F. Comparing Single-SNP, Multi-SNP, and Haplotype-Based Approaches in Association Studies for Major Traits in Barley. Plant Genome 2019, 12, 190036. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Jiang, Y.; Zhao, Y.; Schulthess, A.W.; Reif, J.C. Haplotype-based genome-wide association increases the predictability of leaf rust (Puccinia triticina) resistance in wheat. J. Exp. Bot. 2020, 71, 6958–6968. [Google Scholar] [CrossRef]
- Liu, F.; Schmidt, R.H.; Reif, J.C.; Jiang, Y. Selecting Closely-Linked SNPs Based on Local Epistatic Effects for Haplotype Construction Improves Power of Association Mapping. G3 Genes Genomes Genet. 2019, 9, 4115–4126. [Google Scholar] [CrossRef]
- Pryce, J.; Bolormaa, S.; Chamberlain, A.; Bowman, P.; Savin, K.; Goddard, M.; Hayes, B. A validated genome-wide association study in 2 dairy cattle breeds for milk production and fertility traits using variable length haplotypes. J. Dairy Sci. 2010, 93, 3331–3345. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef]
- Winfield, M.O.; Allen, A.M.; Burridge, A.; Barker, G.L.A.; Benbow, H.R.; Wilkinson, P.A.; Coghill, J.; Waterfall, C.; Davassi, A.; Scopes, G.; et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 2016, 14, 1195–1206. [Google Scholar] [CrossRef]
- Marone, D.; Panio, G.; Ficco, D.B.M.; Russo, M.A.; De Vita, P.; Papa, R.; Rubiales, D.; Cattivelli, L.; Mastrangelo, A.M. Characterization of wheat DArT markers: Genetic and functional features. Mol. Genet. Genom. 2012, 287, 741–753. [Google Scholar] [CrossRef] [PubMed]
- Poland, J.; Endelman, J.; Dawson, J.; Rutkoski, J.; Wu, S.; Manes, Y.; Dreisigacker, S.; Crossa, J.; Sánchez-Villeda, H.; Sorrells, M.; et al. Genomic Selection in Wheat Breeding using Genotyping-by-Sequencing. Plant Genome 2012, 5, 103–113. [Google Scholar] [CrossRef]
- He, F.; Pasam, R.; Shi, F.; Kant, S.; Keeble-Gagnere, G.; Kay, P.; Forrest, K.; Fritz, A.; Hucl, P.; Wiebe, K.; et al. Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet. 2019, 51, 896–904. [Google Scholar] [CrossRef]
- Schmidt, J.; Tricker, P.J.; Eckermann, P.; Kalambettu, P.; Garcia, M.; Fleury, D. Novel Alleles for Combined Drought and Heat Stress Tolerance in Wheat. Front. Plant Sci. 2020, 10, 1800. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Keeble-Gagnère, G.; Hickey, L.T.; Tibbits, J.; Nagornyy, S.; Hayden, M.J.; Pasam, R.K.; Kant, S.; Friedt, W.; Snowdon, R.J.; et al. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Theor. Appl. Genet 2019, 132, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Courtois, B.; Audebert, A.; Dardou, A.; Roques, S.; Herrera, T.G.; Droc, G.; Frouin, J.; Rouan, L.; Gozé, E.; Kilian, A.; et al. Genome-Wide Association Mapping of Root Traits in a Japonica Rice Panel. PLoS ONE 2013, 8, e78037. [Google Scholar] [CrossRef]
- Kaler, A.S.; Abdel-Haleem, H.; Fritschi, F.B.; Gillman, J.D.; Ray, J.D.; Smith, J.R.; Purcell, L.C. Genome-Wide Association Mapping of Dark Green Color Index using a Diverse Panel of Soybean Accessions. Sci. Rep. 2020, 10, 5166. [Google Scholar] [CrossRef]
- Karunarathne, S.D.; Han, Y.; Zhang, X.-Q.; Zhou, G.; Hill, C.B.; Chen, K.; Angessa, T.; Li, C. Genome-Wide Association Study and Identification of Candidate Genes for Nitrogen Use Efficiency in Barley (Hordeum vulgare L.). Front. Plant Sci. 2020, 11, 571912. [Google Scholar] [CrossRef]
- De León, L.J.D.; Escoppinichi, R.; Geraldo, N.; Thelma, C.; Mujeeb-Kazi, A.; Röde, M. Quantitative trait loci associated with salinity tolerance in field grown bread wheat. Euphytica 2011, 181, 371–383. [Google Scholar] [CrossRef]
- Dadshani, S.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Multi-dimensional evaluation of response to salt stress in wheat. PLoS ONE 2019, 14, e0222659. [Google Scholar] [CrossRef]
- Asch, F.; Dingkuhn, M.; Dorffling, K. Salinity increases CO2 assimilation but reduces growth in field-grown, irrigated rice. Plant Soil 2000, 218, 1. [Google Scholar] [CrossRef]
- Horie, T.; Karahara, I.; Katsuhara, M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice 2012, 5, 11. [Google Scholar] [CrossRef]
- Tilbrook, J.; Schilling, R.K.; Berger, B.; Garcia, A.F.; Trittermann, C.; Coventry, S.; Rabie, H.; Brien, C.; Nguyen, M.; Tester, M.; et al. Variation in shoot tolerance mechanisms not related to ion toxicity in barley. Funct. Plant Biol. 2017, 44, 1194–1206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Smith, J.A.C.; Harberd, N.P.; Jiang, C. The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 2016, 91, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Gilliham, M.; Flowers, T.J.; Colmer, T.D. Tissue tolerance: An essential but elusive trait for salt-tolerant crops. Funct. Plant Biol. 2016, 43, 1103–1113. [Google Scholar] [CrossRef]
- Campbell, M.T.; Knecht, A.C.; Berger, B.; Brien, C.J.; Wang, D.; Walia, H. Integrating Image-Based Phenomics and Association Analysis to Dissect the Genetic Architecture of Temporal Salinity Responses in Rice. Plant Physiol. 2015, 168, 1476–1489. [Google Scholar] [CrossRef]
- Saade, S.; Brien, C.; Pailles, Y.; Berger, B.; Shahid, M.; Russell, J.; Waugh, R.; Negrão, S.; Tester, M. Dissecting new genetic components of salinity tolerance in two-row spring barley at the vegetative and reproductive stages. PLoS ONE 2020, 15, e0236037. [Google Scholar] [CrossRef]
- Munns, R. Genes and salt tolerance: Bringing them together. New Phytol. 2005, 167, 645–663. [Google Scholar] [CrossRef] [PubMed]
- Oyiga, B.C.; Ogbonnaya, F.C.; Sharma, R.C.; Baum, M.; Léon, J.; Ballvora, A. Genetic and transcriptional variations in NRAMP-2 and OPAQUE1 genes are associated with salt stress response in wheat. Theor. Appl. Genet. 2019, 132, 323–346. [Google Scholar] [CrossRef] [PubMed]
- Oyiga, B.C.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant Cell Environ. 2018, 41, 919–935. [Google Scholar] [CrossRef] [PubMed]
- Devi, R.; Ram, S.; Rana, V.; Malik, V.K.; Pande, V.; Singh, G.P. QTL mapping for salt tolerance associated traits in wheat (Triticum aestivum L.). Euphytica 2019, 215, 210. [Google Scholar] [CrossRef]
- Genc, Y.; Oldach, K.; Verbyla, A.P.; Lott, G.; Hassan, M.; Tester, M.; Wallwork, H.; McDonald, G.K. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor. Appl. Genet. 2010, 121, 877–894. [Google Scholar] [CrossRef]
- Luo, Q.; Zheng, Q.; Hu, P.; Liu, L.; Yang, G.; Li, H.; Li, B.; Li, Z. Mapping QTL for agronomic traits under two levels of salt stress in a new constructed RIL wheat population. Theor. Appl. Genet. 2021, 134, 171–189. [Google Scholar] [CrossRef]
- Yu, S.; Wu, J.; Wang, M.; Shi, W.; Xia, G.; Jia, J.; Kang, Z.; Han, D. Haplotype variations in QTL for salt tolerance in Chinese wheat accessions identified by marker-based and pedigree-based kinship analyses. Crop J. 2020, 8, 1011–1024. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, H.; Guan, C.; Cui, J.; Zhang, Q.; Liu, M.; Zhang, M.; Guo, Q.; Hou, Y.; Xiang, M.; et al. Physical information of 2705 PCR-based molecular markers and the evaluation of their potential use in wheat. J. Genet. 2019, 98, 69. [Google Scholar] [CrossRef]
- Keeble-Gagnère, G.; Isdale, D.; Suchecki, R.-ł.; Kruger, A.; Lomas, K.; Carroll, D.; Li, S.; Whan, A.; Hayden, M.; Tibbits, J.F.G. Integrating past, present and future wheat research with Pretzel. bioRxiv 2019, 517953. [Google Scholar]
- Nia, S.H.; Zarea, M.J.; Rejali, F.; Varma, A. Yield and yield components of wheat as affected by salinity and inoculation with Azospirillum strains from saline or non-saline soil. J. Saudi Soc. Agric. Sci. 2012, 11, 113–121. [Google Scholar] [CrossRef]
- Cuong, D.M.; Kwon, S.-J.; Nguyen, B.V.; Chun, S.W.; Kim, J.K.; Park, S.U. Effect of Salinity Stress on Phenylpropanoid Genes Expression and Related Gene Expression in Wheat Sprout. Agronomy 2020, 10, 390. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Christensen, S.; Schmelz, E.; Huffaker, A.; McAuslane, H.J.; Alborn, H.T.; Romero, M.; Allen, L.H.; Teal, P.E.A. Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 2015, 38, 2195–2207. [Google Scholar] [CrossRef] [PubMed]
- Boba, A.; Kostyn, K.; Kozak, B.; Wojtasik, W.; Preisner, M.; Prescha, A.; Gola, E.M.; Lysh, D.; Dudek, B.; Szopa, J.; et al. Fusarium oxysporum infection activates the plastidial branch of the terpenoid biosynthesis pathway in flax, leading to increased ABA synthesis. Planta 2020, 251, 50. [Google Scholar] [CrossRef] [PubMed]
- Allan, W.L.; Clark, S.M.; Hoover, G.J.; Shelp, B.J. Role of plant glyoxylate reductases during stress: A hypothesis. Biochem. J. 2009, 423, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Carillo, P. GABA Shunt in Durum Wheat. Front. Plant Sci. 9 2018, 9, 100. [Google Scholar] [CrossRef]
- Woodrow, P.; Ciarmiello, L.F.; Annunziata, M.G.; Pacifico, S.; Iannuzzi, F.; Mirto, A.; D’Amelia, L.; Dell’Aversana, E.; Piccolella, S.; Fuggi, A.; et al. Durum wheat seedling responses to simultaneous high light and salinity involve a fine reconfiguration of amino acids and carbohydrate metabolism. Physiol. Plant. 2017, 159, 290–312. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Guan, Q.; Wang, Z.Y.; Wang, Y.; Zhu, J. A bi-functional xyloglucan galactosyltransferase is an indispensable salt stress tolerance determinant in Arabidopsis. Mol. Plant 2013, 6, 1344–1354. [Google Scholar] [CrossRef]
- Scheible, W.-R.; Pauly, M. Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 2004, 7, 285–295. [Google Scholar] [CrossRef]
- Banerjee, B.P.; Joshi, S.; Thoday-Kennedy, E.; Pasam, R.K.; Tibbits, J.; Hayden, M.; Spangenberg, G.; Kant, S. High-throughput phenotyping using digital and hyperspectral imaging-derived biomarkers for genotypic nitrogen response. J. Exp. Bot. 2020, 71, 4604–4615. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Klukas, C.; Chen, D.; Pape, J.-M. Integrated Analysis Platform: An Open-Source Information System for High-Throughput Plant Phenotyping. Plant Physiol. 2014, 165, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Fischer, R.; Maurer, R. Drought resistance in spring wheat cultivars, I. Grain yield responses. Aust. J. Agric. Res. 1978, 29, 897–912. [Google Scholar] [CrossRef]
- Munns, R.; Wallace, P.A.; Teakle, N.L.; Colmer, T.D. Measuring soluble ion concentrations (Na+, K+, Cl−) in salt-treated plants. Methods Mol. Biol. 2010, 639, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.B.; Schaffner, S.F.; Nguyen, H.; Moore, J.M.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The Structure of Haplotype Blocks in the Human Genome. Science 2002, 296, 2225. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Utsunomiya, Y.T.; Milanesi, M.; Utsunomiya, A.T.H.; Ajmone-Marsan, P.; Garcia, J.F. GHap: An R package for genome-wide haplotyping. Bioinformatics 2016, 32, 2861–2862. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Lee, S.H.; van der Werf, J.H.J. MTG2: An efficient algorithm for multivariate linear mixed model analysis based on genomic information. Bioinformatics 2016, 32, 1420–1422. [Google Scholar] [CrossRef]
- Nicodemus, K.K.; Liu, W.; Chase, G.A.; Tsai, Y.; Fallin, M.D. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet. 2005, 6, S78. [Google Scholar] [CrossRef]
- Johnson, R.C.; Nelson, G.W.; Troyer, J.L.; Lautenberger, J.A.; Kessing, B.D.; Winkler, C.A.; O’Brien, S.J. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genom. 2010, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, A.; Zhu, Z.; Vinkhuyzen, A.A.E.; Hill, W.D.; McRae, A.F.; Visscher, P.M.; Yang, J. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci. Rep. 2016, 6, 32894. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasam, R.K.; Kant, S.; Thoday-Kennedy, E.; Dimech, A.; Joshi, S.; Keeble-Gagnere, G.; Forrest, K.; Tibbits, J.; Hayden, M. Haplotype-Based Genome-Wide Association Analysis Using Exome Capture Assay and Digital Phenotyping Identifies Genetic Loci Underlying Salt Tolerance Mechanisms in Wheat. Plants 2023, 12, 2367. https://doi.org/10.3390/plants12122367
Pasam RK, Kant S, Thoday-Kennedy E, Dimech A, Joshi S, Keeble-Gagnere G, Forrest K, Tibbits J, Hayden M. Haplotype-Based Genome-Wide Association Analysis Using Exome Capture Assay and Digital Phenotyping Identifies Genetic Loci Underlying Salt Tolerance Mechanisms in Wheat. Plants. 2023; 12(12):2367. https://doi.org/10.3390/plants12122367
Chicago/Turabian StylePasam, Raj K., Surya Kant, Emily Thoday-Kennedy, Adam Dimech, Sameer Joshi, Gabriel Keeble-Gagnere, Kerrie Forrest, Josquin Tibbits, and Matthew Hayden. 2023. "Haplotype-Based Genome-Wide Association Analysis Using Exome Capture Assay and Digital Phenotyping Identifies Genetic Loci Underlying Salt Tolerance Mechanisms in Wheat" Plants 12, no. 12: 2367. https://doi.org/10.3390/plants12122367
APA StylePasam, R. K., Kant, S., Thoday-Kennedy, E., Dimech, A., Joshi, S., Keeble-Gagnere, G., Forrest, K., Tibbits, J., & Hayden, M. (2023). Haplotype-Based Genome-Wide Association Analysis Using Exome Capture Assay and Digital Phenotyping Identifies Genetic Loci Underlying Salt Tolerance Mechanisms in Wheat. Plants, 12(12), 2367. https://doi.org/10.3390/plants12122367







