Aerobiological Monitoring and Metabarcoding of Grass Pollen
Abstract
:1. Introduction
2. Results
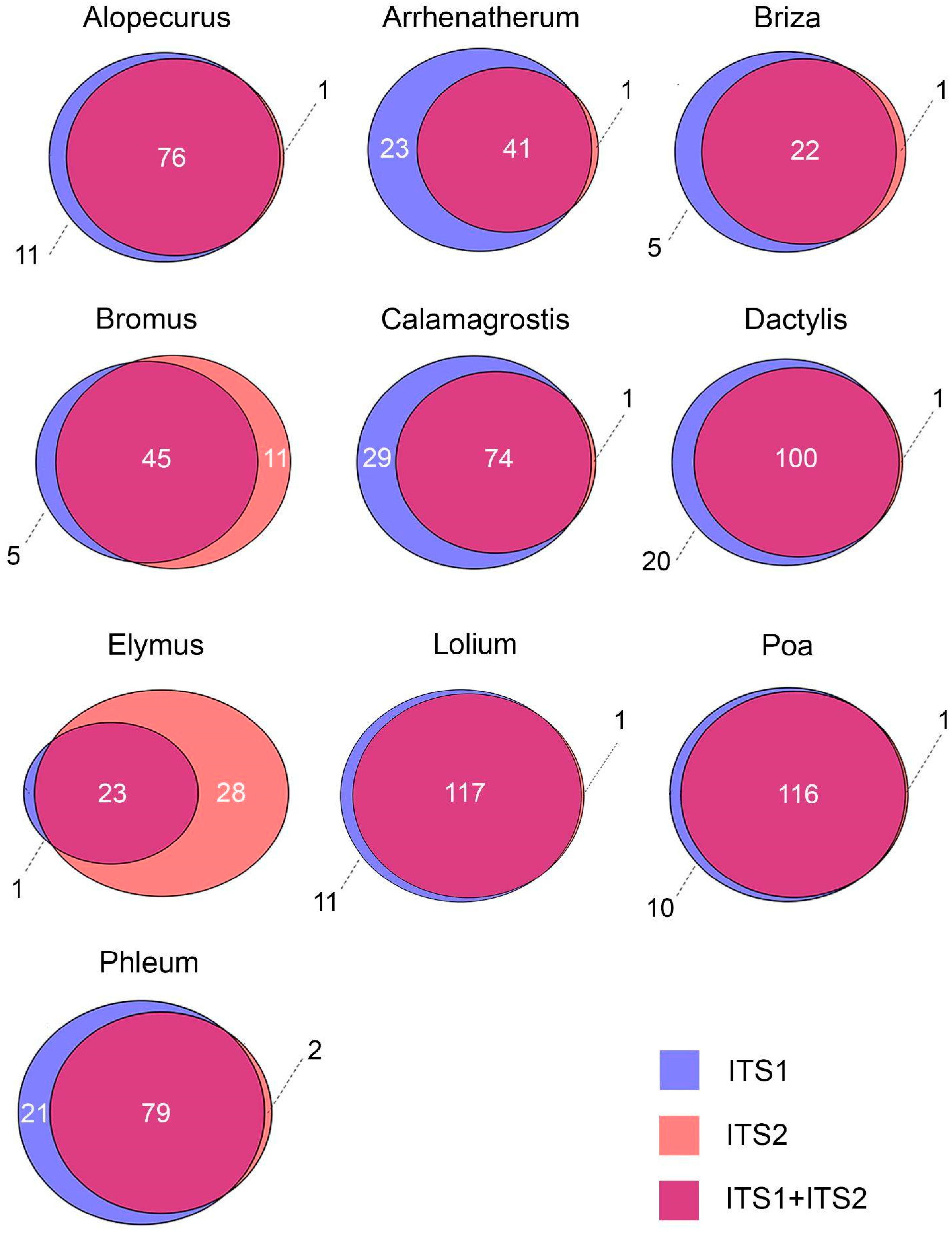
2.1. Sequencing Results
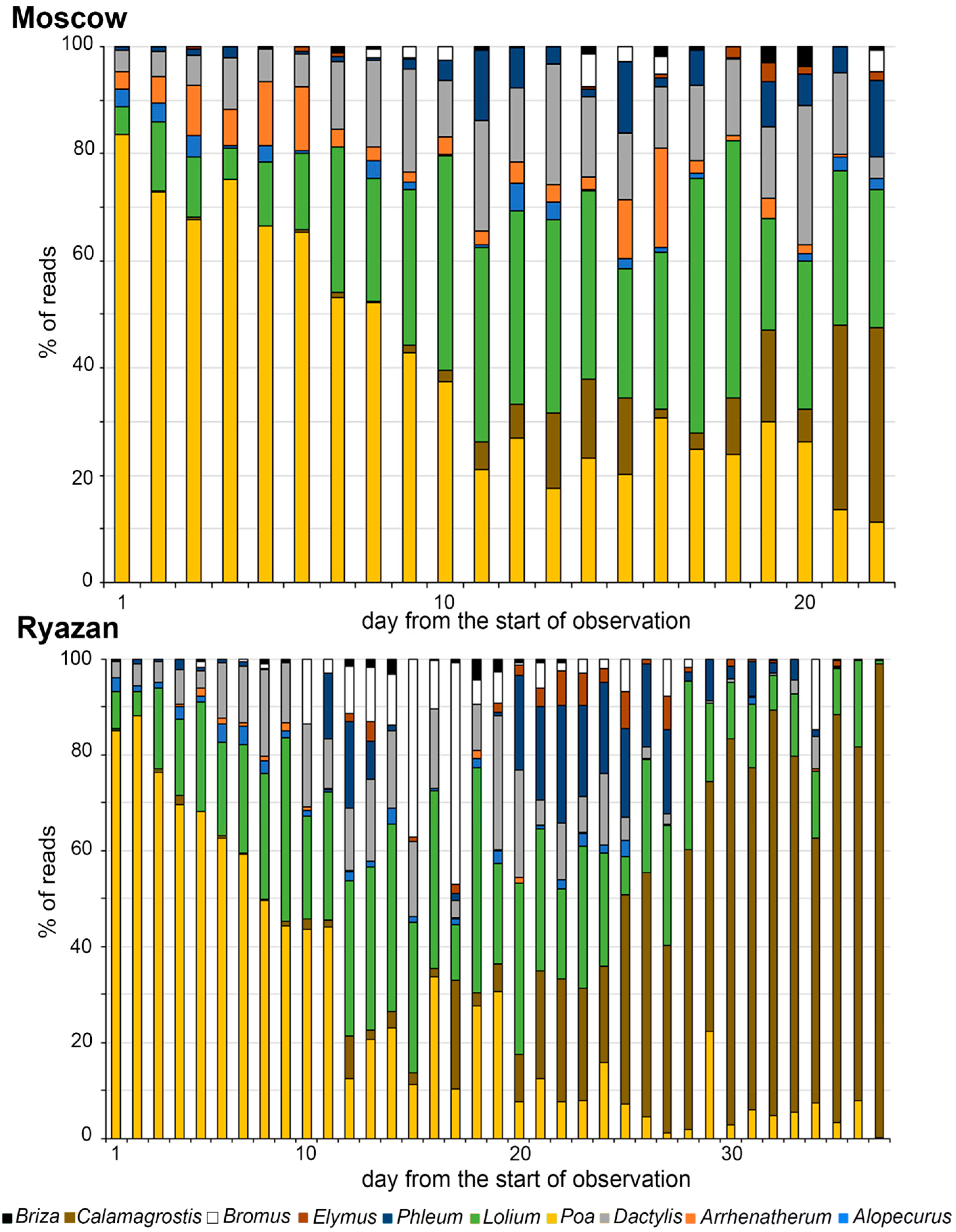
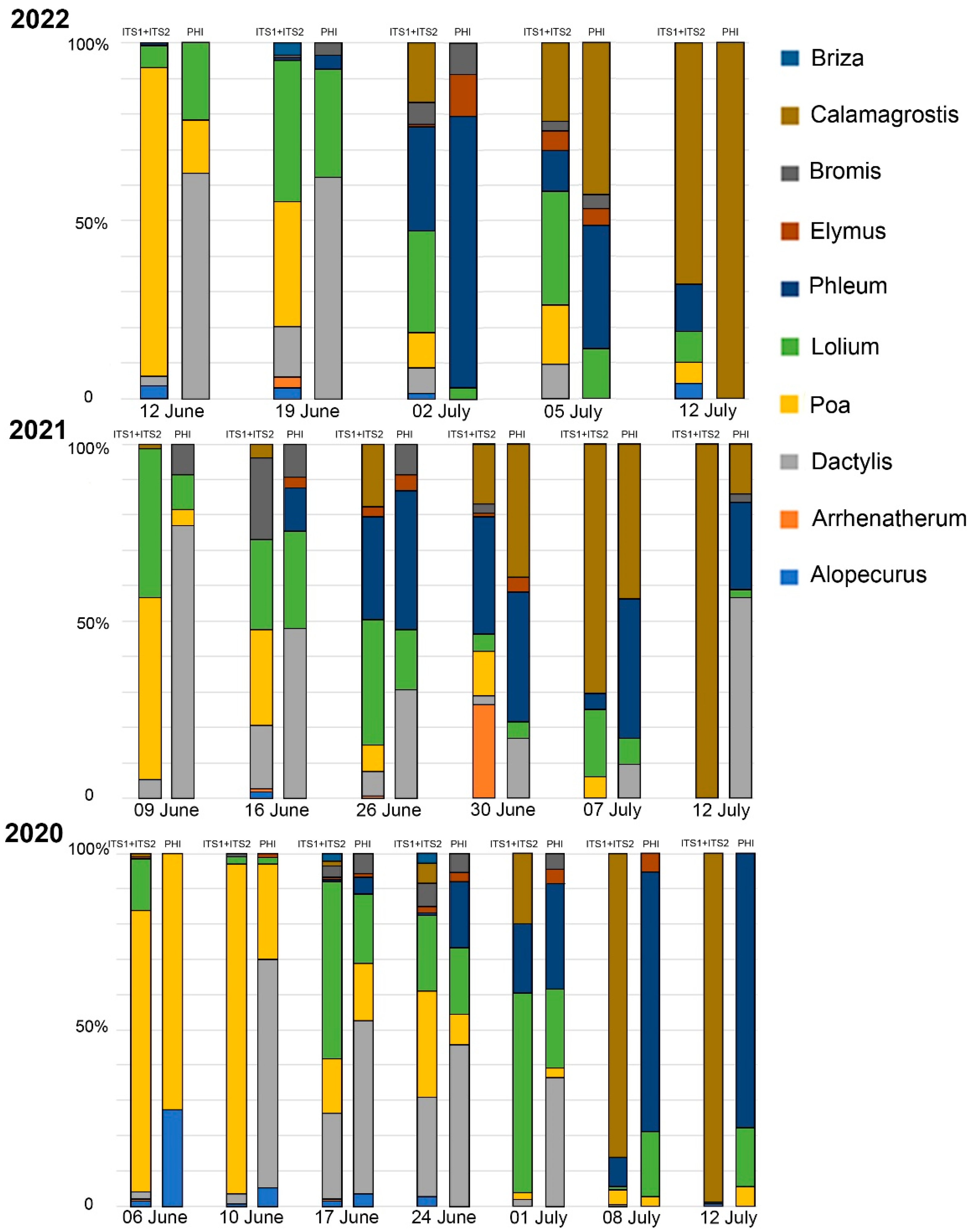
2.2. Metabarcoding Analysis of Airborne Samples from Moscow and Ryazan
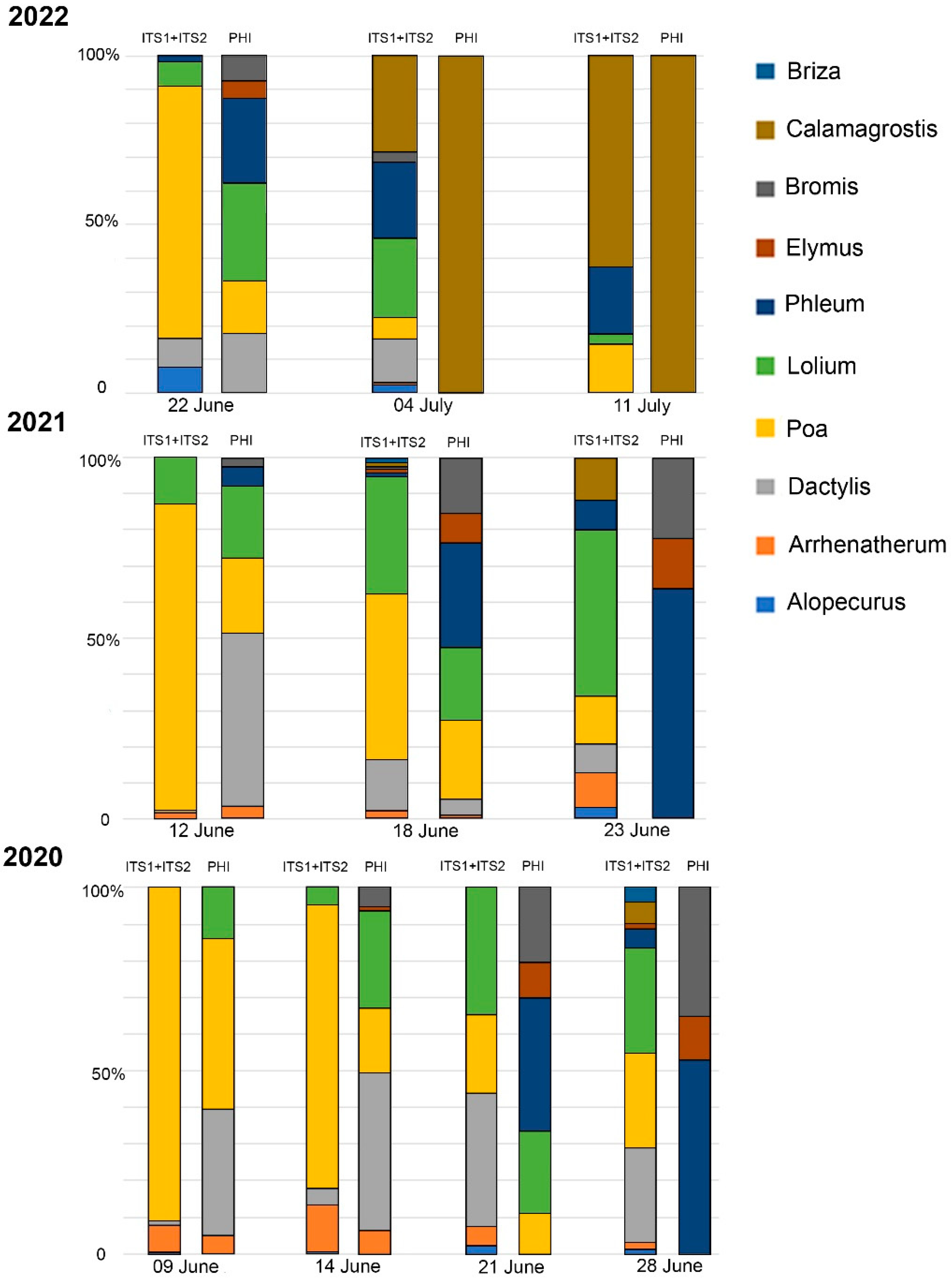
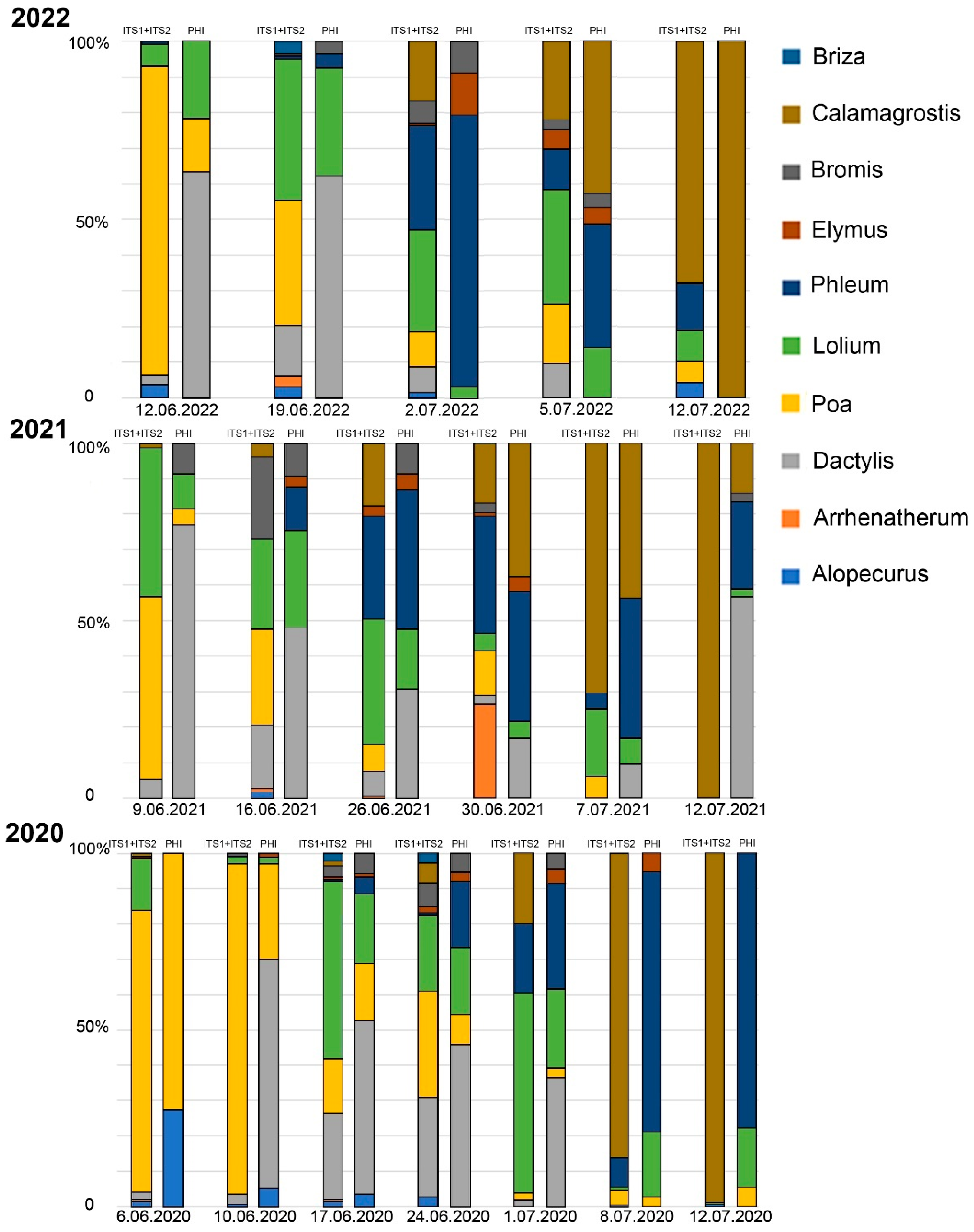
2.3. Comparison of Metabarcoding and Phenological Analysis
3. Discussion
4. Materials and Methods
4.1. Sampling
Phenological Observations
4.2. Statistical Methods
4.3. Optimization of Pollen Wash-off Procedure
4.4. Optimization of Pollen DNA Isolation
- DNA isolation from pollen according to the protocol described in [55]. The approximate time to isolate DNA from six samples is 4 h.
- Enzymatic treatment with lysozyme and zymolysin. First, the enzymatic treatment of pollen was carried out with the addition of 50 μL of lysozyme and 50 μL of zymolysin (1 U each) at a temperature of 37 °C for an hour. Then, 5 µL of proteinase K was added to the solution and incubated at 60 °C for 20 min without the homogenization stage. Next, 800–900 µL of lysis buffer (CTAB, 0.04% SDS) was added up to 1 mL of the final solution and incubated for another hour at a temperature of 60 °C. Then, DNA was extracted by adding a 1× volume of chloroform, stirring, centrifugation at room temperature for 30 min at 13,400 rpm, and collecting the upper aqueous fraction, which was placed in a clean tube. Next, DNA was precipitated by adding an equal volume of isopropanol and 0.1× v/v potassium acetate, followed by centrifugation at 4° C for 30 min at 15,000 rpm. Then, the DNA pellet was washed twice with 700 µL of 70% ethanol and dissolved in 20 µL of nuclease-free water. The approximate time to isolate DNA from six samples is 5 h and 15 min.
- Enzymatic treatment with lysozyme and zymolysin followed theisolation protocol from [55], excluding mechanical treatment (which combines the first and second methods). The approximate time to isolate DNA from six samples is 4 h and 45 min.
4.5. PCR and Sequencing
4.6. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.M.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Soltis, D.E.; Mabberley, D.J.; Sennikov, A.N.; Soltis, P.S.; et al. An Update of the Angiosperm Phylogeny Group Classification for the Orders and Families of Flowering Plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Soreng, R.J.; Peterson, P.M.; Zuloaga, F.O.; Romaschenko, K.; Clark, L.G.; Teisher, J.K.; Gillespie, L.J.; Barberá, P.; Welker, C.A.D.; Kellogg, E.A.; et al. A Worldwide Phylogenetic Classification of the Poaceae (Gramineae) III: An Update. J. Syst. Evol. 2022, 60, 476–521. [Google Scholar] [CrossRef]
- Alexeev, Y.E. The Family Poaceae. In Maevskyi P.F. Flora of the Middle Part of European Russia; KMK: Moscow, Russia, 2014; pp. 509–569. ISBN 5-04-099221-1. [Google Scholar]
- Linder, H.P.; Rudall, P.J. Evolutionary History of Poales. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 107–124. [Google Scholar] [CrossRef]
- Kellogg, E.A. Poaceae. In The Families and Genera of Vascular Plants; Springer: Berlin, Germany, 2015; Volume XIII, pp. 1–416. [Google Scholar]
- Smart, I.J.; Tuddenham, W.G.; Knox, R.B. Aerobiology of Grass Pollen in the City Atmosphere of Melbourne: Effects of Weather Parameters and Pollen Sources. Aust. J. Bot. 1979, 27, 333–342. [Google Scholar] [CrossRef]
- Prieto-Baena, J.C.; Hidalgo, P.J.; Domínguez, E.; Galán, C. Pollen Production in the Poaceae Family. Grana 2003, 42, 153–159. [Google Scholar] [CrossRef]
- Piotrowska, K. Pollen Production in Selected Species of Anemophilous Plants. Acta Agrobot. 2008, 61, 41–52. [Google Scholar] [CrossRef]
- Aboulaich, N.; Bouziane, H.; Kadiri, M.; Mar Trigo, M.; Riadi, H.; Kazzaz, M.; Merzouki, A. Pollen Production in Anemophilous Species of the Poaceae Family in Tetouan (NW Morocco). Aerobiologia 2009, 25, 27–38. [Google Scholar] [CrossRef]
- Tormo-Molina, R.; Maya-Manzano, J.-M.; Silva-Palacios, I.; Fernández-Rodríguez, S.; Gonzalo-Garijo, Á. Flower Production and Phenology in Dactylis glomerata. Aerobiologia 2015, 31, 469–479. [Google Scholar] [CrossRef]
- Severova, E.; Kopylov-Guskov, Y.; Selezneva, Y.; Karaseva, V.; Yadav, S.R.; Sokoloff, D. Pollen Production of Selected Grass Species in Russia and India at the Levels of Anther, Flower and Inflorescence. Plants 2022, 11, 285. [Google Scholar] [CrossRef]
- D’Amato, G.; Cecchi, L.; Bonini, S.; Nunes, C.; Annesi-Maesano, I.; Behrendt, H.; Liccardi, G.; Popov, T.; Van Cauwenberge, P. Allergenic Pollen and Pollen Allergy in Europe. Allergy 2007, 62, 976–990. [Google Scholar] [CrossRef]
- García-Mozo, H. Poaceae Pollen as the Leading Aeroallergen Worldwide: A Review. Allergy 2017, 72, 1849–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damialis, A.; Traidl-Hoffmann, C.; Treudler, R. Climate Change and Pollen Allergies. In Biodiversity and Health in the Face of Climate Change; Springer: Berlin, Germany, 2019; pp. 47–66. [Google Scholar]
- Weber, R.W. Patterns of Pollen Cross-Allergenicity. J. Allergy Clin. Immunol. 2003, 112, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowney, F.M.; Brennan, G.L.; Skjøth, C.A.; Griffith, G.W.; McInnes, R.N.; Clewlow, Y.; Adams-Groom, B.; Barber, A.; de Vere, N.; Economou, T.; et al. Environmental DNA Reveals Links between Abundance and Composition of Airborne Grass Pollen and Respiratory Health. Curr. Biol. 2021, 31, 1995–2003.E4. [Google Scholar] [CrossRef] [PubMed]
- Leiferman, K.M.; Gleich, G.J. The Cross-Reactivity of IgE Antibodies with Pollen Allergens: I. Analyses of Various Species of Grass Pollens. J. Allergy Clin. Immunol. 1976, 58, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hrabina, M.; Peltre, G.; Van Ree, R.; Moingeon, P. Grass Pollen Allergens. Clin. Exp. Allergy Rev. 2008, 8, 7–11. [Google Scholar] [CrossRef]
- Andersson, K.; Lidholm, J. Characteristics and Immunobiology of Grass Pollen Allergens. Int. Arch. Allergy Immunol. 2003, 130, 87–107. [Google Scholar] [CrossRef]
- Valenta, R.; Duchene, M.; Ebner, C.; Valent, P.; Sillaber, C.; Deviller, P.; Ferreira, F.; Tejkl, M.; Edelmann, H.; Kraft, D. Profilins Constitute a Novel Family of Functional Plant Pan-Allergens. J. Exp. Med. 1992, 175, 377–385. [Google Scholar] [CrossRef] [Green Version]
- Laffer, S.; Spitzauer, S.; Susani, M.; Pairleitnera, H.; Schweiger, C.; Grönlundd, H.; Menz, G.; Pauli, G.; Ishii, T.; Nolte, H. Comparison of Recombinant Timothy Grass Pollen Allergens with Natural Extract for Diagnosis of Grass Pollen Allergy in Different Populations. J. Allergy Clin. Immunol. 1996, 98, 652–658. [Google Scholar] [CrossRef]
- Jutel, M.; Jaeger, L.; Suck, R.; Meyer, H.; Fiebig, H.; Cromwell, O. Allergen-Specific Immunotherapy with Recombinant Grass Pollen Allergens. J. Allergy Clin. Immunol. 2005, 116, 608–613. [Google Scholar] [CrossRef]
- Chabre, H.; Gouyon, B.; Huet, A.; Boran-Bodo, V.; Nony, E.; Hrabina, M.; Fenaille, F.; Lautrette, A.; Bonvalet, M.; Maillere, B. Molecular Variability of Group 1 and 5 Grass Pollen Allergens between Pooideae Species: Implications for Immunotherapy. Clin. Exp. Allergy 2010, 40, 505–519. [Google Scholar] [CrossRef]
- Weger, L.A.; Beerthuizen, T.; Gast-Strookman, J.M.; Plas, D.T.; Terreehorst, I.; Hiemstra, P.S.; Sont, J.K. Difference in Symptom Severity between Early and Late Grass Pollen Season in Patients with Seasonal Allergic Rhinitis. Clin. Transl. Allergy 2011, 1, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nony, E.; Timbrell, V.; Hrabina, M.; Boutron, M.; Solley, G.; Moingeon, P.; Davies, J.M. Specific IgE Recognition of Pollen Allergens from Subtropic Grasses in Patients from the Subtropics. Ann. Allergy. Asthma. Immunol. 2015, 114, 214–220.e2. [Google Scholar] [CrossRef] [PubMed]
- Kailaivasan, T.H.; Timbrell, V.L.; Solley, G.; Smith, W.B.; McLean-Tooke, A.; van Nunen, S.; Smith, P.; Upham, J.W.; Langguth, D.; Davies, J.M. Biogeographical Variation in Specific IgE Recognition of Temperate and Subtropical Grass Pollen Allergens in Allergic Rhinitis Patients. Clin. Transl. Immunol. 2020, 9, e01103. [Google Scholar] [CrossRef] [PubMed]
- Galán, C.; Smith, M.; Thibaudon, M.; Frenguelli, G.; Oteros, J.; Gehrig, R.; Berger, U.; Clot, B.; Brandao, R. Pollen Monitoring: Minimum Requirements and Reproducibility of Analysis. Aerobiologia 2014, 30, 385–395. [Google Scholar] [CrossRef]
- Driessen, M.N.B.M.; Willemse, M.T.M.; Van Luijn, J.A.G. Grass Pollen Grain Determination by Light- and UV-Microscopy. Grana 1989, 28, 115–122. [Google Scholar] [CrossRef]
- Latorre, F.; Bianchi, M.M. Relationships between Flowering Development of Ulmus pumila and Fraxinus excelsior and Their Airborne Pollen. Grana 1998, 37, 233–238. [Google Scholar] [CrossRef]
- Jato, V.; Méndez, J.; Rodríguez-Rajo, J.; Seijo, C. The Relationship between the Flowering Phenophase and Airborne Pollen of Betula in Galicia (NW Spain). Aerobiologia 2002, 18, 55–64. [Google Scholar] [CrossRef]
- Orlandi, F.; Ruga, L.; Romano, B.; Fornaciari, M. An Integrated Use of Aerobiological and Phenological Data to Analyse Flowering in Olive Groves. Grana 2005, 44, 51–56. [Google Scholar] [CrossRef]
- Damialis, A.; Charalampopoulos, A.; Lazarina, M.; Diamanti, E.; Almpanidou, V.; Maraidoni, A.M.; Symeonidou, A.; Staikidou, E.; Syropoulou, E.; Leontidou, K.; et al. Plant Flowering Mirrored in Airborne Pollen Seasons? Evidence from Phenological Observations in 14 Woody Taxa. Atmos. Environ. 2020, 240, 117708. [Google Scholar] [CrossRef]
- León-Ruiz, E.; Alcázar, P.; Domínguez-Vilches, E.; Galán, C. Study of Poaceae Phenology in a Mediterranean Climate. Which Species Contribute Most to Airborne Pollen Counts? Aerobiologia 2011, 27, 37–50. [Google Scholar] [CrossRef]
- Frenguelli, G.; Passalacqua, G.; Bonini, S.; Fiocchi, A.; Incorvaia, C.; Marcucci, F.; Tedeschini, E.; Canonica, G.W.; Frati, F. Bridging Allergologic and Botanical Knowledge in Seasonal Allergy: A Role for Phenology. Ann. Allergy. Asthma. Immunol. 2010, 105, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Tormo, R.; Silva, I.; Gonzalo, Á.; Moreno, A.; Pérez, R.; Fernández, S. Phenological Records as a Complement to Aerobiological Data. Int. J. Biometeorol. 2011, 55, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Cebrino, J.; Galán, C.; Domínguez-Vilches, E. Aerobiological and Phenological Study of the Main Poaceae Species in Córdoba City (Spain) and the Surrounding Hills. Aerobiologia 2016, 32, 595–606. [Google Scholar] [CrossRef]
- Kmenta, M.; Bastl, K.; Kramer, M.F.; Hewings, S.J.; Mwange, J.; Zetter, R.; Berger, U. The Grass Pollen Season 2014 in Vienna: A Pilot Study Combining Phenology, Aerobiology and Symptom Data. Sci. Total Environ. 2016, 566–567, 1614–1620. [Google Scholar] [CrossRef]
- Ghitarrini, S.; Galán, C.; Frenguelli, G.; Tedeschini, E. Phenological Analysis of Grasses (Poaceae) as a Support for the Dissection of Their Pollen Season in Perugia (Central Italy). Aerobiologia 2017, 33, 339–349. [Google Scholar] [CrossRef]
- Núñez, A.; Amo de Paz, G.; Rastrojo, A.; García, A.M.; Alcamí, A.; Gutiérrez-Bustillo, A.M.; Moreno, D.A. Monitoring of Airborne Biological Particles in Outdoor Atmosphere. Part 2: Metagenomics Applied to Urban Environments. Int. Microbiol. 2016, 19, 69–80. [Google Scholar] [CrossRef]
- Kraaijeveld, K.; De Weger, L.A.; Ventayol García, M.; Buermans, H.; Frank, J.; Hiemstra, P.S.; Den Dunnen, J.T. Efficient and Sensitive Identification and Quantification of Airborne Pollen Using Next-generation DNA Sequencing. Mol. Ecol. Resour. 2015, 15, 8–16. [Google Scholar] [CrossRef]
- Leontidou, K.; Vernesi, C.; De Groeve, J.; Cristofolini, F.; Vokou, D.; Cristofori, A. DNA Metabarcoding of Airborne Pollen: New Protocols for Improved Taxonomic Identification of Environmental Samples. Aerobiologia 2018, 34, 63–74. [Google Scholar] [CrossRef]
- Brennan, G.L.; Potter, C.; De Vere, N.; Griffith, G.W.; Skjøth, C.A.; Osborne, N.J.; Wheeler, B.W.; McInnes, R.N.; Clewlow, Y.; Barber, A. Temperate Airborne Grass Pollen Defined by Spatio-Temporal Shifts in Community Composition. Nat. Ecol. Evol. 2019, 3, 750–754. [Google Scholar] [CrossRef] [Green Version]
- Banchi, E.; Ametrano, C.G.; Tordoni, E.; Stanković, D.; Ongaro, S.; Tretiach, M.; Pallavicini, A.; Muggia, L.; Verardo, P.; Tassan, F. Environmental DNA Assessment of Airborne Plant and Fungal Seasonal Diversity. Sci. Total Environ. 2020, 738, 140249. [Google Scholar] [CrossRef]
- Longhi, S.; Cristofori, A.; Gatto, P.; Cristofolini, F.; Grando, M.S.; Gottardini, E. Biomolecular Identification of Allergenic Pollen: A New Perspective for Aerobiological Monitoring? Ann. Allergy. Asthma. Immunol. 2009, 103, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Leontidou, K.; Vokou, D.; Sandionigi, A.; Bruno, A.; Lazarina, M.; De Groeve, J.; Li, M.; Varotto, C.; Girardi, M.; Casiraghi, M.; et al. Plant Biodiversity Assessment through Pollen DNA Metabarcoding in Natura 2000 Habitats (Italian Alps). Sci. Rep. 2021, 11, 18226. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, R.P.; Buchheim, M.A.; Anderson, J.; Levetin, E. Molecular Analysis Confirms the Long-Distance Transport of Juniperus ashei Pollen. PLoS ONE 2017, 12, e0173465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghitarrini, S.; Pierboni, E.; Rondini, C.; Tedeschini, E.; Tovo, G.R.; Frenguelli, G.; Albertini, E. New Biomolecular Tools for Aerobiological Monitoring: Identification of Major Allergenic Poaceae Species through Fast Real-time PCR. Ecol. Evol. 2018, 8, 3996–4010. [Google Scholar] [CrossRef]
- Campbell, B.C.; Al Kouba, J.; Timbrell, V.; Noor, M.J.; Massel, K.; Gilding, E.K.; Angel, N.; Kemish, B.; Hugenholtz, P.; Godwin, I.D. Tracking Seasonal Changes in Diversity of Pollen Allergen Exposure: Targeted Metabarcoding of a Subtropical Aerobiome. Sci. Total Environ. 2020, 747, 141189. [Google Scholar] [CrossRef]
- Uetake, J.; Tobo, Y.; Kobayashi, S.; Tanaka, K.; Watanabe, S.; DeMott, P.J.; Kreidenweis, S.M. Visualization of the Seasonal Shift of a Variety of Airborne Pollens in Western Tokyo. Sci. Total Environ. 2021, 788, 147623. [Google Scholar] [CrossRef]
- Korpelainen, H.; Pietiläinen, M. Biodiversity of Pollen in Indoor Air Samples as Revealed by DNA Metabarcoding. Nord. J. Bot. 2017, 35, 602–608. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.D.; Fokar, M.; Cox, R.D.; Barnes, M.A. Airborne Environmental DNA Metabarcoding Detects More Diversity, with Less Sampling Effort, than a Traditional Plant Community Survey. BMC Ecol. Evol. 2021, 21, 218. [Google Scholar] [CrossRef]
- Polling, M.; Sin, M.; de Weger, L.A.; Speksnijder, A.G.C.L.; Koenders, M.J.F.; de Boer, H.; Gravendeel, B. DNA Metabarcoding Using NrITS2 Provides Highly Qualitative and Quantitative Results for Airborne Pollen Monitoring. Sci. Total Environ. 2022, 806, 150468. [Google Scholar] [CrossRef]
- Alonso, A.; Bull, R.D.; Acedo, C.; Gillespie, L.J. Design of Plant-Specific PCR Primers for the ETS Region with Enhanced Specificity for Tribe Bromeae and Their Application to Other Grasses (Poaceae). Botany 2014, 92, 693–699. [Google Scholar] [CrossRef]
- Wang, A.; Gopurenko, D.; Wu, H.; Lepschi, B. Evaluation of Six Candidate DNA Barcode Loci for Identification of Five Important Invasive Grasses in Eastern Australia. PloS ONE 2017, 12, e0175338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omelchenko, D.O.; Krinitsina, A.A.; Kasianov, A.S.; Speranskaya, A.S.; Chesnokova, O.V.; Polevova, S.V.; Severova, E.E. Assessment of ITS1, ITS2, 5′-ETS, and TrnL-F DNA Barcodes for Metabarcoding of Poaceae Pollen. Diversity 2022, 14, 191. [Google Scholar] [CrossRef]
- Bell, K.L.; Burgess, K.S.; Botsch, J.C.; Dobbs, E.K.; Read, T.D.; Brosi, B.J. Quantitative and Qualitative Assessment of Pollen DNA Metabarcoding Using Constructed Species Mixtures. Mol. Ecol. 2019, 28, 431–455. [Google Scholar] [CrossRef]
- Severova, E.; Volkova, O. Sampling Height in Aerobiological Monitoring. In Proceedings of the 11th International Congress on Aerobiology, Parma, Italy, 3–7 September 2018; Programme & Abstract book; Parma Chamber of Commerce Congress: Parma, OH, USA, 2008; p. 74. [Google Scholar]
- Hirst, J. An Automatic Volumetric Spore Trap. Ann. Appl. Biol. 1952, 39, 257–265. [Google Scholar] [CrossRef]
- Meier, U. Growth Stages of Mono-and Dicotyledonous Plants; Blackwell Wissenschafts-Verlag: Berlin/Heidelberg, Germany, 1997; ISBN 3-8263-3152-4. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Speranskaya, A.S.; Khafizov, K.; Ayginin, A.A.; Krinitsina, A.A.; Omelchenko, D.O.; Nilova, M.V.; Severova, E.E.; Samokhina, E.N.; Shipulin, G.A.; Logacheva, M.D. Comparative Analysis of Illumina and Ion Torrent High-Throughput Sequencing Platforms for Identification of Plant Components in Herbal Teas. Food Control 2018, 93, 315–324. [Google Scholar] [CrossRef]
- Omelchenko, D.O.; Speranskaya, A.S.; Ayginin, A.A.; Khafizov, K.; Krinitsina, A.A.; Fedotova, A.V.; Pozdyshev, D.V.; Shtratnikova, V.Y.; Kupriyanova, E.V.; Shipulin, G.A. Improved Protocols of ITS1-Based Metabarcoding and Their Application in the Analysis of Plant-Containing Products. Genes 2019, 10, 122. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics. 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]

| Method | C, ng/µL | OD 260/280 | OD 260/230 |
|---|---|---|---|
| 1 | 0.6 ± 0.12 | 1.78 ± 0.28 | 0.24 ± 0.09 |
| 2 | 0.75 ± 0.52 | 1.62 ± 0.16 | 0.56 ± 0.27 |
| 3 | 0.71 ± 0.13 | 1.75 ± 0.19 | 0.57 ± 0.48 |
| 4a | 0.42 ± 0.02 | 1.16 ± 0.67 | 0.68 ± 0.55 |
| 4b | 0.94 ± 0.69 | 1.77 ± 0.2 | 0.78 ± 0.52 |
| 4c | 1.13 ± 0.23 | 1.89 ± 0.13 | 2.09 ± 0.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krinitsina, A.A.; Omelchenko, D.O.; Kasianov, A.S.; Karaseva, V.S.; Selezneva, Y.M.; Chesnokova, O.V.; Shirobokov, V.A.; Polevova, S.V.; Severova, E.E. Aerobiological Monitoring and Metabarcoding of Grass Pollen. Plants 2023, 12, 2351. https://doi.org/10.3390/plants12122351
Krinitsina AA, Omelchenko DO, Kasianov AS, Karaseva VS, Selezneva YM, Chesnokova OV, Shirobokov VA, Polevova SV, Severova EE. Aerobiological Monitoring and Metabarcoding of Grass Pollen. Plants. 2023; 12(12):2351. https://doi.org/10.3390/plants12122351
Chicago/Turabian StyleKrinitsina, Anastasia A., Denis O. Omelchenko, Artem S. Kasianov, Vera S. Karaseva, Yulia M. Selezneva, Olga V. Chesnokova, Vitaly A. Shirobokov, Svetlana V. Polevova, and Elena E. Severova. 2023. "Aerobiological Monitoring and Metabarcoding of Grass Pollen" Plants 12, no. 12: 2351. https://doi.org/10.3390/plants12122351
APA StyleKrinitsina, A. A., Omelchenko, D. O., Kasianov, A. S., Karaseva, V. S., Selezneva, Y. M., Chesnokova, O. V., Shirobokov, V. A., Polevova, S. V., & Severova, E. E. (2023). Aerobiological Monitoring and Metabarcoding of Grass Pollen. Plants, 12(12), 2351. https://doi.org/10.3390/plants12122351






