Leaf Proteomic Analysis in Seedlings of Two Maize Landraces with Different Tolerance to Boron Toxicity
Abstract
1. Introduction
2. Results
2.1. Classification into Several Functional Categories of Differentially Accumulated Proteins in Both Maize Landraces and B Treatments
2.2. Differentially Expressed Proteins in Sama and Pachía in Response to B Toxicity
3. Discussion
3.1. Several Proteases and Translation-Related Proteins Allow Pachía to Survive in Media with B Excess
3.2. Proteins That Can Confer More B Toxicity Tolerance to Sama
Lower Repression of Photosynthesis-Related Proteins Can Enhance the B Toxicity Tolerance of Sama
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Protein Extraction and Digestion
4.3. Shotgun-DDA-LC-MS/MS Analysis
4.4. Protein Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warington, K. The effect of boric acid and borax on the broad bean and certain other plants. Ann. Bot. 1923, 37, 629–672. [Google Scholar] [CrossRef]
- Princi, M.P.; Lupini, A.; Araniti, F.; Longo, C.; Mauceri, A.; Sunseri, F.; Abenavoli, M.R. Boron toxicity and tolerance in plants: Recent advances and future perspectives. In Plant Metal Interaction: Emerging Remediation Techniques; Ahmad, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 115–147. [Google Scholar]
- González-Fontes, A.; Fujiwara, T. Advances in plant boron. Int. J. Mol. Sci. 2020, 21, 4107. [Google Scholar] [CrossRef] [PubMed]
- González-Fontes, A. Why boron is an essential element for vascular plants. New Phytol. 2020, 226, 1228–1230. [Google Scholar] [CrossRef]
- Wimmer, M.A.; Abreu, I.; Bell, R.W.; Bienert, M.D.; Brown, P.H.; Dell, B.; Fujiwara, T.; Goldbach, H.E.; Lehto, T.; Mock, H.-P.; et al. Boron: An essential element for vascular plants. New Phytol. 2020, 226, 1232–1237. [Google Scholar] [CrossRef]
- Ishii, T.; Matsunaga, T. Isolation and characterization of a boron-rhamnogalacturonan-II complex from cell walls of sugar beet pulp. Carbohydr. Res. 1996, 284, 1–9. [Google Scholar] [CrossRef]
- Kobayashi, M.; Matoh, T.; Azuma, J. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1996, 110, 1017–1020. [Google Scholar] [CrossRef]
- O’Neill, M.A.; Warrenfeltz, D.; Kates, K.; Pellerin, P.; Doco, T.; Darvill, A.G.; Albersheim, P. Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. J. Biol. Chem. 1996, 271, 22923–22930. [Google Scholar] [CrossRef]
- Cakmak, I.; Römheld, V. Boron deficiency-induced impairments of cellular functions in plants. Plant Soil 1997, 193, 71–83. [Google Scholar] [CrossRef]
- Goldbach, H.E.; Yu, Q.; Wingender, R.; Schulz, M.; Wimmer, M.; Findeklee, P.; Baluška, F. Rapid response reactions of roots to boron deprivation. J. Plant Nutr. Soil Sci. 2001, 164, 173–181. [Google Scholar] [CrossRef]
- Brown, P.H.; Bellaloui, N.; Wimmer, M.A.; Bassil, E.S.; Ruiz, J.; Hu, H.; Pfeffer, H.; Dannel, F.; Römheld, V. Boron in plant biology. Plant Biol. 2002, 4, 205–223. [Google Scholar] [CrossRef]
- Voxeur, A.; Fry, S.C. Glycosylinositol phosphorylceramides from Rosa cell cultures are boron-bridged in the plasma membrane and form complexes with rhamnogalacturonan II. Plant J. 2014, 79, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yang, C.; Pan, Z.; Liu, Y.; Peng, S. Boron deficiency in woody plants: Various responses and tolerance mechanisms. Front. Plant Sci. 2015, 6, 916. [Google Scholar] [CrossRef]
- Blevins, D.G.; Lukaszewski, K.M. Boron in plant structure and function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 481–500. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Cristóbal, J.J.; Navarro-Gochicoa, M.T.; Rexach, J.; González-Fontes, A.; Herrera-Rodríguez, M.B. Plant response to boron deficiency and boron use efficiency in crop plants. In Plant Micronutrient Use Efficiency. Molecular and Genomic Perspectives in Crop Plants; Hossain, M.A., Kamiya, T., Burritt, D.J., Phan Tran, L.-S., Fujiwara, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 109–121. [Google Scholar]
- Shireen, F.; Nawaz, M.A.; Chen, C.; Zhang, Q.; Zheng, Z.; Sohail, H.; Sun, J.; Cao, H.; Huang, Y.; Bie, Z. Boron: Functions and approaches to enhance its availability in plants for sustainable agriculture. Int. J. Mol. Sci. 2018, 19, 1856. [Google Scholar] [CrossRef]
- Reid, R.J.; Hayes, J.E.; Post, A.; Stangoulis, J.C.R.; Graham, R.D. A critical analysis of the causes of boron toxicity in plants. Plant Cell Environ. 2004, 27, 1405–1414. [Google Scholar] [CrossRef]
- Camacho-Cristóbal, J.J.; Rexach, J.; González-Fontes, A. Boron in plants: Deficiency and toxicity. J. Integr. Plant Biol. 2008, 50, 1247–1255. [Google Scholar] [CrossRef]
- Beato, V.M.; Rexach, J.; Navarro-Gochicoa, M.T.; Camacho-Cristóbal, J.J.; Herrera-Rodríguez, M.B.; Maldonado, J.M.; González-Fontes, A. A tobacco asparagine synthetase gene responds to carbon and nitrogen status and its root expression is affected under boron stress. Plant Sci. 2010, 178, 289–298. [Google Scholar] [CrossRef]
- Beato, V.M.; Navarro-Gochicoa, M.T.; Rexach, J.; Herrera-Rodríguez, M.B.; Camacho-Cristóbal, J.J.; Kempa, S.; Weckwerth, W.; González-Fontes, A. Expression of root glutamate dehydrogenase genes in tobacco plants subjected to boron deprivation. Plant Physiol. Biochem. 2011, 49, 1350–1354. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.-W.; Guo, L.-X.; Liu, Y.-Z.; Jin, L.-F.; Hussain, S.B.; Du, W.; Deng, Z.; Peng, S.-A. Transcriptome changes associated with boron deficiency in leaves of two citrus scion-rootstock combinations. Front. Plant Sci. 2017, 8, 317. [Google Scholar] [CrossRef]
- Landi, M.; Margaritopoulou, T.; Papadakis, I.E.; Araniti, F. Boron toxicity in higher plants: An update. Planta 2019, 250, 1011–1032. [Google Scholar] [CrossRef]
- Brdar-Jokanovi’c, M. Boron toxicity and deficiency in agricultural plants. Int. J. Mol. Sci. 2020, 21, 1424. [Google Scholar] [CrossRef]
- Kabay, N.; Güler, E.; Bryjak, M. Boron in seawater and methods for its separation—A review. Desalination 2010, 261, 212–217. [Google Scholar] [CrossRef]
- Chatzistathis, T.; Fanourakis, D.; Aliniaeifard, S.; Kotsiras, A.; Delis, C.; Tsaniklidis, G. Leaf age-dependent effects of boron toxicity in two Cucumis melo varieties. Agronomy 2021, 11, 759. [Google Scholar] [CrossRef]
- Uluisik, I.; Karakaya, H.C.; Koc, A. The importance of boron in biological systems. J. Trace Elem. Med. Biol. 2018, 45, 156–162. [Google Scholar] [CrossRef]
- Mamani-Huarcaya, B.M.; González-Fontes, A.; Navarro-Gochicoa, M.T.; Camacho-Cristóbal, J.J.; Ceacero, C.J.; Herrera-Rodríguez, M.B.; Fernández Cutire, Ó.; Rexach, J. Characterization of two Peruvian maize landraces differing in boron toxicity tolerance. Plant Physiol. Biochem. 2022, 185, 167–177. [Google Scholar] [CrossRef]
- Behera, B.; Kancheti, M.; Raza, M.B.; Shiv, A.; Mangal, V.; Rathod, G.; Altaf, M.A.; Kumar, A.; Aftab, T.; Kumar, R.; et al. Mechanistic insight on boron-mediated toxicity in plant vis-a-vis its mitigation strategies: A review. Int. J. Phytoremediat. 2023, 25, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Ralston, N.V.C.; Hunt, C.D. Diadenosine phosphates and S-adenosylmethionine: Novel boron binding biomolecules detected by capillary electrophoresis. Biochim. Biophys. Acta 2001, 1527, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Pandey, P.; Stoerger, V.; Xu, Y.; Qiu, Y.; Ge, Y.; Schnable, J.C. Conventional and hyperspectral timeseries imaging of maize lines widely used in field trials. GigaScience 2018, 72, gix117. [Google Scholar]
- Andorf, C.; Beavis, W.D.; Hufford, M.; Smith, S.; Suza, W.P.; Wang, K.; Woodhouse, M.; Yu, J.; Lübberstedt, T. Technological advances in maize breeding: Past, present, and future. Theor. Appl. Genet. 2019, 132, 817–849. [Google Scholar] [CrossRef]
- Kausch, A.P.; Wang, K.; Kaeppler, H.F.; Gordon-Kamm, W. Maize transformation: History, progress, and perspectives. Mol. Breed. 2021, 41, 38. [Google Scholar] [CrossRef]
- Xi, Y.; Hu, W.; Zhou, Y.; Liu, X.; Qian, Y. Genome-wide identification and functional analysis of polyamine oxidase genes in maize reveal essential roles in abiotic stress tolerance. Front. Plant Sci. 2022, 13, 950064. [Google Scholar] [CrossRef]
- Ogunwole, A.A.; Otusanya, O.O.; Oloyede, F.A.; Olabamiji, T.M. Comparative effects of boron toxicity and deficiency on the growth, chlorophyll, protein and some cations accumulation in Zea mays seedlings. Int. J. Sci. Res. Innov. 2015, 17, 316–335. [Google Scholar]
- Gotz, L.F.; Silvestrin, F.; Motta, A.C.V.; Pauletti, V. Response to application and tissue diagnosis of boron deficiency and toxicity in maize. Commun. Soil Sci. Plant Anal. 2021, 52, 2898–2911. [Google Scholar] [CrossRef]
- de Abreu Neto, J.B.; Hurtado-Perez, M.C.; Wimmer, M.A.; Frei, M. Genetic factors underlying boron toxicity tolerance in rice: Genome-wide association study and transcriptomic analysis. J. Exp. Bot. 2017, 68, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Kayıhan, C.; Öz, M.T.; Eyidoğan, F.; Yücel, M.; Öktem, H.A. Physiological, biochemical, and transcriptomic responses to boron toxicity in leaf and root tissues of contrasting wheat cultivars. Plant Mol. Biol. Rep. 2017, 35, 97–109. [Google Scholar] [CrossRef]
- Pandey, A.; Khan, M.K.; Hamurcu, M.; Brestic, M.; Topal, A.; Gezgin, S. Insight into the root transcriptome of a boron-tolerant Triticum zhukovskyi genotype grown under boron toxicity. Agronomy 2022, 12, 2421. [Google Scholar] [CrossRef]
- Sang, W.; Huang, Z.-R.; Yang, L.-T.; Guo, P.; Ye, X.; Chen, L.-S. Effects of high toxic boron concentration on protein profiles in roots of two citrus species differing in boron-tolerance revealed by a 2-DE based MS approach. Front. Plant Sci. 2017, 8, 180. [Google Scholar] [CrossRef]
- Fan, T.; Bykova, N.V.; Rampitsch, C.; Xing, T. Identification and characterization of a serine protease from wheat leaves. Eur. J. Plant Pathol. 2016, 146, 293–304. [Google Scholar] [CrossRef]
- Malefo, M.B.; Mathibela, E.O.; Crampton, B.G.; Makgopa, M.E. Investigating the role of Bowman-Birk serine protease inhibitor in Arabidopsis plants under drought stress. Plant Physiol. Biochem. 2020, 149, 286–293. [Google Scholar] [CrossRef]
- D’Ippolito, S.; Rey-Burusco, M.F.; Feingold, S.E.; Guevara, M.G. Role of proteases in the response of plants to drought. Plant Physiol. Biochem. 2021, 168, 1–9. [Google Scholar] [CrossRef]
- Sharma, P.; Gayen, D. Plant protease as regulator and signaling molecule for enhancing environmental stress-tolerance. Plant Cell Rep. 2021, 40, 2081–2095. [Google Scholar] [CrossRef]
- Moin, M.; Bakshi, A.; Saha, A.; Dutta, M.; Madhav, S.M.; Kirti, P.B. Rice ribosomal protein large subunit genes and their spatio-temporal and stress regulation. Front. Plant Sci. 2016, 7, 1284. [Google Scholar] [CrossRef]
- Nozawa, A.; Miwa, K.; Kobayashi, M.; Fujiwara, T. Isolation of Arabidopsis thaliana cDNAs that confer yeast boric acid tolerance. Biosci. Biotechnol. Biochem. 2006, 70, 1724–1730. [Google Scholar] [CrossRef]
- Tanaka, M.; Sotta, N.; Yamazumi, Y.; Yamashita, Y.; Miwa, K.; Murota, K.; Chiba, Y.; Hirai, M.Y.; Akiyama, T.; Onouchi, H.; et al. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 2016, 28, 2830–2849. [Google Scholar] [CrossRef]
- Sotta, N.; Chiba, Y.; Miwa, K.; Takamatsu, S.; Tanaka, M.; Yamashita, Y.; Naito, S.; Fujiwara, T. Global analysis of boron-induced ribosome stalling reveals its effects on translation termination and unique regulation by AUG-stops in Arabidopsis shoots. Plant J. 2021, 106, 1455–1467. [Google Scholar] [CrossRef]
- Saidi, A.; Hajibarat, Z. In-silico analysis of eukaryotic translation initiation factors (eIFs) in response to environmental stresses in rice (Oryza sativa). Biologia 2020, 75, 1731–1738. [Google Scholar] [CrossRef]
- Pakdel, H.; Hassani, S.B.; Ghotbi-Ravandi, A.A.; Bernard, F. Contrasting the expression pattern change of polyamine oxidase genes and photosynthetic efficiency of maize (Zea mays L.) genotypes under drought stress. J. Biosci. 2020, 45, 73. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Guo, S.-R.; Sun, J.; Yuan, L.-Y. Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. Physiol. Plant 2012, 146, 285–296. [Google Scholar] [CrossRef]
- Hamdani, S.; Yaakoubi, H.; Carpentier, R. Polyamines interaction with thylakoid proteins during stress. J. Photochem. Photobiol. B Biol. 2011, 104, 314–331. [Google Scholar] [CrossRef]
- Chen, M.; Blankenship, R.E. Expanding the solar spectrum used by photosynthesis. Trends Plant Sci. 2011, 16, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chang, E.; Yu, X.; Chen, Y.; Yang, Q.; Cao, Y.; Li, X.; Wang, Y.; Fu, A.; Xu, M. Molecular characterization of magnesium chelatase in soybean [Glycine max (L.) Merr.]. Front. Plant Sci. 2018, 9, 720. [Google Scholar] [CrossRef]
- Sasi, S.; Venkatesh, J.; Daneshi, R.F.; Gururani, M.A. Photosystem II extrinsic proteins and their putative role in abiotic stress tolerance in higher plants. Plants 2018, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Portis, A.R. A novel nucleus-encoded chloroplast protein, PIFI, is involved in NAD(P)H dehydrogenase complex-mediated chlororespiratory electron transport in Arabidopsis. Plant Physiol. 2007, 144, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, N.; Takabayashi, A.; Noguchi, K.; Tazoe, Y.; Yamamoto, H.; von Caemmerer, S.; Sato, F.; Endo, T. NDH-mediated cyclic electron flow around photosystem I is crucial for C4 photosynthesis. Plant Cell Physiol. 2016, 57, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, Y.; Bai, C.; Yong, J.W.H. The significance of chloroplast NAD(P)H dehydrogenase complex and its dependent cyclic electron transport in photosynthesis. Front. Plant Sci. 2021, 12, 661863. [Google Scholar] [CrossRef]
- Yamori, W.; Shikanai, T. Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu. Rev. Plant Biol. 2016, 67, 81–106. [Google Scholar] [CrossRef]
- Zhu, D.; Luo, F.; Zou, R.; Liu, J.; Yan, Y. Integrated physiological and chloroplast proteome analysis of wheat seedling leaves under salt and osmotic stresses. J. Proteom. 2021, 234, 104097. [Google Scholar] [CrossRef]
- Murata, N.; Nishiyama, Y. ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ. 2018, 41, 285–299. [Google Scholar] [CrossRef]
- Araniti, F.; Miras-Moreno, B.; Lucini, L.; Landi, M.; Abenavoli, M.R. Metabolomic, proteomic and physiological insights into the potential mode of action of thymol, a phytotoxic natural monoterpenoid phenol. Plant Physiol. Biochem. 2020, 153, 141–153. [Google Scholar] [CrossRef]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
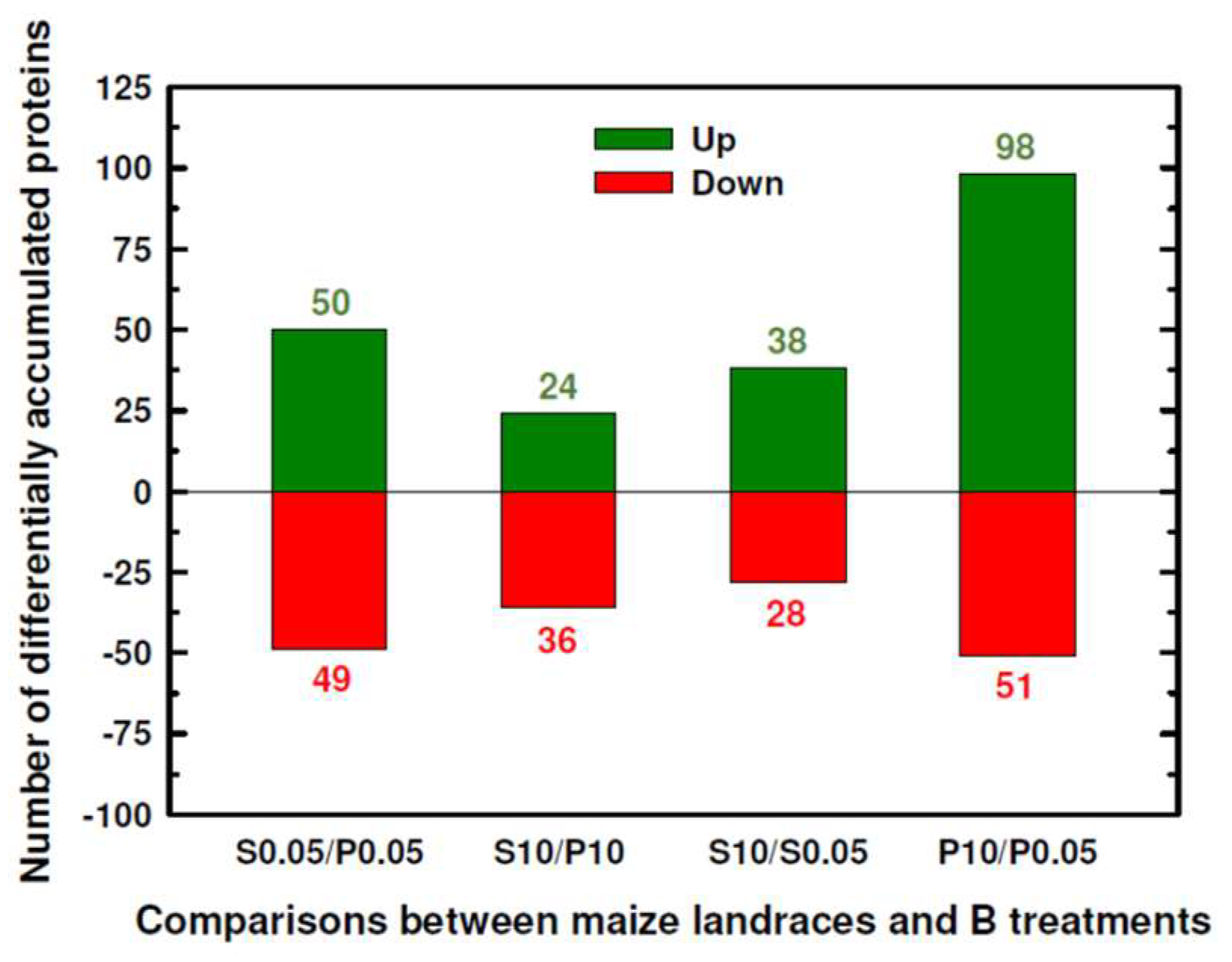
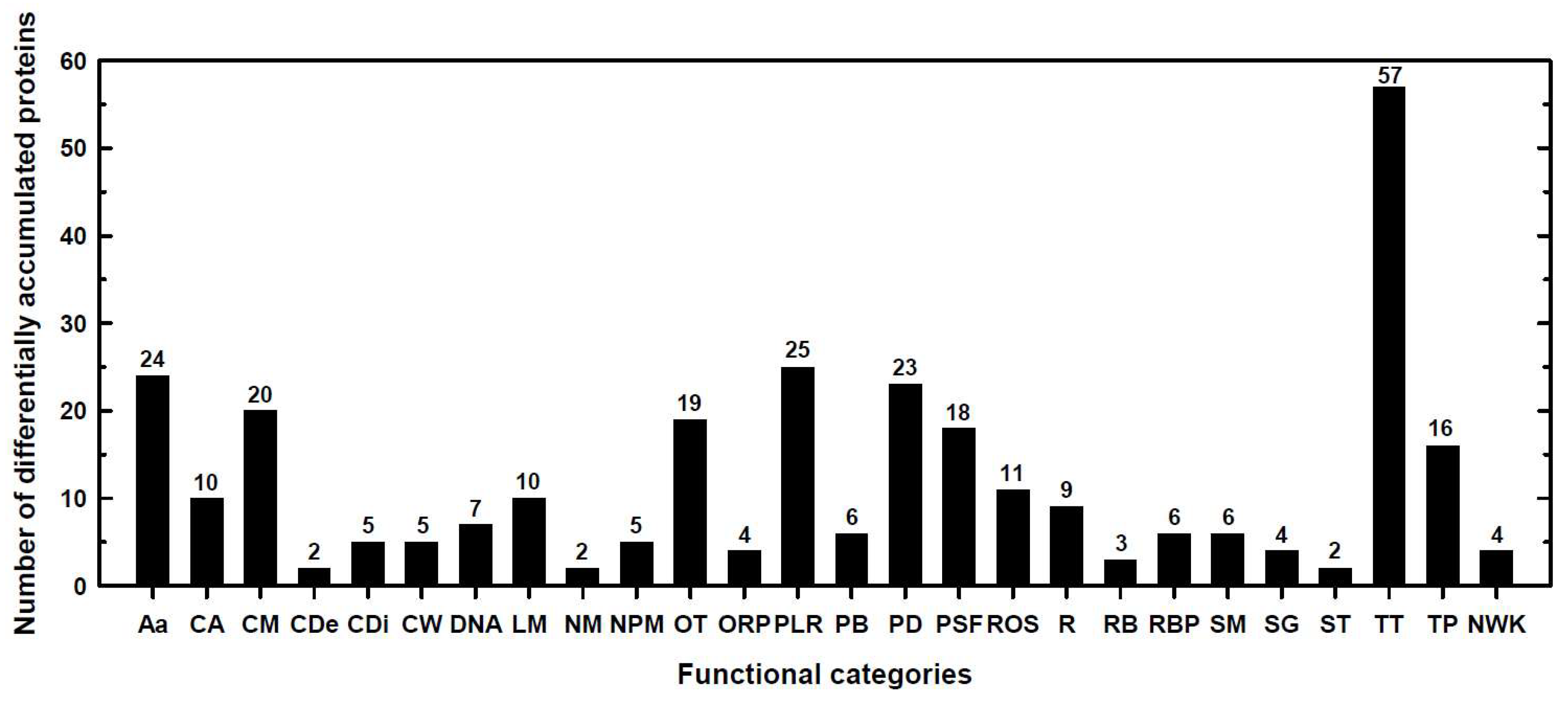
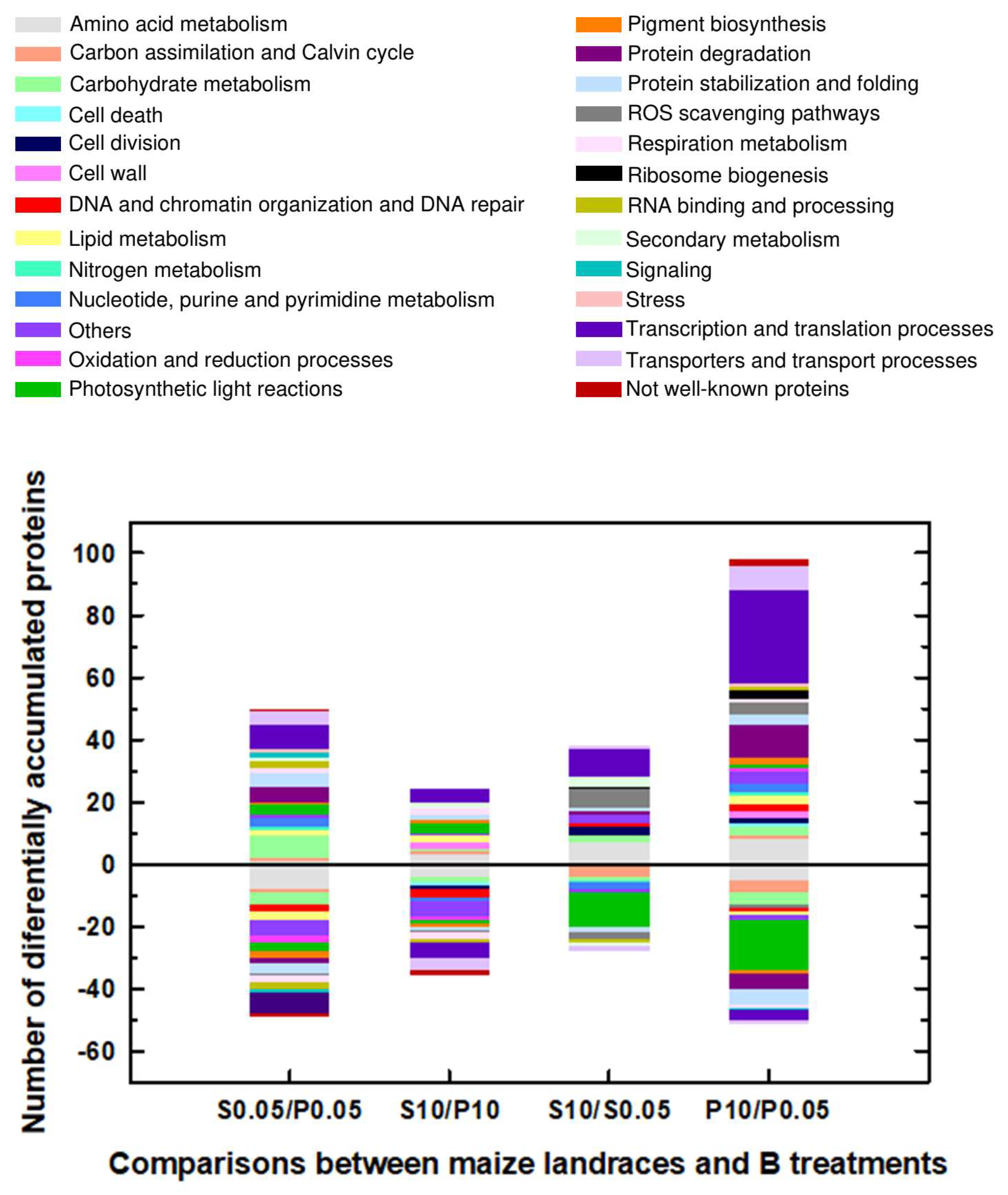
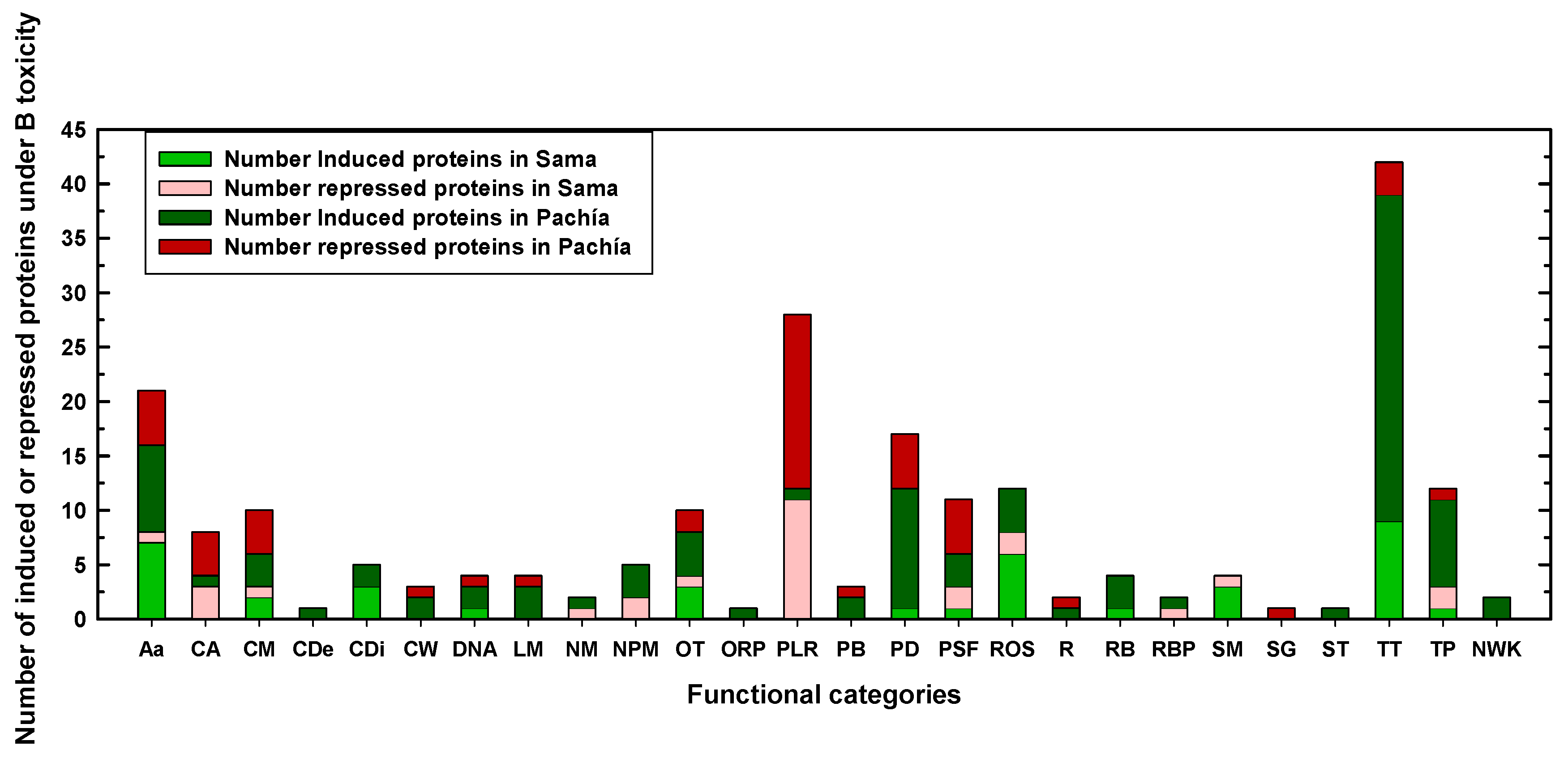
| P0.05 mM (Control) | P10 mM B (B Toxicity) | S0.05 mM (Control) | S10 mM (B Toxicity) | |
|---|---|---|---|---|
| Number of detected proteins 1 | 1100 | 1040 | 1111 | 1145 |
| S0.05 versus P0.05 (control conditions) | S10 versus P10 (B toxicity conditions) | |||
| Number of significant DAPs between Sama and Pachía | 99 | 60 | ||
| Sama S10 versus S0.05 | Pachía P10 versus P0.05 | |||
| Number of significant DAPs by B toxicity | 66 | 149 | ||
| Pachía | Sama | |||||||
|---|---|---|---|---|---|---|---|---|
| Protein ID 1 | Gene Name/ID 2 | Protein Name/ Annotation | FC 3 | p-Value 4 | FC 3 | p-Value 4 | FCSA/ FCPA 5 | Function/Biological Process 6 |
| AMINO ACID METABOLISM | ||||||||
| B6SKB7 | Zm00001d031013 | Methylcrotonoyl-CoA carboxylase subunit α | 4.44 | 0.0022 | 3.56 | 0.0049 | 0.80 | Leucine degradation |
| A0A1D6K836 | Zm00001d029848 | Branched-chain amino-acid aminotransferase | 2.35 | 0.0272 | 1.65 | 0.0241 | 0.70 | Branched-chain amino acid biosynthesis |
| B4G011 | Zm00001d046923 | d-3-phosphoglycerate dehydrogenase chloroplastic | 2.31 | 0.0154 | 1.52 | 0.0202 | 0.66 | Serine biosynthesis |
| A0A1D6DW07 | Zm00001d002051 | d-3-phosphoglycerate dehydrogenase | 1.78 | 0.0494 | 1.69 | 0.0175 | 0.95 | Serine biosynthesis |
| CARBON ASSIMILATION/CALVIN CYCLE | ||||||||
| O24574 | Zm00001d004894 | Ribulose bisphosphate carboxylase small chain | 0.38 | 0.0113 | 0.33 | 0.0466 | 0.87 | Carbon dioxide fixation |
| CARBOHYDRATE METABOLISM | ||||||||
| Q9FQ11 | Zm00001d010523 | Sucrose-phosphatase 1 | 1.50 | 0.0154 | 1.58 | 0.0420 | 1.05 | Sucrose biosynthesis |
| A0A1D6IJ76 | Zm00001d022107 | Glyceraldehyde-3-phosphate dehydrogenase A | 0.34 | 0.0319 | 0.51 | 0.0019 | 1.52 | Carbon metabolism |
| CELL DIVISION | ||||||||
| A0A1D6FRI4 | Zm00001d010500 | ERBB-3 binding protein 1 | 1.89 | 0.0387 | 1.58 | 0.0266 | 0.84 | Cell division and cell growth regulation |
| PHOTOSYNTHETIC LIGHT REACTIONS | ||||||||
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 0.27 | 0.0110 | 0.48 | 0.0354 | 1.78 | Photosynthesis. Photosystem II oxygen-evolving complex |
| B4FWG2 | Zm00001d048422 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.25 | 0.0047 | 0.41 | 0.0200 | 1.62 | Photosynthetic electron transport flow around photosystem I to produce ATP |
| A0A1X7YHG9 | AtpA | ATP synthase subunit α chloroplastic (ATPα) | 0.20 | 0.0166 | 0.61 | 0.0163 | 2.99 | Chloroplast ATP synthesis-coupled proton transport |
| P46617 | PetA | Cytochrome f | 0.18 | 0.0193 | 0.29 | 0.0161 | 1.59 | Photosynthetic electron transport activity |
| P00827 | Zm00001d006403 | ATP synthase subunit β chloroplastic (ATPβ) | 0.15 | 0.0076 | 0.52 | 0.0274 | 3.45 | Chloroplast ATP synthesis-coupled proton transport |
| REACTIVE OXYGEN SPECIES (ROS) SCAVENGING PATHWAYS/RESPONSE TO OXIDATIVE STRESS | ||||||||
| A0A1D6MSE3 | Zm00001d040721 | Dihydrolipoyl dehydrogenase | 2.30 | 0.0273 | 1.80 | 0.0205 | 0.78 | Cell redox homeostasis |
| A0A1D6JPH3 | Zm00001d027769 | Glutathione reductase | 2.21 | 0.0053 | 1.71 | 0.0436 | 0.77 | Cell redox homeostasis. Glutathione metabolic process. Cellular oxidant detoxification |
| RIBOSOME BIOGENESIS | ||||||||
| K7UTH7 | Zm00001d009596 | GTPase ERA1 chloroplastic | 2.61 | 0.0108 | 1.81 | 0.0126 | 0.69 | Ribosome biogenesis. Ribosomal small subunit assembly. rRNA processing |
| TRANSCRIPTION AND TRANSLATION PROCESSES | ||||||||
| A0A1D6LIV5 | Zm00001d035802 | Phenylalanine–tRNA ligase beta subunit cytoplasmic | 2.56 | 0.0314 | 2.23 | 0.0093 | 0.87 | Translation. Phenylalanyl-tRNA aminoacylation |
| TRANSPORTERS AND TRANSPORT PROCESSES | ||||||||
| B6SP43 | Zm00001d007597 | ABC family1 | 4.54 | 0.0103 | 2.69 | 0.0125 | 0.59 | ATPase-coupled transmembrane transporter activity |
| Protein ID 1 | Gene Name/ID 2 | Protein Name/Annotation | FC 3 | p-Value 4 | Function/Biological Process 5 |
|---|---|---|---|---|---|
| AMINO ACID AND PEPTIDE METABOLISMS | |||||
| Proteins very strongly induced by B toxicity in Pachía | |||||
| B6SKB7 | Zm00001d031013 | Methylcrotonoyl-CoA carboxylase subunit α | 4.44 | 0.0022 | Leucine degradation |
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| A0A1D6ICL3 | Zm00001d021596 | Adenosine 5-phosphosulfate reductase-like1 | 0.29 | 0.0140 | Cysteine biosynthetic process. Sulfate reduction |
| B6TZD1 | Zm00001eb168430 | Methylthioribose-1-phosphate isomerase | 0.24 | 0.0461 | Methionine biosynthesis |
| CARBON ASSIMILATION AND CALVIN CYCLE | |||||
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| B4FQ59 | Zm00001d017711 | Phosphoribulokinase | 0.33 | 0.0004 | Calvin–Benson cycle |
| Q9ZT00 | Zm00001eb164390 | Ribulose bisphosphate carboxylase/oxygenase activase chloroplastic | 0.26 | 0.0090 | Carbon dioxide fixation. Rubisco activator activity |
| CELL WALL | |||||
| Proteins very strongly induced by B toxicity in Pachía | |||||
| B4F9J1 | Zm00001d046357 | β-galactosidase | 3.17 | 0.0092 | Xyloglucan degradation |
| DNA AND CHROMATIN ORGANIZATION AND DNA REPAIR | |||||
| Proteins very strongly induced by B toxicity in Pachía | |||||
| B6TGH8 | Zm00001d034479 | Histone H1 | 3.60 | 0.0349 | Chromosome condensation. Nucleosome assembly. Nucleosome positioning |
| C0P6Q6 | Zm00001d040416 | DNA gyrase subunit B | 3.48 | 0.0007 | DNA topological change |
| LIPID METABOLISM | |||||
| Proteins very strongly repressed by B toxicity | |||||
| B4FLS8 | Zm00001d003584 | 12-oxo-phytodienoic acid reductase 5 | 0.33 | 0.0436 | Fatty acid and oxylipin biosynthesis |
| OTHERS | |||||
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| C0PE12 | Zm00001d009877 | Protein plastid transcriptionally active 16 chloroplastic | 0.24 | 0.0121 | Circadian rhythm |
| PHOTOSYNTHETIC LIGHT REACTIONS | |||||
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| B6SP99 | Zm00001d024148 | Photosynthetic NDH subunit of subcomplex B 1 chloroplastic | 0.33 | 0.0137 | Photosynthetic electron transport in photosystem I |
| B4FJP7 | Zm00001d027729 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.32 | 0.0169 | Photosynthetic electron transport in photosystem I |
| B4FR80 | Zm00001d033098 | Post-illumination chlorophyll fluorescence increase (ZmPIFI) | 0.28 | 0.0270 | Chlororespiration |
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 0.27 | 0.0110 | Photosynthesis. Photosystem II oxygen-evolving complex |
| B4FWG2 | Zm00001d048422 | Photosynthetic NDH subunit of subcomplex B 2 chloroplastic | 0.25 | 0.0047 | Photosynthetic electron transport flow around photosystem I to produce ATP |
| P19124 | NdhJ | NAD(P)H-quinone oxidoreductase subunit J, chloroplastic | 0.22 | 0.0147 | Photosynthesis, light reaction, photosynthetic electron transport chain. Couples the photosynthetic redox reaction to proton translocation |
| A0A1X7YHG9 | AtpA | ATP synthase subunit α (ATPα) | 0.20 | 0.0166 | Chloroplast ATP synthesis-coupled proton transport |
| P46617 | PetA | Cytochrome f | 0.18 | 0.0193 | Photosynthetic electron transport chain |
| P00827 | Zm00001d009488 | ATP synthase subunit β, chloroplastic (ATPβ) | 0.15 | 0.0076 | Chloroplast ATP synthesis-coupled proton transport |
| A0A1D6JYG6 | Zm00001d028670 | Photosynthetic NDH subunit of lumenal location 1 chloroplastic | 0.13 | 0.0134 | Part of photosystem II oxygen-evolving complex |
| PROTEIN DEGRADATION | |||||
| Proteins very strongly induced by B toxicity in Pachía | |||||
| B4FS65 | Zm00001d005391 | Cysteine protease 14 | 4.38 | 0.0146 | Proteolysis. Proteolysis involved in protein catabolic process |
| A0A1D6HM49 | Zm00001d018282 | Subtilisin-like protease SBT1.4 | 3.70 | 0.0399 | Serine protease. Serine-type endopeptidase activity. Proteolysis |
| A0A1D6H4R4 | Zm00001d015962 | Prolyl oligopeptidase family protein | 3.58 | 0.0080 | Proteolysis. Serine protease. Serine-type peptidase activity |
| PROTEIN STABILIZATION AND FOLDING | |||||
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| G2XK63 | Zm00001d040257 | T-complex protein 1 subunit β | 0.27 | 0.0065 | Protein folding. Chaperone |
| B4FR04 | Zm00001d019052 | Peptidylprolyl isomerase | 0.23 | 0.0205 | Protein folding. Rotamase |
| SIGNALING | |||||
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| P49235 | Zm00001eb411380 | 4-hydroxy-7-methoxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-2-yl glucoside beta-d-glucosidase 1, chloroplastic | 0.19 | 0.0090 | Cytokinin signaling pathway |
| TRANSCRIPTION AND TRANSLATION PROCESSES | |||||
| Proteins very strongly induced by B toxicity in Pachía | |||||
| A0A1D6LEN8 | Zm00001d035139 | MA3 domain-containing protein | 4.95 | 0.0073 | Negative regulation of transcription, DNA-templated. Regulation of translation |
| Q6R9D1 | GRMZM5G806488 | Ribosomal protein S7 | 3.89 | 0.0202 | Translation. Ribosomal small subunit assembly. Structural constituent of ribosomes |
| A0A1D6IAN8 | Zm00001d021400 | Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein/tetratricopeptide repeat (TPR)-containing protein | 3.47 | 0.0323 | RNA processing |
| C0P456 | Zm00001d002789 | Pentatricopeptide repeat-containing protein | 3.26 | 0.0259 | Likely involved in post-transcriptional control of gene expression in organelles |
| Proteins very strongly repressed by B toxicity in Pachía | |||||
| O50018 | Zm00001d046449 | Elongation factor 1-α | 0.29 | 0.0269 | Translation. Translation elongation factor activity |
| TRANSPORTERS AND TRANSPORT PROCESSES | |||||
| Proteins very strongly induced by B toxicity in Pachía | |||||
| B6SP43 | Zm00001d007597 | ABC family1 | 4.54 | 0.0103 | ATPase-coupled transmembrane transporter activity |
| A0A1D6H2R4 | Zm00001d015569 | H+-exporting diphosphatase | 4.34 | 0.0050 | Ion transport. Pyrophosphate hydrolysis-driven proton transmembrane transporter activity |
| A0A1D6MS70 | Zm00001d040686 | Protein translocase subunit SECA1 chloroplastic | 4.12 | 0.0173 | Protein transport |
| A0A1D6DSW6 | Zm00001d001788 | K+ efflux antiporter 2 chloroplastic | 3.79 | 0.0414 | Chloroplast potassium ion trans-port |
| B6T5R1 | Zm00001d010504 | Ran-binding protein 1 | 3.16 | 0.0492 | Intracellular transport. Protein and mRNA transport. Nucleocytoplasmic transport |
| Protein ID 1 | Gene Name/ID 2 | Protein Name/Annotation | FC 3 | p-Value 4 | Function/Biological Process 5 |
|---|---|---|---|---|---|
| AMINO ACID AND PEPTIDE METABOLISMS | |||||
| Proteins very strongly induced by B toxicity in Sama | |||||
| B6SKB7 | Zm00001d031013 | Methylcrotonoyl-CoA carboxylase subunit alpha | 3.56 | 0.0049 | Leucine degradation |
| CARBON ASSIMILATION AND CALVIN CYCLE | |||||
| Proteins very strongly repressed by B toxicity in Sama | |||||
| O24574 | Zm00001d004894 | Ribulose bisphosphate carboxylase small chain | 0.33 | 0.0466 | Carbon dioxide fixation |
| P05348 | Rbcs | Ribulose bisphosphate carboxylase small chain, chloroplastic | 0.13 | 0.0096 | Carbon dioxide fixation |
| CELL DIVISION | |||||
| Proteins very strongly induced by B toxicity in Sama | |||||
| C0P4T2 | Zm00001d042664 | Patellin-1 | 3.05 | 0.0149 | Cell division and cell cycle |
| PHOTOSYNTHETIC LIGHT REACTIONS | |||||
| Proteins very strongly repressed by B toxicity in Sama | |||||
| P46617 | PetA | Cytochrome f | 0.29 | 0.0161 | Photosynthetic electron transport chain |
| B6SQV5 | Zm00001d049387 | Photosystem II 10 kDa polypeptide | 0.14 | 0.0438 | Photosynthesis. Photosystem II oxygen-evolving complex |
| PROTEIN STABILIZATION AND FOLDING | |||||
| Proteins very strongly repressed by B toxicity in Sama | |||||
| C4J6Y2 | Zm00001d018077 | Peptidylprolyl isomerase | 0.18 | 0.0422 | Protein folding. Rotamase |
| REACTIVE OXYGEN SPECIES (ROS) SCAVENGING PATHWAYS/RESPONSE TO OXIDATIVE STRESS | |||||
| Proteins very strongly repressed by B toxicity in Sama | |||||
| B4FZ35 | Zm00001d002240 | CHL-Zea mays chloroplastic lipocalin | 0.31 | 0.0272 | Response to oxidative stress. Violaxanthin, antheraxanthin, and zeaxanthin interconversion |
| SECONDARY METABOLISM | |||||
| Proteins very strongly induced by B toxicity in Sama | |||||
| O64411 | Zm00001d024281 | Polyamine oxidase 1 (PAO1) | 3.34 | 0.0108 | Spermine degradation. Amine and polyamine degradation |
| Protein ID 1 | Gene Name/ID 2 | Protein Name/Annotation | FC 3 | p-Value 4 | Function/Biological Process 5 |
|---|---|---|---|---|---|
| AMINO ACID AND PEPTIDE METABOLISMS | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6ICL3 | Zm00001d021596 | Adenosine 5-phosphosulfate reductase-like1 | 2.33 | 0.0417 | Cysteine biosynthetic process. Sulfate reduction |
| CARBON ASSIMILATION AND CALVIN CYCLE | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6EXF1 | Zm00001d006520 | PDK regulatory protein1 | 2.16 | 0.0167 | Regulation of C4 photosynthetic carbon assimilation cycle |
| CARBOHYDRATE METABOLISM | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| Q9SYS1 | Zm00001d021702 | β-amylase | 2.63 | 0.0499 | β-amylase activity. Starch degradation |
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6K5L6 | Zm00001d029502 | Glucose-6-phosphate 1-dehydrogenase | 0.36 | 0.0411 | Pentose phosphate pathway |
| A0A1D6LY56 | Zm00001d037480 | Alkaline α galactosidase 2 | 0.33 | 0.0438 | Carbohydrate metabolic process |
| CELL DEATH | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6JNJ8 | Zm00001d027656 | Lethal leaf-spot 1 | 0.32 | 0.0016 | Cell death. Chlorophyll catabolic process |
| CELL DIVISION | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6JH24 | Zm00001d026532 | Protein RCC2 | 0.42 | 0.0214 | Cell division |
| CELL WALL | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6MWZ7 | Zm00001d041578 | Glossy6 | 3.27 | 0.0403 | Epicuticular wax accumulation. Intracellular trafficking of cuticular waxes |
| DNA AND CHROMATIN ORGANIZATION AND DNA REPAIR | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| B4FQA5 | Zm00001d018981 | Histone1a | 0.35 | 0.0318 | Chromosome condensation. Nucleosome assembly |
| B6TGH8 | Zm00001d034479 | Histone H1 | 0.31 | 0.0138 | Chromosome condensation. Nucleosome assembly. Nucleosome positioning |
| LIPID METABOLISM | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| Q8W0V2 | Zm00001d033623 | Lipoxygenase 3 | 5.06 | 0.0455 | Fatty acid and oxylipin biosynthesis |
| Q06XS3 | Zm00001d053675 | Lipoxygenase 10 | 3.44 | 0.0247 | Fatty acid and oxylipin biosynthesis |
| OTHERS | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| B6TY16 | Zm00001d040331 | SUN domain protein2 | 0.41 | 0.0262 | Nuclear envelope organization |
| B4F7V3 | Zm00001d021582 | Protein phosphatase 2C isoform ε | 0.39 | 0.0214 | Protein dephosphorylation |
| A0A1D6HUN3 | Zm00001d019040 | D-2-hydroxyglutarate dehydrogenase mitochondrial | 0.33 | 0.0024 | Photorespiration |
| OXIDATION AND REDUCTION PROCESSES | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| B4F987 | Zm00001d020984 | Putative sarcosine oxidase | 0.23 | 0.0321 | Sarcosine oxidase activity |
| PHOTOSYNTHETIC LIGHT REACTIONS | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| B4FR80 | Zm00001d033098 | Post-illumination chlorophyll fluorescence increase (ZmPIFI) | 2.52 | 0.0097 | Chlororespiration |
| A0A1D6HS38 | Zm00001d018779 | Oxygen-evolving enhancer protein 2-1 chloroplastic (OEE2-1) | 2.31 | 0.0325 | Photosynthesis. Photosystem II oxygen-evolving complex |
| PIGMENT BIOSYNTHESIS | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6JHX0 | Zm00001d026603 | Magnesium-chelatase subunit ChlH1 chloroplastic (ChlH1) | 2.90 | 0.0484 | Chlorophyll biosynthetic process |
| PROTEIN STABILIZATION AND FOLDING | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6KE29 | Zm00001d030725 | Heat shock protein 70 | 0.43 | 0.0406 | Protein refolding. Protein folding chaperone. Cellular response to unfolded protein |
| RESPIRATION (GLYCOLISIS, TCA CYCLE AND MITOCHONDRIAL ELECTRON TRANSFER) | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| B4G1C9 | Zm00001d023606 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | 2.04 | 0.0332 | Acetyl-CoA biosynthetic process from pyruvate |
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6MAK9 | Zm00001d038792 | Phosphotransferase | 0.49 | 0.0331 | Glycolysis |
| SECONDARY METABOLISM | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| O64411 | Zm00001d024281 | Polyamine oxidase 1 (PAO1) | 5.15 | 0.0007 | Spermine degradation. Amine and polyamine degradation |
| TRANSCRIPTION AND TRANSLATION | |||||
| Strongly up-accumulated proteins in Sama in media with 10 mM B | |||||
| B4FP25 | Zm00001d047296 | 40S ribosomal protein S19 | 6.38 | 0.0289 | Translation. Structural constituent of ribosome. Ribosomal small subunit assembly |
| B6TDF7 | Zm00001d019898 | Plastid-specific 30S ribosomal protein 2 | 2.31 | 0.0243 | Ribosomal protein. Ribonucleoprotein complex. RNA-binding |
| C0PEC4 | Zm00001d032420 | 30S ribosomal protein S5 chloroplastic | 2.12 | 0.0487 | Translation. Structural constituent of ribosome |
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| B6SX73 | Zm00001d016549 | 60S ribosomal protein L35 | 0.42 | 0.0284 | Translation. Structural constituent of ribosome |
| Q6R9D1 | GRMZM5G806488 | Ribosomal protein S7 | 0.35 | 0.0426 | Translation. Structural constituent of ribosome. Ribosomal small subunit assembly |
| TRANSPORTER AND TRANSPORT PROCESSES | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6H2R4 | Zm00001d015569 | H+-exporting diphosphatase | 0.33 | 0.0169 | Ion transport. Pyrophosphate hydrolysis-driven proton transmembrane transporter activity |
| A0A1D6K7N5 | Zm00001d029762 | Hexose transporter | 0.20 | 0.0439 | Hexose transporter |
| NOT WELL-KNOWN PROTEINS | |||||
| Strongly down-accumulated proteins in Sama in media with 10 mM B | |||||
| A0A1D6KKK1 | Zm00001d031677 | MtN19-like protein | 0.23 | 0.0121 | Not well determined |
| Protein ID 1 | Gene Name/ID 2 | Protein Name/Annotation | Function/Biological Process 3 |
|---|---|---|---|
| DNA AND CHROMATIN ORGANIZATION AND DNA REPAIR | |||
| Proteins exclusively detected in Pachía in 10 mM B | |||
| A0A1D6KX75 | Zm00001d033247 | Nfc103a–nucleosome/chromatin assembly factor C | Nucleosome/chromatin assembly. DNA repair. Chromatin remodeling, regulation of DNA-templated transcription |
| OTHERS | |||
| Proteins exclusively detected in Sama in both B treatments | |||
| K7VAT7 | Zm00001d046569 | Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain | Protein serine/threonine kinase activity. Protein phosphorylation |
| REACTIVE OXYGEN SPECIES (ROS) SCAVENGING PATHWAYS/RESPONSE TO OXIDATIVE STRESS | |||
| Proteins exclusively detected in Pachía in both B treatments | |||
| B4FKV6 | Zm00001d014341 | Peroxidase 54 | Response to oxidative stress. Peroxidase activity |
| TRANSCRIPTION AND TRANSLATION | |||
| Proteins exclusively detected in Pachía in 10 mM B | |||
| A0A096RFR6 | Zm00001d039518 | Eukaryotic translation initiation factor 3 subunit A (eIF3a) | Translation initiation factor activity. Protein synthesis. Formation of cytoplasmic translation initiation complex |
| TRANSPORTER AND TRANSPORT PROCESSES | |||
| Proteins exclusively detected in Pachía in both B treatments | |||
| A0A1D6EU13 | Zm00001d006238 | Calcium lipid binding protein-like | Lipid transport |
| A0A1D6JN64 | Zm00001d027580 | Outer mitochondrial membrane porin1 (ommp1) | Voltage-gated anion channel activity. Inorganic anion transport, transmembrane transport, anion transmembrane transport |
| Proteins exclusively detected in Sama in both B treatments | |||
| Q7Y1W6 | Zm00001d018693 | Pentatricopeptide repeat 2 (PPR2) | Chloroplast translation |
| NOT WELL-KNOWN PROTEINS | |||
| Proteins exclusively detected in Sama in both B treatments | |||
| A0A1D6DWG9 | Zm00001d002089 | Tetratricopeptide repeat (TPR)-like superfamily protein | Unknown |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mamani-Huarcaya, B.M.; Navarro-Gochicoa, M.T.; Herrera-Rodríguez, M.B.; Camacho-Cristóbal, J.J.; Ceacero, C.J.; Fernández Cutire, Ó.; González-Fontes, A.; Rexach, J. Leaf Proteomic Analysis in Seedlings of Two Maize Landraces with Different Tolerance to Boron Toxicity. Plants 2023, 12, 2322. https://doi.org/10.3390/plants12122322
Mamani-Huarcaya BM, Navarro-Gochicoa MT, Herrera-Rodríguez MB, Camacho-Cristóbal JJ, Ceacero CJ, Fernández Cutire Ó, González-Fontes A, Rexach J. Leaf Proteomic Analysis in Seedlings of Two Maize Landraces with Different Tolerance to Boron Toxicity. Plants. 2023; 12(12):2322. https://doi.org/10.3390/plants12122322
Chicago/Turabian StyleMamani-Huarcaya, Betty Maribel, María Teresa Navarro-Gochicoa, María Begoña Herrera-Rodríguez, Juan José Camacho-Cristóbal, Carlos Juan Ceacero, Óscar Fernández Cutire, Agustín González-Fontes, and Jesús Rexach. 2023. "Leaf Proteomic Analysis in Seedlings of Two Maize Landraces with Different Tolerance to Boron Toxicity" Plants 12, no. 12: 2322. https://doi.org/10.3390/plants12122322
APA StyleMamani-Huarcaya, B. M., Navarro-Gochicoa, M. T., Herrera-Rodríguez, M. B., Camacho-Cristóbal, J. J., Ceacero, C. J., Fernández Cutire, Ó., González-Fontes, A., & Rexach, J. (2023). Leaf Proteomic Analysis in Seedlings of Two Maize Landraces with Different Tolerance to Boron Toxicity. Plants, 12(12), 2322. https://doi.org/10.3390/plants12122322






