Pueraria montana Population Structure and Genetic Diversity Based on Chloroplast Genome Data
Abstract
:1. Introduction
2. Results
2.1. Pueraria Montana Plastid Genome Assembly
2.2. Chloroplast Genome Sequence Variation in Pueraria Montana
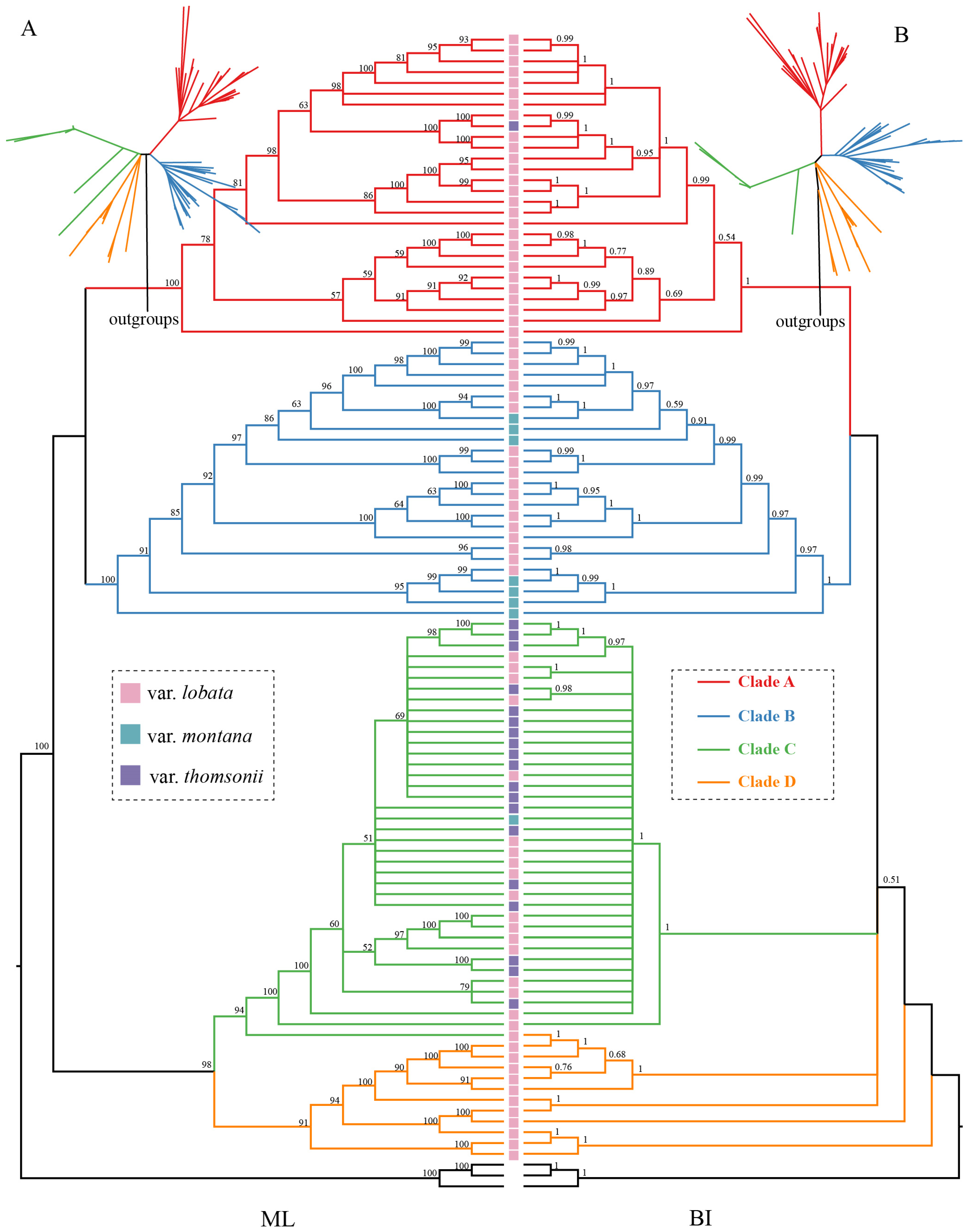
2.3. Pueraria Montana Phylogeny based on the Chloroplast Genome
2.4. Population Structure and PCA of Pueraria Montana
2.5. Pueraria Montana Divergence Time
2.6. Genetic Evolution in Different Groups
3. Discussion
3.1. Highly Variable Pueraria Montana Chloroplast Genome Sequences
3.2. Pueraria Montana Genetic Divergence and Diversity
4. Materials and Methods
4.1. Samples and Chloroplast Genome Sequencing
4.2. Variation Identification and Statistics
4.3. Population Structure and Principal Component Analysis
4.4. Genetic Diversity in P. montana Subclades
4.5. Divergence Time Estimation and Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [Green Version]
- Wicke, S.; Schneeweiss, G.M.; Depamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Chen, Y.-P.; Salmaki, Y.; Drew, B.T.; Wilson, T.C.; Scheen, A.-C.; Celep, F.; Bräuchler, C.; Bendiksby, M.; Wang, Q.; et al. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 2021, 19, 2. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Li, E.; Liu, Y.; Xu, C.; Wang, Y.; Liu, K.; Cui, X.; Sun, J.; Suo, Z.; Zhang, Z.; et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 2022, 20, 92. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, S.; Liu, Y.; Yuan, Q.; Sun, J.; Guo, L. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genom. 2021, 22, 103. [Google Scholar] [CrossRef]
- Li, E.; Liu, K.; Deng, R.; Gao, Y.; Liu, X.; Dong, W.; Zhang, Z. Insights into the phylogeny and chloroplast genome evolution of Eriocaulon (Eriocaulaceae). BMC Plant Biol. 2023, 23, 32. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, K.; Li, E.; Wang, Y.; Xu, C.; Zhao, L.; Dong, W. Dynamic evolution of the plastome in the Elm family (Ulmaceae). Planta 2023, 257, 14. [Google Scholar] [CrossRef]
- Dong, W.; Sun, J.; Liu, Y.; Xu, C.; Wang, Y.; Suo, Z.; Zhou, S.; Zhang, Z.; Wen, J. Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae). J. Syst. Evol. 2022, 60, 1263–1280. [Google Scholar] [CrossRef]
- Dong, W.; Liu, Y.; Li, E.; Xu, C.; Sun, J.; Li, W.; Zhou, S.; Zhang, Z.; Suo, Z. Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mol. Phylogenet. Evol. 2022, 166, 107330. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Garran, T.; Liu, H.; Lin, H.; Luo, J.; Yuan, Q.; Sun, J.; Dong, W.; Guo, L. Genetic diversity and population divergence of Leonurus japonicus and its distribution dynamic changes from the last interglacial to the present in China. BMC Plant Biol. 2023, 23, 276. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- China Plant BOLG; Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Nat. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed] [Green Version]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.S.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef] [Green Version]
- Shang, C.; Li, E.; Yu, Z.; Lian, M.; Chen, Z.; Liu, K.; Xu, L.; Tong, Z.; Wang, M.; Dong, W. Chloroplast Genomic Resources and Genetic Divergence of Endangered Species Bretschneidera sinensis (Bretschneideraceae). Front. Ecol. Evol. 2022, 10, 873100. [Google Scholar] [CrossRef]
- Torre, S.; Sebastiani, F.; Burbui, G.; Pecori, F.; Pepori, A.L.; Passeri, I.; Ghelardini, L.; Selvaggi, A.; Santini, A. Novel Insights Into Refugia at the Southern Margin of the Distribution Range of the Endangered Species Ulmus laevis. Front. Plant Sci. 2022, 13, 826158. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Wang, J.; Mao, Q.; Dong, W.; Yuan, Q.; Guo, L.; Huang, L. Coptis huanjiangensis, a new species of Ranunculaceae from Guangxi, China. PhytoKeys 2022, 213, 131–141. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Zhao, Z.; Xu, C.; Qiao, P.; Wang, S.; Wang, M.; Xu, Z.; Yuan, Q.; Guo, L.; et al. Multiplexed Massively Parallel Sequencing of Plastomes Provides Insights Into the Genetic Diversity, Population Structure, and Phylogeography of Wild and Cultivated Coptis chinensis. Front. Plant Sci. 2022, 13, 923600. [Google Scholar] [CrossRef]
- Perdereau, A.; Klaas, M.; Barth, S.; Hodkinson, T.R. Plastid genome sequencing reveals biogeographical structure and extensive population genetic variation in wild populations of Phalaris arundinacea L. in north-western Europe. GCB Bioenergy 2017, 9, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Geng, F.D.; Li, J.J.; Zhang, D.Q.; Gao, F.; Huang, L.; Zhang, X.H.; Kang, J.Q.; Zhang, J.Q.; Ren, Y. Divergence in the Aquilegia ecalcarata complex is correlated with geography and climate oscillations: Evidence from plastid genome data. Mol. Ecol. 2021, 30, 5796–5813. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, J.; Qiao, P.; Wang, J.; Wang, M.; Du, Y.; Xiong, F.; Luo, J.; Yuan, Q.; Dong, W.; et al. Evolutionary history of genus Coptis and its dynamic changes in the potential suitable distribution area. Front. Plant Sci. 2022, 13, 1003368. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.I.; Hefer, C.A.; Kolosova, N.; Douglas, C.J.; Cronk, Q.C. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 2014, 204, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Ding, Z.; Zhu, Q.; Wu, Y.; Gao, P. Population structure and genetic diversity of watermelon (Citrullus lanatus) based on SNP of chloroplast genome. 3 Biotech 2020, 10, 374. [Google Scholar] [CrossRef]
- Xu, Y.; Liao, B.; Ostevik, K.L.; Zhou, H.; Wang, F.; Wang, B.; Xia, H. The Maternal Donor of Chrysanthemum Cultivars Revealed by Comparative Analysis of the Chloroplast Genome. Front. Plant Sci. 2022, 13, 923442. [Google Scholar] [CrossRef]
- Nock, C.J.; Hardner, C.M.; Montenegro, J.D.; Ahmad Termizi, A.A.; Hayashi, S.; Playford, J.; Edwards, D.; Batley, J. Wild Origins of Macadamia Domestication Identified Through Intraspecific Chloroplast Genome Sequencing. Front. Plant Sci. 2019, 10, 334. [Google Scholar] [CrossRef] [Green Version]
- Tong, W.; Kim, T.-S.; Park, Y.-J. Rice Chloroplast Genome Variation Architecture and Phylogenetic Dissection in Diverse Oryza Species Assessed by Whole-Genome Resequencing. Rice 2016, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Van der Maesen, L. Revision of the Genus Pueraria DC with Some Notes on Teyleria Backer (Leguminosae); Taylor & Francis: London, UK, 1985. [Google Scholar]
- Egan, A.N.; Vatanparast, M.; Cagle, W. Parsing polyphyletic Pueraria: Delimiting distinct evolutionary lineages through phylogeny. Mol. Phylogenet. Evol. 2016, 104, 44–59. [Google Scholar] [CrossRef] [Green Version]
- Haynsen, M.S. Population Genetics of Pueraria montana var. Lobata; The George Washington University: Washington, DC, USA, 2018. [Google Scholar]
- Sun, Y.; Shaw, P.-C.; Fung, K.-P. Molecular Authentication of Radix Puerariae Lobatae and Radix Puerariae Thomsonii by ITS and 5S rRNA Spacer Sequencing. Biol. Pharm. Bull. 2007, 30, 173–175. [Google Scholar] [CrossRef] [Green Version]
- Kartzinel, T.R.; Hamrick, J.L.; Wang, C.; Bowsher, A.W.; Quigley, B.G.P. Heterogeneity of clonal patterns among patches of kudzu, Pueraria montana var. lobata, an invasive plant. Ann. Bot. 2015, 116, 739–750. [Google Scholar] [CrossRef] [Green Version]
- Pappert, R.A.; Hamrick, J.L.; Donovan, L.A. Genetic variation in Pueraria lobata (Fabaceae), an introduced, clonal, invasive plant of the southeastern United States. Am. J. Bot. 2000, 87, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Heider, B.; Fischer, E.; Berndl, T.; Schultze-Kraft, R. Analysis of Genetic Variation Among Accessions of Pueraria montana (Lour.) Merr. var. lobata and Pueraria phaseoloides (Roxb.) Benth. based on RAPD Markers. Genet. Resour. Crop. Evol. 2007, 54, 529–542. [Google Scholar] [CrossRef]
- Chen, D.; Peng, R.; Li, L.; Zhang, X.; Wang, Y. Analysis of genetic relationships of Pueraria thomsonii based on SRAP markers. Zhongguo Zhong Yao Za Zhi China J. Chin. Mater. Med. 2011, 36, 538–541. [Google Scholar]
- Sun, J.H.; Li, Z.C.; Jewett, D.K.; Britton, K.O.; Ye, W.H.; Ge, X.J. Genetic diversity of Pueraria lobata (kudzu) and closely related taxa as revealed by inter-simple sequence repeat analysis. Weed Res. 2005, 45, 255–260. [Google Scholar] [CrossRef]
- Chen, J.; Tan, L.; Zhang, Q.; Guan, Q.; Yang, Y.; Tian, S.; Yang, Z.; Zhu, Z.; Xu, L. Genetic diversity of Pueraria lobata varieties and differences of puerarin analysis in Three Gorges Reservoir Area. Southwest China J. Agric. Sci. 2015, 28, 2334–2336. [Google Scholar]
- Tsia, P.; Ho, S.; Hou, C.; Lin, J.; Li, T. The analysis of genetic diversity of Pueraria montana in Taiwan revealed by ISSR method. J. Taiwan Livest. Res. 2017, 50, 294–302. [Google Scholar]
- Hoffberg, S.L.; Bentley, K.E.; Lee, J.B.; Myhre, K.E.; Iwao, K.; Glenn, T.C.; Mauricio, R. Characterization of 15 microsatellite loci in kudzu (Pueraria montana var. lobata) from the native and introduced ranges. Conserv. Genet. Resour. 2015, 7, 403–405. [Google Scholar] [CrossRef]
- Bentley, K.E.; Mauricio, R. High degree of clonal reproduction and lack of large-scale geographic patterning mark the introduced range of the invasive vine, kudzu (Pueraria montana var. lobata), in North America. Am. J. Bot. 2016, 103, 1499–1507. [Google Scholar] [CrossRef] [Green Version]
- Haynsen, M.S.; Vatanparast, M.; Mahadwar, G.; Zhu, D.; Moger-Reischer, R.Z.; Doyle, J.J.; Crandall, K.A.; Egan, A.N. De novo transcriptome assembly of Pueraria montana var. lobata and Neustanthus phaseoloides for the development of eSSR and SNP markers: Narrowing the US origin(s) of the invasive kudzu. BMC Genom. 2018, 19, 439. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Gupta, V.K.; Misra, A.K.; Modi, D.R.; Pandey, B.K. Potential of Molecular Markers in Plant Biotechnology. Plant Omics 2009, 2, 141–162. [Google Scholar]
- Li, J.; Yang, M.; Li, Y.; Jiang, M.; Liu, C.; He, M.; Wu, B. Chloroplast genomes of two Pueraria DC. species: Sequencing, comparative analysis and molecular marker development. FEBS Open Bio 2022, 12, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Wu, Z.; Shang, X.; Shi, P.; Wei, M.; Wang, H.; Xiao, L.; Cao, S.; Lu, L.; Zeng, W.; et al. Chromosome-level and graphic genomes provide insights into metabolism of bioactive metabolites and cold-adaption of Pueraria lobata var. montana. DNA Res. 2022, 29, dsac030. [Google Scholar] [CrossRef]
- Shang, X.; Yi, X.; Xiao, L.; Zhang, Y.; Huang, D.; Xia, Z.; Ou, K.; Ming, R.; Zeng, W.; Wu, D.; et al. Chromosomal-level genome and multi-omics dataset of Pueraria lobata var. thomsonii provide new insights into legume family and the isoflavone and puerarin biosynthesis pathways. Hortic. Res. 2022, 9, uhab035. [Google Scholar] [CrossRef]
- Zachos, J.C.; Dickens, G.R.; Zeebe, R.E. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature 2008, 451, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Mohamoud, Y.A.; Mathew, L.S.; Torres, M.F.; Younuskunju, S.; Krueger, R.; Suhre, K.; Malek, J.A. Novel subpopulations in date palm (Phoenix dactylifera) identified by population-wide organellar genome sequencing. BMC Genom. 2019, 20, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariotti, R.; Cultrera, N.G.M.; Díez, C.M.; Baldoni, L.; Rubini, A. Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biol. 2010, 10, 211. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Wang, S.; Wang, Y.; Wang, R.; Liu, K.; Li, E.; Qiao, P.; Shi, L.; Dong, W.; Huang, L.; et al. Phylogenomics and Genetic Diversity of Arnebiae Radix and Its Allies (Arnebia, Boraginaceae) in China. Front. Plant Sci. 2022, 13, 920826. [Google Scholar] [CrossRef]
- Sancho, R.; Cantalapiedra, C.P.; Lopez-Alvarez, D.; Gordon, S.P.; Vogel, J.P.; Catalan, P.; Contreras-Moreira, B. Comparative plastome genomics and phylogenomics of Brachypodium: Flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 2018, 218, 1631–1644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohmann, N.; Wolf, E.M.; Rigault, P.; Zhou, W.; Kiefer, M.; Zhao, Y.; Fu, C.-X.; Koch, M.A. Ginkgo biloba’s footprint of dynamic Pleistocene history dates back only 390,000 years ago. BMC Genom. 2018, 19, 299. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Li, J.; Sun, X.; Li, Z.; Jiang, B. Polymorphism analysis of the chloroplast and mitochondrial genomes in soybean. BMC Plant Biol. 2023, 23, 15. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Zhang, P.; Wang, E.; Wang, S.; Qiang, X.; Li, L.; Song, Y.; Chang, H.; Liu, X.; Zhou, W. Changes of the monsoon-arid environment in China and growth of the Tibetan Plateau since the Miocene. Quat. Sci. 2006, 26, 678–693. [Google Scholar]
- An, Z. Late Cenozoic Climate Change in Asia: Loess, Monsoon and Monsoon-Arid Environment Evolution; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Chang, Z.; Xiao, J.; Lü, L.; Yao, H. Abrupt shifts in the Indian monsoon during the Pliocene marked by high-resolution terrestrial records from the Yuanmou Basin in southwest China. J. Asian Earth Sci. 2010, 37, 166–175. [Google Scholar] [CrossRef]
- Corriveau, J.L.; Coleman, A.W. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988, 75, 1443–1458. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.S. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003, 44, 941–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarz, E.N.; Ruhlman, T.A.; Weng, M.-L.; Khiyami, M.A.; Sabir, J.S.M.; Hajarah, N.H.; Alharbi, N.S.; Rabah, S.O.; Jansen, R.K. Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J. Mol. Evol. 2017, 84, 187–203. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J.N.V.; Boore, J.L.; Jansen, R.K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Nat. Acad. Sci. USA 2008, 105, 18424–18429. [Google Scholar] [CrossRef] [Green Version]
- Weng, M.-L.; Blazier, J.C.; Govindu, M.; Jansen, R.K. Reconstruction of the Ancestral Plastid Genome in Geraniaceae Reveals a Correlation between Genome Rearrangements, Repeats, and Nucleotide Substitution Rates. Mol. Biol. Evol. 2014, 31, 645–659. [Google Scholar] [CrossRef] [Green Version]
- Barnard-Kubow, K.B.; Sloan, D.B.; Galloway, L.F. Correlation between sequence divergence and polymorphism reveals similar evolutionary mechanisms acting across multiple timescales in a rapidly evolving plastid genome. BMC Evol. Biol. 2014, 14, 268. [Google Scholar] [CrossRef] [Green Version]
- Keung, W.M. Pueraria: The genus Pueraria; CRC Press: Boca Raton, FL, USA, 2002. [Google Scholar]
- Callen, S.T.; Miller, A.J. Signatures of niche conservatism and niche shift in the North American kudzu (Pueraria montana) invasion. Divers. Distrib. 2015, 21, 853–863. [Google Scholar] [CrossRef]
- Dlugosch, K.M.; Parker, I.M. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008, 17, 431–449. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Huang, D.I.; Cronk, Q.C.B. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015, 3, 1500026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Se-Al: Sequence Alignment Editor; Version 2.0; University of Oxford: Oxford, UK, 1996.
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Vu, V.Q. ggbiplot: A ggplot2 Based biplot. R Package Version 0.55. 2011. Available online: https://github.com/vqv/ggbiplot (accessed on 17 November 2022).
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Suchard, M.; Xie, D.; Drummond, A. Tracer v1. 6. 2014. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 6 December 2022).
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: Estimating gene genealogies. Parallel Distrib. Process. Symp. Int. Proc. 2002, 2, 184. [Google Scholar]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kuhnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comp. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]




| Group | Accessions | All Variations | |||
|---|---|---|---|---|---|
| SNPs | Indels | Total | Density/kb | ||
| Clade A | 28 | 464 | 190 | 654 | 4.09 |
| Clade B | 26 | 422 | 225 | 647 | 4.04 |
| Clade C | 39 | 221 | 104 | 325 | 2.03 |
| Clade D | 11 | 232 | 151 | 383 | 2.39 |
| Total | 104 | 1118 | 556 | 1674 | 10.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Wang, Y.; Qiao, P.; Zhang, L.; Li, E.; Dong, W.; Zhao, Y.; Huang, L. Pueraria montana Population Structure and Genetic Diversity Based on Chloroplast Genome Data. Plants 2023, 12, 2231. https://doi.org/10.3390/plants12122231
Sun J, Wang Y, Qiao P, Zhang L, Li E, Dong W, Zhao Y, Huang L. Pueraria montana Population Structure and Genetic Diversity Based on Chloroplast Genome Data. Plants. 2023; 12(12):2231. https://doi.org/10.3390/plants12122231
Chicago/Turabian StyleSun, Jiahui, Yiheng Wang, Ping Qiao, Lei Zhang, Enze Li, Wenpan Dong, Yuping Zhao, and Luqi Huang. 2023. "Pueraria montana Population Structure and Genetic Diversity Based on Chloroplast Genome Data" Plants 12, no. 12: 2231. https://doi.org/10.3390/plants12122231
APA StyleSun, J., Wang, Y., Qiao, P., Zhang, L., Li, E., Dong, W., Zhao, Y., & Huang, L. (2023). Pueraria montana Population Structure and Genetic Diversity Based on Chloroplast Genome Data. Plants, 12(12), 2231. https://doi.org/10.3390/plants12122231








