Analysis of the Differentially Expressed Proteins and Metabolic Pathways of Honeybush (Cyclopia subternata) in Response to Water Deficit Stress
Abstract
1. Introduction
2. Results
2.1. 1D-SDS-PAGE of C. subternata Protein Samples
2.2. Identification of Induced Proteins in C. subternata Using LC-MS/MS Analysis
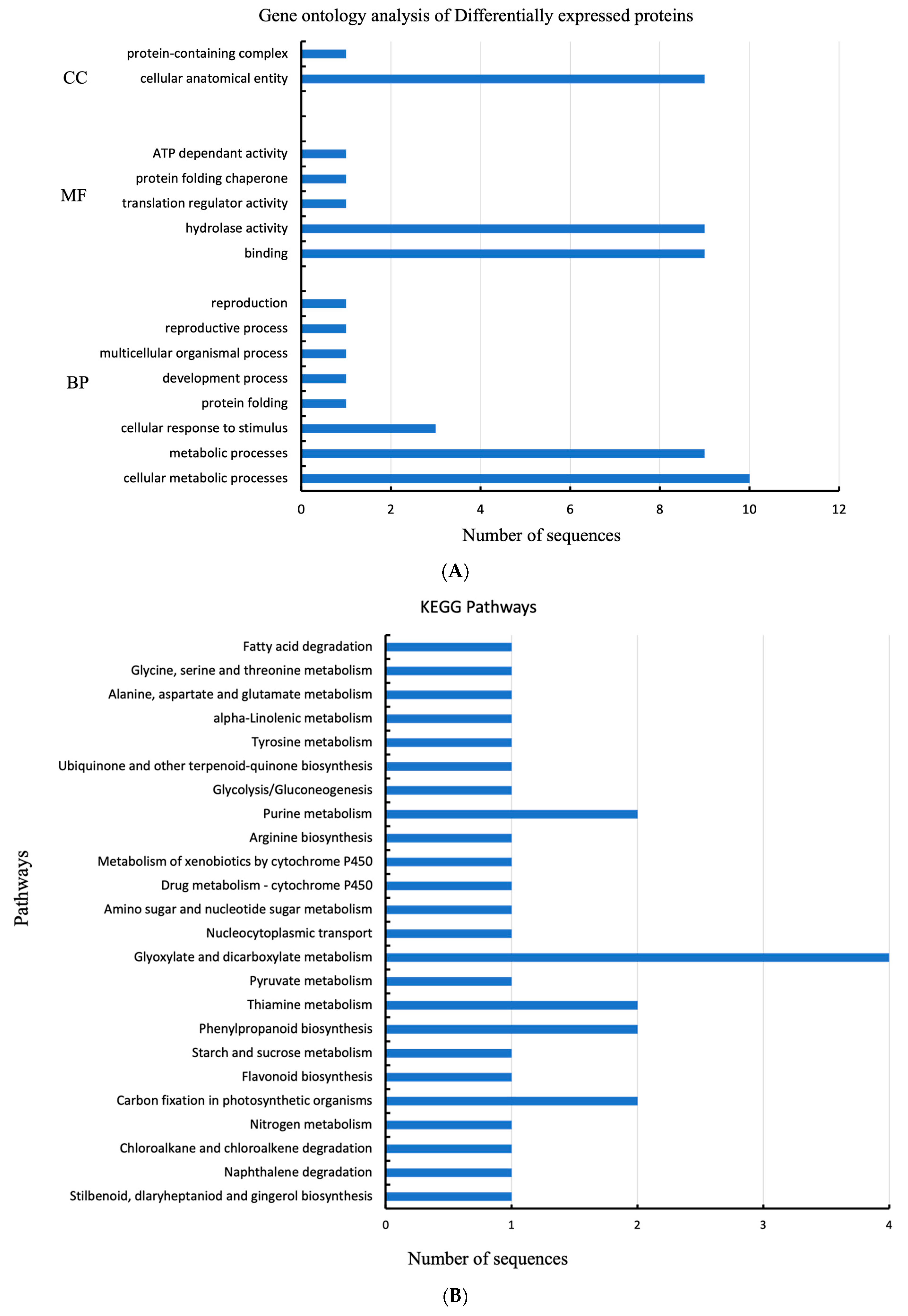
2.3. GO Analysis of Gene Ontology Enrichment
2.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
3. Discussion
3.1. 1D-SDS-PAGE and LC-MS/MS Analysis
3.2. Gene Ontology Enrichment and KEGG Pathway Analysis
3.3. Regulation of Biosynthesis of Secondary-Metabolites-Related Pathways
3.3.1. Phenylpropanoids Pathway
3.3.2. Carbon Fixation in Photosynthetic Organism
3.3.3. Purine Metabolism Pathways
4. Materials and Methods
4.1. Plant Material and Sample Collection
4.2. Sample Preparation
4.3. Protein Extraction and Pellet Solubilization
4.4. Quality Control Using SDS–PAGE Analysis
4.5. Protein Pellet Solubilization
4.6. On-Bead Digest
4.7. LC–MS/MS Analysis—Dionex Nano-RSLC
4.7.1. Mass Spectrometry
4.7.2. MS Data Analysis
4.8. Gene Ontology and KEGG Analysis Pipeline
4.9. Experimental Design and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schutte, A.L.; Van Wyk, B.E. Evolutionary relationships in the Podalyrieae and Liparieae (Fabaceae) based on morphological, cytological, and chemical evidence. Plant Syst. Evol. 1998, 209, 1–31. [Google Scholar] [CrossRef]
- Ntlhokwe, G.E. Application of Comprehensive Two-Dimensional Gas Chromatography for the Characterization of the Volatile Composition of Honeybush tea. Ph.D. Thesis, Stellenbosch University, Stellenbosch, South Africa, 2016. [Google Scholar]
- Mabizela, G.S. Metabolic and Quality Profiling of Cyclopia subternata and C. genistoides in Response to Seasonal Variation and Drought Stress. Ph.D. Thesis, Tshwane University of Technology, Pretoria, South Africa, 2020. [Google Scholar]
- Koen, J.; Slabbert, M.M.; Booyse, M.; Bester, C. Honeybush (Cyclopia spp.) anther-stigma distance and intraspecies cross compatibilities. S. Afr. J. Plant Soil 2020, 38, 13–18. [Google Scholar] [CrossRef]
- Le Roux, M.; Cronje, J.C.; Burger, B.V.; Joubert, E. Characterization of Volatiles and Aroma-Active Compounds in Honeybush (Cyclopia subternata) by GC-MS and GC-O Analysis. J. Agric. Food Chem. 2012, 60, 2657–2664. [Google Scholar] [CrossRef]
- Soni, R.P.; Katoch, M.; Kumar, A.; Ladohiya, R.; Verma, P. Tea: Production, Composition, Consumption and its Potential as an Antioxidant and Antimicrobial Agent. Int. J. Food Ferment. Technol. 2015, 5, 95–106. [Google Scholar] [CrossRef]
- Joubert, E.; De Beer, D.; Malherbe, C.J.; Muller, M.; Louw, A.; Gelderblom, W.C.A. Formal honeybush tea industry reaches 20-year milestone–progress of product research targeting phenolic composition, quality, and bioactivity. S. Afr. J. Bot. 2019, 127, 58–79. [Google Scholar] [CrossRef]
- Jankun, J.; Selman, S.H.; Swiercz, R.; Skrzypczak-Jankun, E. Why drinking green tea could prevent cancer. Nature 1997, 387, 561. [Google Scholar] [CrossRef]
- Kanwar, J.; Taskeen, M.; Mohammad, I. Recent advances on tea polyphenols. Front. Biosci. 2012, 4, 11131. [Google Scholar]
- Joubert, E.; Joubert, M.E.; Bester, C.; De Beer, D.; De Lange, J.H. Honeybush (Cyclopia spp.): From local cottage industry to global markets—The catalytic and supporting role of research. S. Afr. J. Bot. 2011, 77, 887–907. [Google Scholar] [CrossRef]
- McGregor, G. An Overview of the Honeybush Industry; Department of Environmental Affairs and Development Planning: Cape Town, South Africa, 2017. [Google Scholar]
- Karsen, P.A.; Lötze, E.; Valentine, A.J.; Hoffman, E.W. Propagation and cultivation practices of honeybush (Cyclopia spp.) for the sustainable production of an export quality indigenous South African tea. Crop. Sci. 2022, 62, 1702–1733. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Komatsu, S.; Mock, H.-P.; Yang, P.; Svensson, B. Application of proteomics for improving crop protection/artificial regulation. Front. Plant Sci. 2013, 4, 522. [Google Scholar] [CrossRef]
- Zahid, G.; Iftikhar, S.; Shimira, F.; Ahmad, H.M.; Kaçar, Y.A. An overview and recent progress of plant growth regulators (PGRs) in the mitigation of abiotic stresses in fruits: A review. Sci. Hortic. 2023, 309, 111621. [Google Scholar] [CrossRef]
- Henry, W.B.; Krutz, L.J. Water in Agriculture: Improving Corn Production Practices to Minimize Climate Risk and Optimize Profitability. Curr. Clim. Chang. Rep. 2016, 2, 49–54. [Google Scholar] [CrossRef]
- Farooqi, Z.U.R.; Ayub, M.A.; Rehman, M.Z.U.; Sohail, M.I.; Usman, M.; Khalid, H.; Naz, K. Regulation of drought stress in plants. In Plant Life under Changing Environment; Chapter 4; Academic Press: Cambridge, MA, USA, 2020; pp. 77–104. [Google Scholar] [CrossRef]
- Metwaly, E.-S.E.; Al-Yasi, H.M.; Ali, E.F.; Farouk, H.A.; Farouk, S. Deteriorating Harmful Effects of Drought in Cucumber by Spraying Glycinebetaine. Agriculture 2022, 12, 2166. [Google Scholar] [CrossRef]
- Farouk, S.; Al-Huqail, A.A. Sodium nitroprusside application regulates antioxidant capacity, improves phytopharmaceutical production and essential oil yield of marjoram herb under drought. Ind. Crop. Prod. 2020, 158, 113034. [Google Scholar] [CrossRef]
- Farouk, S.; Al-Ghamdi, A.A.M. Sodium nitroprusside application enhances drought tolerance in marjoram herb by promoting chlorophyll biosynthesis and enhancing osmotic adjustment capacity. Arab. J. Geosci. 2021, 14, 430. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, X.; Ottosen, C.O.; Rosenqvist, E.; Zhao, L.; Wang, Y.; Yu, W.; Zhao, T.; Wu, Z. Drought stress had a predominant effect overheat stress on three tomato cultivars subjected to combined stress. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 1–8. [Google Scholar] [CrossRef]
- Wojtyla, L.; Paluch-Lubawa, E.; Sobieszczuk-Nowicka, E.; Garnczarska, M. Drought stress memory and subsequent drought stress tolerance in plants. In Priming-Mediated Stress and Cross-Stress Tolerance in Plants; Chapter 7; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 115–131. [Google Scholar] [CrossRef]
- Belay, Z.A.; Caleb, O.J. Role of integrated omics in unravelling fruit stress and defence responses during postharvest: A review. Food Chem. Mol. Sci. 2022, 5, 100118. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, S.; Liu, K.; Wang, S.; Huang, L.; Guo, L. Proteomics: A powerful tool to study plant responses to biotic stress. Plant Methods 2019, 15, 1–20. [Google Scholar] [CrossRef]
- Jorrin-Novo, J.V.; Komatsu, S.; Sanchez-Lucas, R.; de Francisco, L.E.R. Gel electrophoresis-based plant proteomics: Past, present, and future. Happy 10th anniversary Journal of Proteomics! J. Proteom. 2019, 198, 1–10. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Dileep, A.; Lebonah, D.E.; Pramoda Kumari, J. A short review on proteomics and its applications. Int. Lett. Nat. Sci. 2014, 12, 77–84. [Google Scholar] [CrossRef]
- Lyu, S.; Gao, L.; Zhang, R.; Zhang, C.; Hou, X. Correlation analysis of expression profile and quantitative iTRAQ-LC-MS/MS proteomics reveals resistance mechanism against TuMV in Chinese Cabbage (Brassica rapa ssp. pekinensis). Front. Genet. 2020, 11, 963. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, L.; Zhao, M.; Gu, S.; Wang, C.; Zhao, J.; Tang, Z.; Gao, H.; Zhang, L.; Fu, L.; et al. iTRAQ proteomics reveals the regulatory response to Magnaporthe oryzae in durable resistant vs. susceptible rice genotypes. PLoS ONE 2020, 15, e0227470. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Komatsu, S. Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean. J. Proteom. 2018, 172, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Katam, R.; Shokri, S.; Murthy, N.; Singh, S.K.; Suravajhala, P.; Khan, M.N.; Bahmani, M.; Sakata, K.; Reddy, K.R. Proteomics, physiological, and biochemical analysis of cross tolerance mechanisms in response to heat and water stresses in soybean. PLoS ONE 2020, 15, e0233905. [Google Scholar] [CrossRef]
- Haddoudi, L.; Hdira, S.; Hanana, M.; Romero, I.; Haddoudi, I.; Mahjoub, A.; Ben Jouira, H.; Djébali, N.; Ludidi, N.; Sanchez-Ballesta, M.T.; et al. Evaluation of the morpho-physiological, biochemical, and molecular responses of contrasting Medicago truncatula lines under water deficit stress. Plants 2021, 10, 2114. [Google Scholar] [CrossRef]
- Gokul, A.; Carelse, M.F.; Niekerk, L.-A.; Klein, A.; Ludidi, N.; Mendoza-Cozatl, D.; Keyster, M. Exogenous 3,3′-Diindolylmethane Improves Vanadium Stress Tolerance in Brassica napus Seedling Shoots by Modulating Antioxidant Enzyme Activities. Biomolecules 2021, 11, 436. [Google Scholar] [CrossRef]
- Nsumpi, A.N.; Belay, Z.A.; Caleb, O.J. Good intentions, bad outcomes: Impact of mixed-fruit loading on banana fruit protein expression, physiological responses and quality. Food Packag. Shelf Life 2020, 26, 100594. [Google Scholar] [CrossRef]
- Ubiparip, Z.; Beerens, K.; Franceus, J.; Vercauteren, R.; Desmet, T. Thermostable alpha-glucan phosphorylases: Characteristics and industrial applications. Appl. Microbiol. Biotechnol. 2018, 102, 8187–8202. [Google Scholar] [CrossRef] [PubMed]
- Haworth, M.; Marino, G.; Brunetti, C.; Killi, D.; De Carlo, A.; Centritto, M. The impact of heat stress and water deficit on the photosynthetic and stomatal physiology of olive (Olea europaea L.)—A case study of the 2017 heat wave. Plants 2018, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Chu, Y.; Ma, G.; Zhang, Y.; Zhang, X.; Wang, M.; Lu, H.; Wang, L.; Kang, G.; Ma, D.; et al. Physiological mechanisms underlying reduced photosynthesis in wheat leaves grown in the field under conditions of nitrogen and water deficiency. Crop. J. 2022, in press. [CrossRef]
- Morales, F.; Ancín, M.; Fakhet, D.; González-Torralba, J.; Gámez, A.L.; Seminario, A.; Soba, D.; Ben Mariem, S.; Garriga, M.; Aranjuelo, I. Photosynthetic Metabolism under Stressful Growth Conditions as a Bases for Crop Breeding and Yield Improvement. Plants 2019, 9, 88. [Google Scholar] [CrossRef]
- Wang, L.-F.; Lu, K.-K.; Li, T.-T.; Zhang, Y.; Guo, J.-X.; Song, R.-F.; Liu, W.-C. Maize phytomelatonin receptor1 functions in plant tolerance to osmotic and drought stress. J. Exp. Bot. 2022, 73, 5961–5973. [Google Scholar] [CrossRef]
- Mansell, R.L.; Gross, G.G.; Stöckigt, J.; Franke, H.; Zenk, M.H. Purification and properties of cinnamyl alcohol dehydrogenase from higher plants involved in lignin biosynthesis. Phytochemistry 1974, 13, 2427–2435. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef]
- Saqib, A.; Scheller, H.V.; Fredslund, F.; Welner, D.H. Molecular characteristics of plant UDP-arabinopyranose mutases. Glycobiology 2019, 29, 839–846. [Google Scholar] [CrossRef]
- Otto, M.; Wynands, B.; Lenzen, C.; Filbig, M.; Blank, L.M.; Wierckx, N. Rational Engineering of Phenylalanine Accumulation in Pseudomonas taiwanensis to Enable High-Yield Production of Trans-Cinnamate. Front. Bioeng. Biotechnol. 2019, 7, 312. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Hu, Y.; Luo, C. The metabolites involved in phenylpropanoid biosynthesis increase the susceptibility of octoploid strawberry to crown rot caused by Colletotrichum siamense. Sci. Hortic. 2022, 306, 111447. [Google Scholar] [CrossRef]
- van Heeswijk, W.C.; Westerhoff, H.V.; Boogerd, F.C. Nitrogen Assimilation in Escherichia coli: Putting Molecular Data into a Systems Perspective. Microbiol. Mol. Biol. Rev. 2013, 77, 628–695. [Google Scholar] [CrossRef] [PubMed]
- Brink, C.; Postma, K.; Jacobs, K. Rhizobial diversity and functions in rooibos (Asphalathus linearis) and honeybush (Cyclopia spp.) plants, a review. S. Afr. J. Bot. 2017, 110, 80–86. [Google Scholar] [CrossRef]
- Bester, C. A model for commercialization of honeybush tea, an indigenous crop. Acta Hortic 2013, 1007, 889–894. [Google Scholar] [CrossRef]
- Postma, A.; Slabbert, E.; Postma, F.; Jacobs, K. Soil bacterial communities associated with natural and commercial Cyclopia spp. FEMS Microbiol. Ecol. 2016, 92, fiw016. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Xu, Z.; Jiang, Y.; Jia, B.; Zhou, G. Elevated-CO2 Response of Stomata and Its Dependence on Environmental Factors. Front. Plant Sci. 2016, 7, 657. [Google Scholar] [CrossRef]
- Benešová, M.; Holá, D.; Fischer, L.; Jedelsky, P.; Hnilička, F.; Wilhelmova, N.; Rothová, O.; Kočová, M.; Procházková, D.; Honnerová, J.; et al. The Physiology and Proteomics of Drought Tolerance in Maize: Early Stomatal Closure as a Cause of Lower Tolerance to Short-Term Dehydration? PLoS ONE 2012, 7, e38017. [Google Scholar] [CrossRef]
- Chaki, M.; Begara-Morales, J.C.; Barroso, J.B. Oxidative stress in plants. Antioxidants 2020, 9, 481. [Google Scholar] [CrossRef]
- Biała, W.; Jasiński, M. The phenylpropanoid case–it is transport that matters. Front. Plant Sci. 2018, 9, 1610. [Google Scholar] [CrossRef]
- Yang, L.; Wen, K.-S.; Ruan, X.; Zhao, Y.-X.; Wei, F.; Wang, Q. Response of Plant Secondary Metabolites to Environmental Factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef]
- Clemens, S.; Weber, M. The essential role of coumarin secretion for Fe acquisition from alkaline soil. Plant Signal. Behav. 2015, 11, e1114197. [Google Scholar] [CrossRef]
- Yu, A.; Zhao, J.; Wang, Z.; Cheng, K.; Zhang, P.; Tian, G.; Liu, X.; Guo, E.; Du, Y.; Wang, Y. Transcriptome and metabolite analysis reveal the drought tolerance of foxtail millet significantly correlated with phenylpropanoids-related pathways during germination process under PEG stress. BMC Plant Biol. 2020, 20, 274. [Google Scholar] [CrossRef]
- Ducat, D.C.; Silver, P.A. Improving carbon fixation pathways. Curr. Opin. Chem. Biol. 2012, 16, 337–344. [Google Scholar] [CrossRef]
- Stitt, M.; Lunn, J.; Usadel, B. Arabidopsis and primary photosynthetic metabolism—More than the icing on the cake. Plant J. 2010, 61, 1067–1091. [Google Scholar] [CrossRef]
- Khalil, A.M.; Murchie, E.H.; Mooney, S.J. Quantifying the influence of water deficit on root and shoot growth in wheat using X-ray Computed Tomography. AoB Plants 2020, 12, plaa036. [Google Scholar] [CrossRef] [PubMed]
- von Caemmerer, S. Rubisco carboxylase/oxygenase: From the enzyme to the globe: A gas exchange perspective. J. Plant Physiol. 2020, 252, 153240. [Google Scholar] [CrossRef] [PubMed]
- Stasolla, C.; Katahira, R.; Thorpe, T.A.; Ashihara, H. Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 2003, 160, 1271–1295. [Google Scholar] [CrossRef]
- Wasternack, C. Metabolism of pyrimidines and purines. In Encyclopedia of Plant Physiology; Pirson, A., Zimmermann, M.H., Eds.; New Series; Springer: Berlin, Germany, 1982; Volume 14B, pp. 263–301. [Google Scholar]
- Bray, C.M. Nitrogen Metabolism in Plants; Longman: London, UK, 1983. [Google Scholar]
- Beukers, M.W.; Pirovano, I.M.; van Weert, A.; Kerkhof, C.J.; Ijzerman, A.P.; Soudijn, W. Characterization of ECTO-ATPase on human blood cells: A physiological role in platelet aggregation? Biochem. Pharmacol. 1993, 46, 1959–1966. [Google Scholar] [CrossRef] [PubMed]
- Tuberosa, R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012, 3, 347. [Google Scholar] [CrossRef] [PubMed]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Ranatunge, I.; Adikary, S.; Dasanayake, P.; Fernando, C.D.; Soysa, P. Development of a Rapid and Simple Method to Remove Polyphenols from Plant Extracts. Int. J. Anal. Chem. 2017, 2017, 7230145. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef]
- Hooijberg, E.H.; Miller, M.; Cray, C.; Buss, P.; Steenkamp, G.; Goddard, A. Serum protein electrophoresis in healthy and injured southern white rhinoceros (Ceratotherium simum simum). PLoS ONE 2018, 13, e0200347. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]



| Protein Name | Cellular Component | Molecular Functions | Biological Processes | MW (kDa) | Tax. ID | FET | * FC (log2) | FC | Expression Change |
|---|---|---|---|---|---|---|---|---|---|
| Alpha-glucan phosphorylase, H isozyme | Cytoplasm | Glycogen phosphorylase activity, pyridoxal phosphate binding, SHG alpha-glucan phosphorylase activity, linear malto-oligosaccharide phosphorylase activity | Carbohydrate metabolism | 95.9 | 3906 | 0.00070 | 0.5 | 1.41 | Upregulated in T17 and downregulated in T19 |
| Ribulose bisphosphate carboxylase large chain | Plastid, Chloroplast | Magnesium ion binding, monooxygenase activity, ribulose-bisphosphate carboxylase activity | Photorespiration, reductive pentose-phosphate cycle | 50 | 49830 | 0.00070 | 1.1 | 2.14 | Upregulated in T1 and downregulated in T19 |
| Trans-cinnamate 4-monooxygenase | Integral component of membrane | Heme binding, iron ion binding, trans-cinnamate 4-monooxygenase activity, | Lignin metabolic process | 58 | 3847 | 0.00029 | 0.0 | 1.00 | Downregulated in T1 and upregulated in T19 |
| Probable UDP-arabinopyranose mutase 1 | Extracellular region (Secreted, cell wall, cell junction, plasmodesma, Golgi apparatus) | UDP-arabinopyranose mutase activity | Cell wall organization, cellulose biosynthetic process, plant-type cell wall organization or biogenesis, protein glycosylation | - | 3888 | 0.00081 | 0.1 | 1.07 | Downregulated in T1 and upregulated in T19 |
| Probable cinnamyl alcohol dehydrogenase | Stem, hypocotyl, root tissue | Cinnamyl-alcohol dehydrogenase activity, sinapyl alcohol dehydrogenase activity, zinc ion binding | Lignin biosynthetic process | - | 3879 | 0.0037 | 0.09 | 1.06 | Downregulated in T1 and upregulated in T19 |
| Ribulose bisphosphate carboxylase large chain | Plastid, Chloroplast | Magnesium ion binding, monooxygenase activity, ribulose-bisphosphate carboxylase activity | Photorespiration, reductive pentose phosphate cycle | 50 | 49830 | 0.00074 | 1.2 | 2.30 | Downregulated in T19 and upregulated in T3 |
| Ribulose bisphosphate carboxylase large chain | Plastid, Chloroplast | Magnesium ion binding, monooxygenase activity, ribulose-bisphosphate carboxylase activity | Photorespiration, reductive pentose phosphate cycle | 50 | 49830 | 0.00081 | 1.2 | 2.30 | Downregulated in T19 and upregulated in T3 |
| Ribulose bisphosphate carboxylase large chain | Plastid, Chloroplast | Magnesium ion binding, monooxygenase activity, ribulose-bisphosphate carboxylase activity | Photorespiration, reductive pentose-phosphate cycle | 53 | 49830 | 0.0015 | 1.2 | 2.30 | Downregulated in T19 and upregulated in T3 |
| Glutamine synthetase nodule isozyme | Cytoplasm | ATP binding, glutamate-ammonia ligase activity | Glutamine biosynthetic process | 39 | 3918 | 0.0018 | 0.7 | 1.62 | Upregulated in T19 and downregulated in T3 |
| Elongation factor 1-alpha | Cytoplasm | GTP binding, translation elongation factor activity, GTPase activity | - | 49 | 3918 | 0.0037 | 1.6 | 3.03 | Downregulated in T19 and upregulated in T3 |
| Chlorophyll a-b binding protein, chloroplastic | Chloroplast thylakoid membrane, integral component of membrane, photosystem I, photosystem II | Chlorophyll binding, metal ion binding | Photosynthesis, light harvesting in photosystem I, response to light stimulus | 26 | 3847 | 0.0041 | 0.6 | 1.52 | Upregulated in T19 and downregulated in T3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahlare, M.-J.S.; Husselmann, L.; Lewu, M.N.; Bester, C.; Lewu, F.B.; Caleb, O.J. Analysis of the Differentially Expressed Proteins and Metabolic Pathways of Honeybush (Cyclopia subternata) in Response to Water Deficit Stress. Plants 2023, 12, 2181. https://doi.org/10.3390/plants12112181
Mahlare M-JS, Husselmann L, Lewu MN, Bester C, Lewu FB, Caleb OJ. Analysis of the Differentially Expressed Proteins and Metabolic Pathways of Honeybush (Cyclopia subternata) in Response to Water Deficit Stress. Plants. 2023; 12(11):2181. https://doi.org/10.3390/plants12112181
Chicago/Turabian StyleMahlare, Mary-Jane S., Lizex Husselmann, Muinat N. Lewu, Cecilia Bester, Francis B. Lewu, and Oluwafemi James Caleb. 2023. "Analysis of the Differentially Expressed Proteins and Metabolic Pathways of Honeybush (Cyclopia subternata) in Response to Water Deficit Stress" Plants 12, no. 11: 2181. https://doi.org/10.3390/plants12112181
APA StyleMahlare, M.-J. S., Husselmann, L., Lewu, M. N., Bester, C., Lewu, F. B., & Caleb, O. J. (2023). Analysis of the Differentially Expressed Proteins and Metabolic Pathways of Honeybush (Cyclopia subternata) in Response to Water Deficit Stress. Plants, 12(11), 2181. https://doi.org/10.3390/plants12112181








