Composition, Abundance, and Diversity of the Soil Microbiome Associated with the Halophytic Plants Tamarix aphylla and Halopeplis perfoliata on Jeddah Seacoast, Saudi Arabia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. PCR Amplification and Next-Generation Sequencing
2.3. Statistical Analysis
3. Results
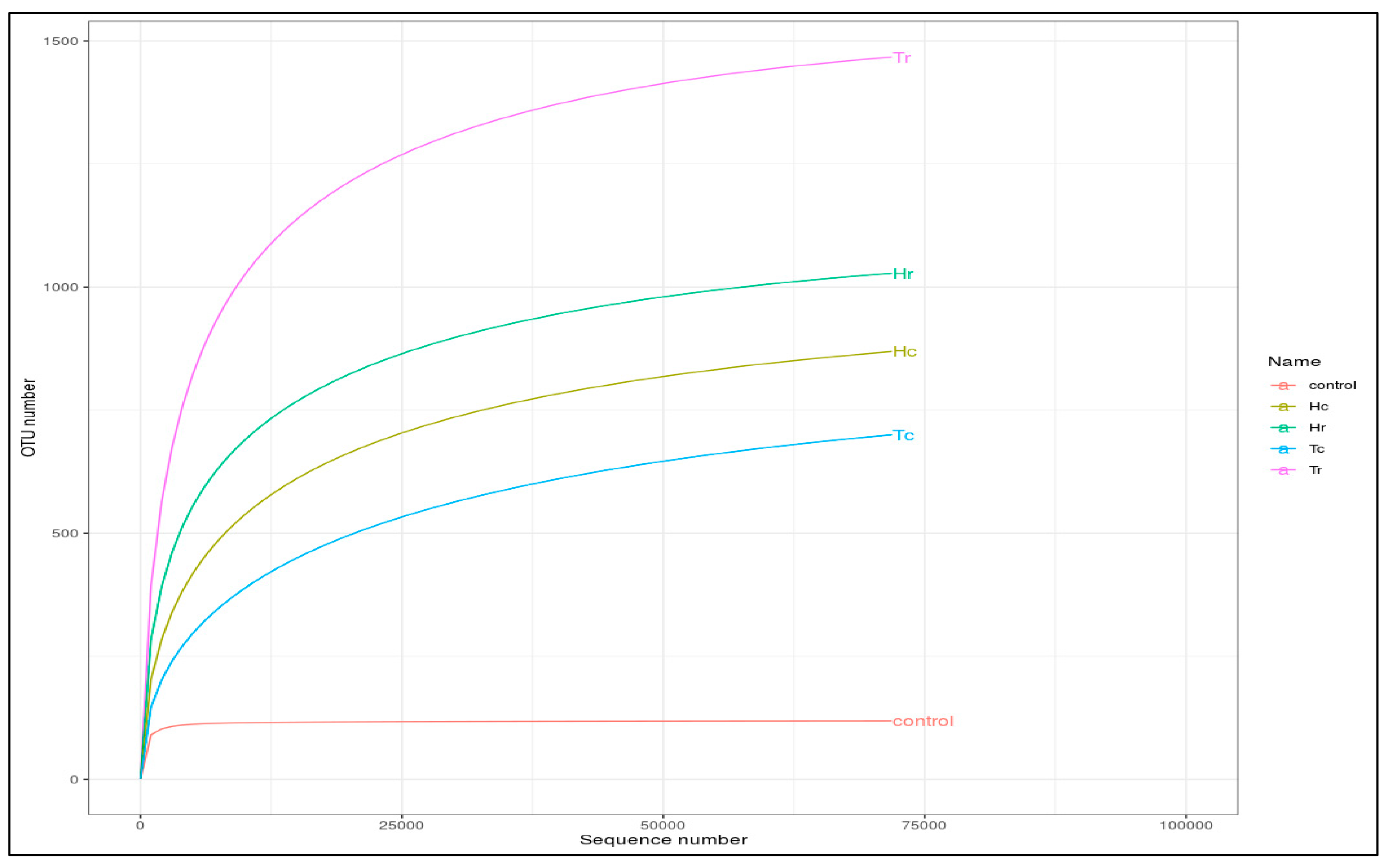
3.1. 16S rRNA Sequence Assembly and the Total Number of OTU Reads
3.2. ITS rRNA Sequence Assembly and the Total Number of OTU Reads
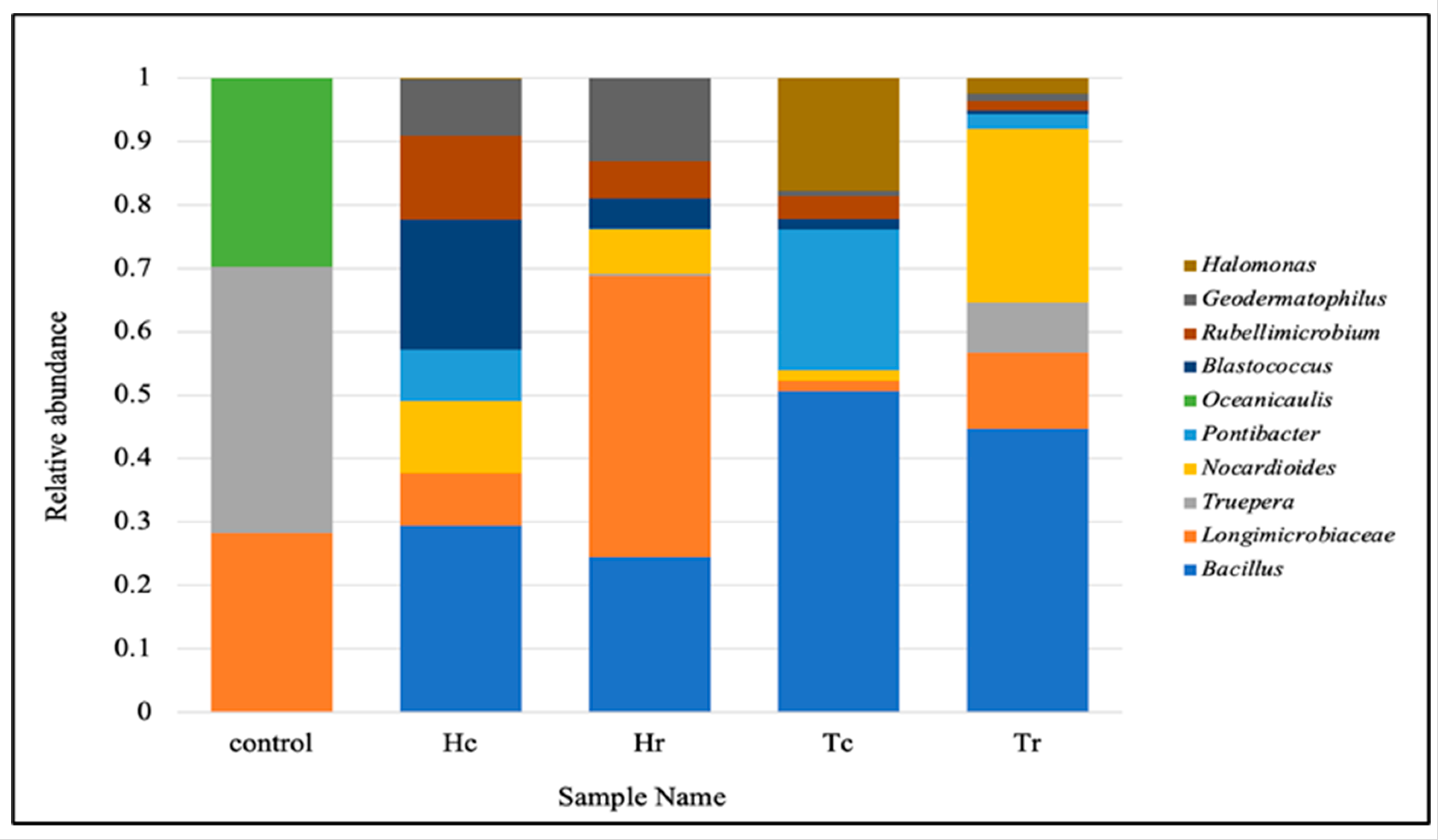
3.3. 16S rRNA Taxonomic Classification at the Phylum and Genus Levels
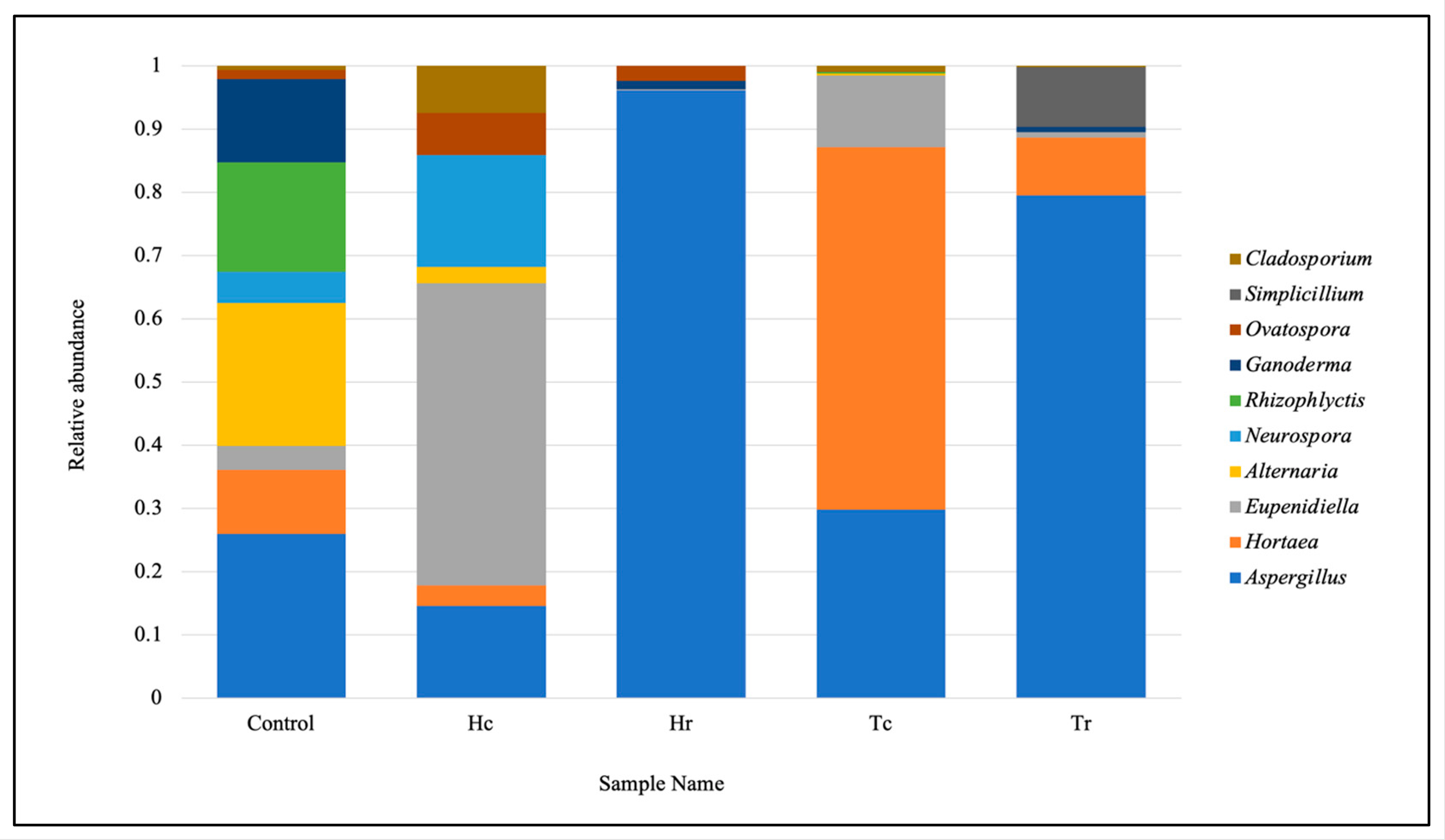
3.4. ITS rRNA Gene-Based Taxonomic Classification and the Relative Abundance of the Fungal Community
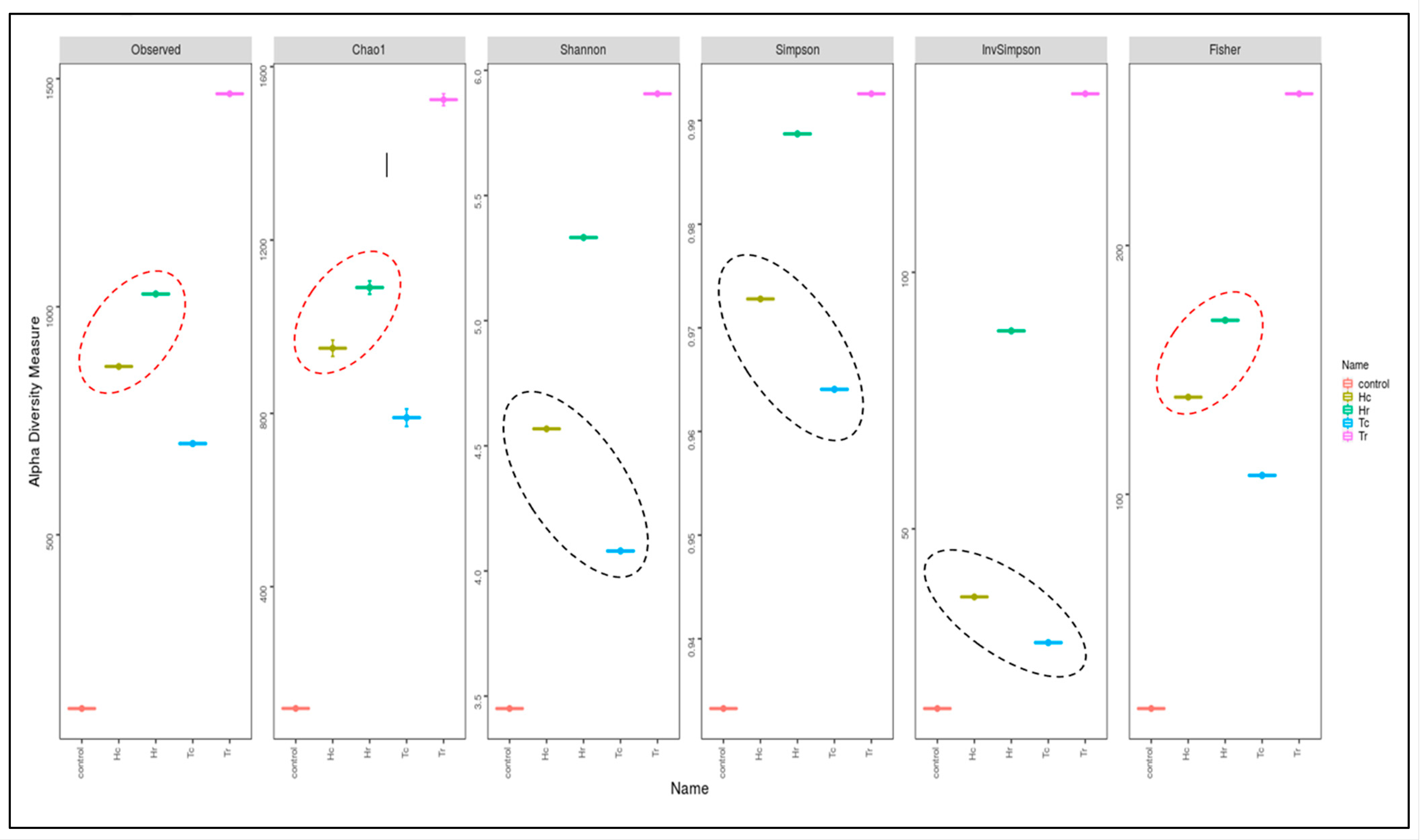
3.5. 16S rRNA Community Richness and Diversity (Alpha and Beta)
3.6. ITS-rRNA Fungal Community Richness and (Alpha/Beta) Diversity
3.7. Taxonomic Classification and Community Structure of the Soil Microbiome
3.7.1. 16S rRNA Heat-Map Analysis
3.7.2. ITS-rRNA Heat-Map Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, L.; Zhu, K.; Wurzburger, N.; Zhang, J. Relationships between plant diversity and soil microbial diversity vary across taxonomic groups and spatial scales. Ecosphere 2020, 11, e02999. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G. Plant-Soil Interactions. In Encyclopedia of Biodiversity; Levin, S.A., Ed.; Elsevier: New York, NY, USA, 2001; pp. 689–709. [Google Scholar]
- Hu, L.; Robert, C.A.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; Van Der Heijden, M.G. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Bennett, J.A.; Maherali, H.; Reinhart, K.O.; Lekberg, Y.; Hart, M.M.; Klironomos, J. Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 2017, 355, 181–184. [Google Scholar] [CrossRef]
- Lowry, C.J.; Smith, R.G. Weed control through crop plant manipulations. In Non-Chemical Weed Control; Elsevier: Amsterdam, The Netherlands, 2018; pp. 73–96. [Google Scholar]
- Moussa, T.A.; Al-Zahrani, H.S.; Almaghrabi, O.A.; Abdelmoneim, T.S.; Fuller, M.P. Comparative metagenomics approaches to characterize the soil fungal communities of western coastal region, Saudi Arabia. PLoS ONE 2017, 12, e0185096. [Google Scholar] [CrossRef] [PubMed]
- Hemida, S.; Abdel-Sater, M. Biodiversity of soil and air-borne fungi in the northern border region of Saudi Arabia. J. Pure Appl. Microbiol. 2016, 10, 967–979. [Google Scholar]
- Alotaibi, M.O.; Sonbol, H.S.; Alwakeel, S.S.; Suliman, R.S.; Fodah, R.A.; Jaffal, A.S.A.; AlOthman, N.I.; Mohammed, A.E. Microbial diversity of some sabkha and desert sites in Saudi Arabia. Saudi J. Biol. Sci. 2020, 27, 2778–2789. [Google Scholar] [CrossRef] [PubMed]
- Baeshen, M.; Moussa, T.A.A.; Ahmed, F.; Abulfaraj, A.A.; Jalal, R.S.; Majeed, M.A.; Baeshen, N.A.; Huelsenbeck, J.P. Diversity Profiling of Associated Bacteria from The Soils of Stress Tolerant Plants from Seacoast of Jeddah, Saudi Arabia. Appl. Ecol. Environ. Res. 2020, 18, 8217–8231. [Google Scholar] [CrossRef]
- MJB, A.-O. The Halophytic Flora of Syria; International Center for Agricultural Research in the Dry Areas: Aleppo, Syria, 2011. [Google Scholar]
- Rasool, S.G.; Hameed, A.; Khan, M.A.; Gul, B. Seeds of Halopeplis perfoliata display plastic responses to various abiotic factors during germination. Flora 2017, 236–237, 76–83. [Google Scholar] [CrossRef]
- Noor, S.O.; Zahrani, D.A.A.; Hussein, R.M.; Jalal, R.S.; Abulfaraj, A.A.; Majeed, M.A.; Baeshen, M.N.; Hejin, A.M.A.; Baeshen, N.A.; Huelsenbeck, J.P. Biodiversity in Bacterial Phyla Composite in Arid Soils of the Community of Desert Medicinal Plant Rhazya stricta. J. Pharm. Res. Int. 2020, 32, 88–98. [Google Scholar] [CrossRef]
- Behairi, S.; Baha, N.; Barakat, M.; Ortet, P.; Achouak, W.; Heulin, T.; Kaci, Y. Bacterial diversity and community structure in the rhizosphere of the halophyte Halocnemum strobilaceum in an Algerian arid saline soil. Extremophiles 2022, 26, 18. [Google Scholar] [CrossRef]
- Alhaddad, F.A.; Abu-Dieyeh, M.H.; ElAzazi, E.M.; Ahmed, T.A. Salt tolerance of selected halophytes at the two initial growth stages for future management options. Sci. Rep. 2021, 11, 10194. [Google Scholar] [CrossRef] [PubMed]
- Usyk, M.; Zolnik, C.P.; Patel, H.; Levi, M.H.; Burk, R.D. Novel ITS1 Fungal Primers for Characterization of the Mycobiome. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Tran, P.Q.; Kieft, K.; Anantharaman, K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020, 14, 2060–2077. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Bardgett, R.D.; Vitousek, P.M.; Maestre, F.T.; Williams, M.A.; Eldridge, D.J.; Lambers, H.; Neuhauser, S.; Gallardo, A.; García-Velázquez, L.; et al. Changes in belowground biodiversity during ecosystem development. Proc. Natl. Acad. Sci. USA 2019, 116, 6891–6896. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.; Lianyan, B.; Mingxiang, Z.; Jiawei, Y.; Yinglong, Z.; Gehong, W.; Honglei, W. Soil bacteria with distinct diversity and functions mediates the soil nutrients after introducing leguminous shrub in desert ecosystems. Glob. Ecol. Conserv. 2021, 31, e01841. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Rawat, N.; Joshi, G.K. Bacterial community structure analysis of a hot spring soil by next generation sequencing of ribosomal RNA. Genomics 2019, 111, 1053–1058. [Google Scholar] [CrossRef]
- Hadziavdic, K.; Lekang, K.; Lanzen, A.; Jonassen, I.; Thompson, E.M.; Troedsson, C. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS ONE 2014, 9, e87624. [Google Scholar] [CrossRef]
- Arfi, Y.; Buée, M.; Marchand, C.; Levasseur, A.; Record, E. Multiple markers pyrosequencing reveals highly diverse and host-specific fungal communities on the mangrove trees Avicennia marina and Rhizophora stylosa. FEMS Microbiol. Ecol. 2012, 79, 433–444. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. Soil microbiomes and climate change. Nat. Rev. Microbiol. 2020, 18, 35–46. [Google Scholar] [CrossRef]
- Sutaria, D.; Kamlesh, R.S.; Sudipti, A.; Sonika, S. Actinomycetes as An Environmental Scrubber. In Crude Oil; Manar Elsayed, A.-R., Mohamed Hasan, E.-K., Eds.; IntechOpen: Rijeka, Republic of Croatia, 2021; p. 6. [Google Scholar]
- Girão, M.; Ribeiro, I.; Carvalho, M.d.F. Actinobacteria from Marine Environments: A Unique Source of Natural Products. In Natural Products from Actinomycetes: Diversity, Ecology and Drug Discovery; Rai, R.V., Bai, J.A., Eds.; Springer: Singapore, 2022; pp. 1–45. [Google Scholar]
- Bao, Y.; Dolfing, J.; Guo, Z.; Chen, R.; Wu, M.; Li, Z.; Lin, X.; Feng, Y. Important ecophysiological roles of non-dominant Actinobacteria in plant residue decomposition, especially in less fertile soils. Microbiome 2021, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Puttaswamygowda, G.H.; Olakkaran, S.; Antony, A.; Kizhakke Purayil, A. Chapter 22—Present Status and Future Perspectives of Marine Actinobacterial Metabolites. In Recent Developments in Applied Microbiology and Biochemistry; Buddolla, V., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 307–319. [Google Scholar]
- Sarkar, G.; Suthindhiran, K. Diversity and Biotechnological Potential of Marine Actinomycetes from India. Indian J. Microbiol. 2022, 62, 475–493. [Google Scholar] [CrossRef] [PubMed]
- Arístegui, J.; Gasol, J.M.; Duarte, C.M.; Herndld, G.J. Microbial oceanography of the dark ocean’s pelagic realm. Limnol. Oceanogr. 2009, 54, 1501–1529. [Google Scholar] [CrossRef]
- Swan, B.K.; Martinez-Garcia, M.; Preston, C.M.; Sczyrba, A.; Woyke, T.; Lamy, D.; Reinthaler, T.; Poulton, N.J.; Masland, E.D.; Gomez, M.L.; et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 2011, 333, 1296–1300. [Google Scholar] [CrossRef]
- Prieto-Barajas, C.M.; Valencia-Cantero, E.; Santoyo, G. Microbial mat ecosystems: Structure types, functional diversity, and biotechnological application. Electron. Journal. Biotechnol. 2018, 31, 48–56. [Google Scholar] [CrossRef]
- Kiama, C.W.; Njire, M.M.; Kambura, A.K.; Mugweru, J.N.; Matiru, V.N.; Wafula, E.N.; Kagali, R.N.; Kuja, J.O. Prokaryotic diversity and composition within equatorial lakes Olbolosat and Oloiden in Kenya (Africa). Curr. Res. Microb. Sci. 2021, 2, 100066. [Google Scholar] [CrossRef]
- Simões, M.F.; Antunes, A.; Ottoni, C.A.; Amini, M.S.; Alam, I.; Alzubaidy, H.; Mokhtar, N.A.; Archer, J.A.; Bajic, V.B. Soil and Rhizosphere Associated Fungi in Gray Mangroves (Avicennia marina) from the Red Sea--A Metagenomic Approach. Genom. Proteom. Bioinform. 2015, 13, 310–320. [Google Scholar] [CrossRef]
- Porras-Alfaro, A.; Muniania, C.N.; Hamm, P.S.; Torres-Cruz, T.J.; Kuske, C.R. 6. Fungal Diversity, Community Structure and Their Functional Roles in Desert Soils. In The Biology of Arid Soils; Blaire, S., Ed.; De Gruyter: Berlin, Germany; Boston, MA, USA, 2017; pp. 97–122. [Google Scholar]
- Challacombe, J.F.; Hesse, C.N.; Bramer, L.M.; McCue, L.A.; Lipton, M.; Purvine, S.; Nicora, C.; Gallegos-Graves, V.; Porras-Alfaro, A.; Kuske, C.R. Genomes and secretomes of Ascomycota fungi reveal diverse functions in plant biomass decomposition and pathogenesis. BMC Genomics 2019, 20, 976. [Google Scholar] [CrossRef]
- Yang, T.; Chen, Q.; Yang, M.; Wang, G.; Zheng, C.; Zhou, J.; Jia, M.; Peng, X. Soil microbial community under bryophytes in different substrates and its potential to degraded karst ecosystem restoration. Int. Biodeterior. Biodegrad. 2022, 175, 105493. [Google Scholar] [CrossRef]
- Bai, Y.; Wang, Q.; Liao, K.; Jian, Z.; Zhao, C.; Qu, J. Fungal Community as a Bioindicator to Reflect Anthropogenic Activities in a River Ecosystem. Front. Microbiol. 2018, 9, 3152. [Google Scholar] [CrossRef]
- Breyer, E.; Zhao, Z.; Herndl, G.J.; Baltar, F. Global contribution of pelagic fungi to protein degradation in the ocean. Microbiome 2022, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Florio Furno, M.; Poli, A.; Ferrero, D.; Tardelli, F.; Manzini, C.; Oliva, M.; Pretti, C.; Campani, T.; Casini, S.; Fossi, M.C.; et al. The Culturable Mycobiota of Sediments and Associated Microplastics: From a Harbor to a Marine Protected Area, a Comparative Study. J. Fungi 2022, 8, 927. [Google Scholar] [CrossRef] [PubMed]
- Luis, P.; Saint-Genis, G.; Vallon, L.; Bourgeois, C.; Bruto, M.; Marchand, C.; Record, E.; Hugoni, M. Contrasted ecological niches shape fungal and prokaryotic community structure in mangroves sediments. Environ. Microbiol. 2019, 21, 1407–1424. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, M.; Burgaud, G.; Yu, H.; Cai, L. Fungal Community Composition and Potential Depth-Related Driving Factors Impacting Distribution Pattern and Trophic Modes from Epi- to Abyssopelagic Zones of the Western Pacific Ocean. Microb. Ecol. 2019, 78, 820–831. [Google Scholar] [CrossRef]
- Baltar, F.; Zhao, Z.; Herndl, G.J. Potential and expression of carbohydrate utilization by marine fungi in the global ocean. Microbiome 2021, 9, 106. [Google Scholar] [CrossRef]


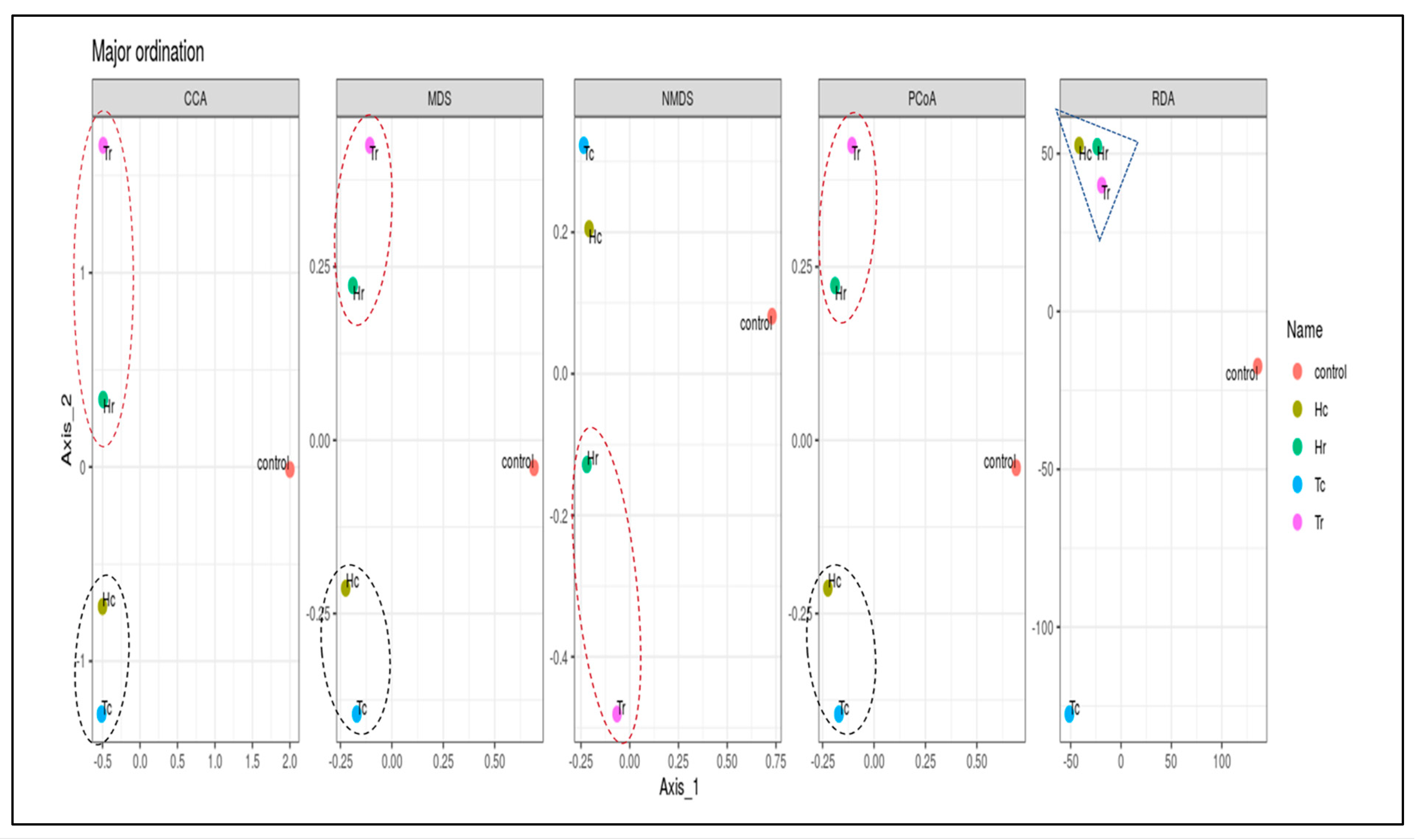
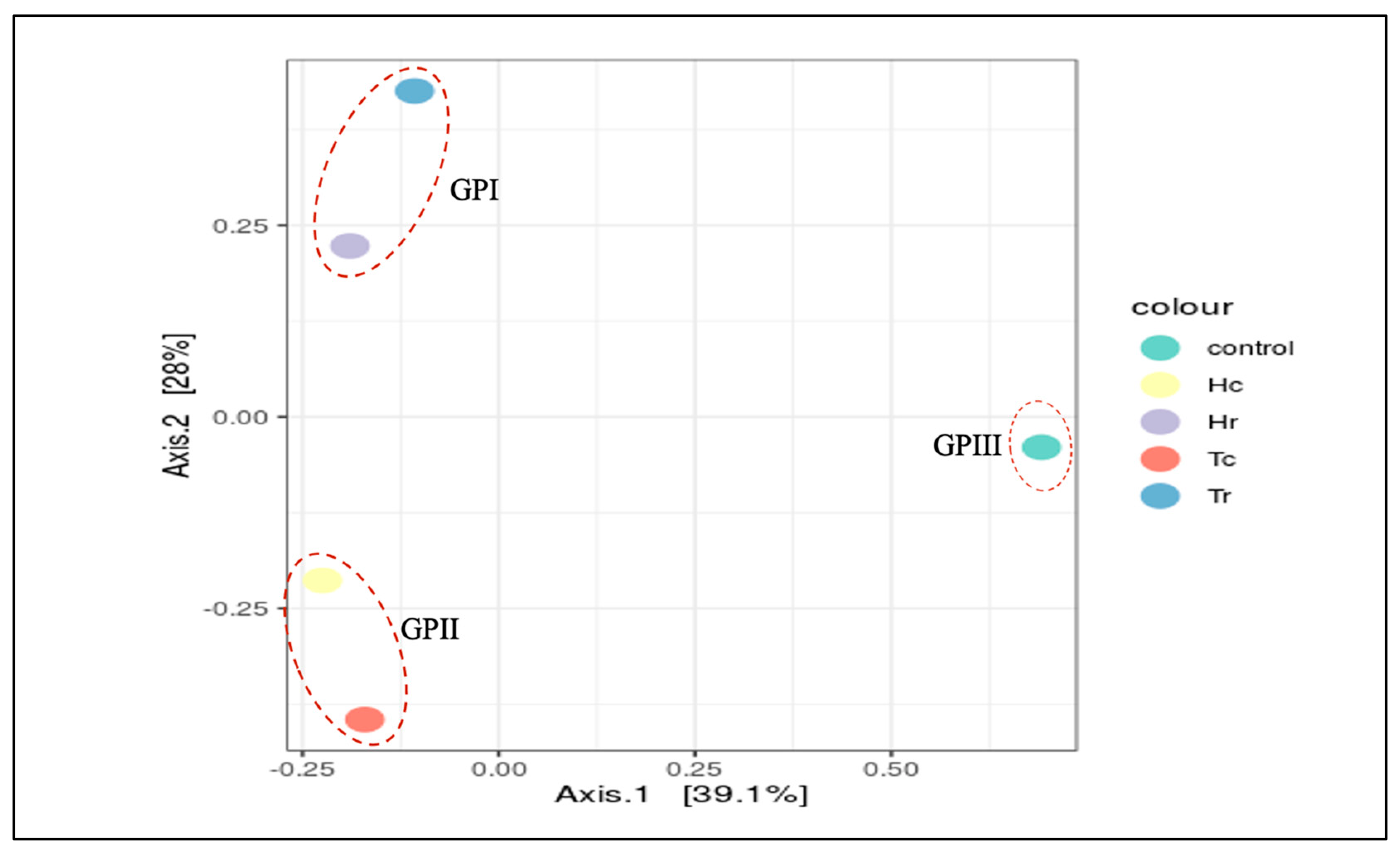



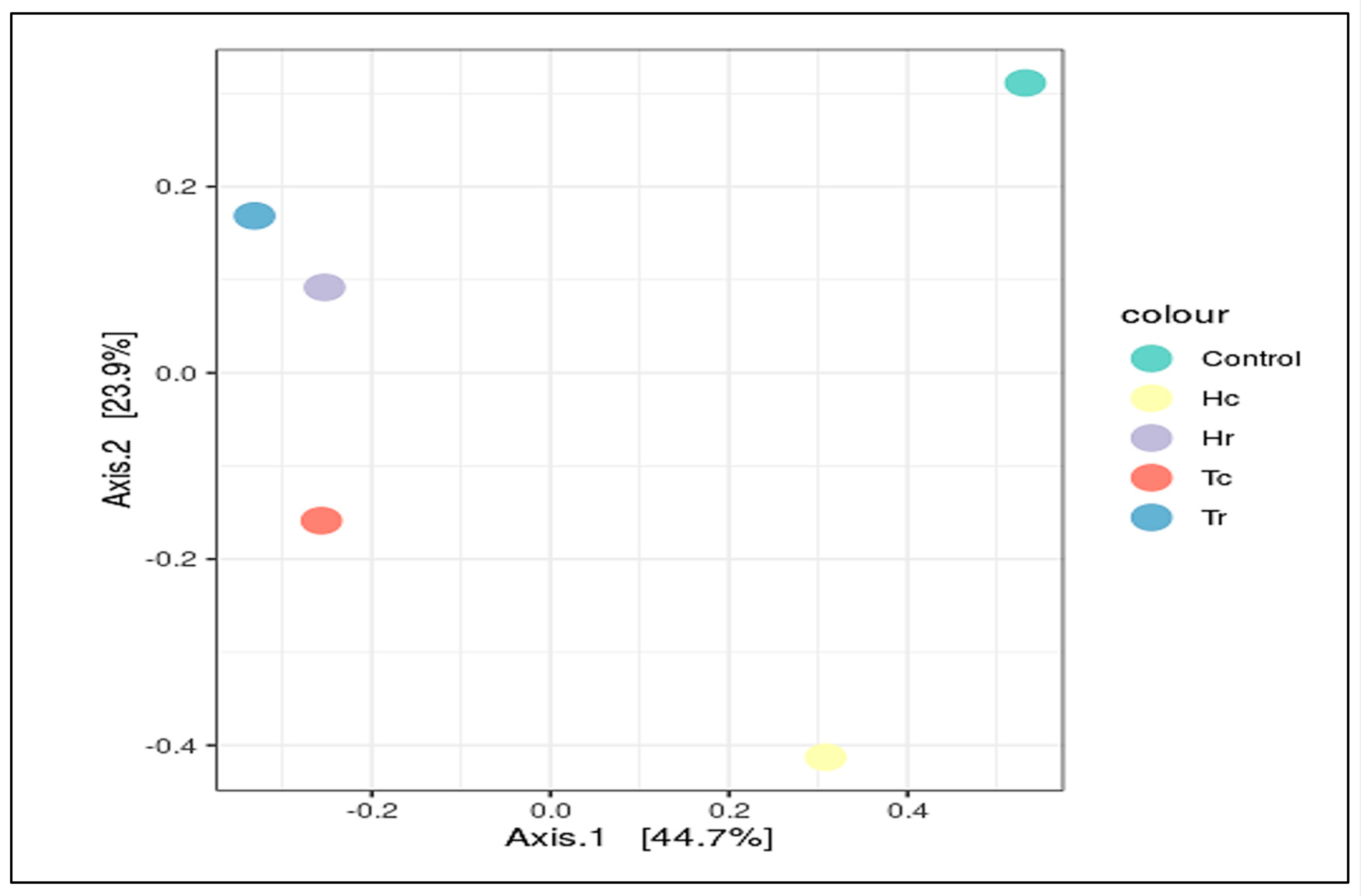

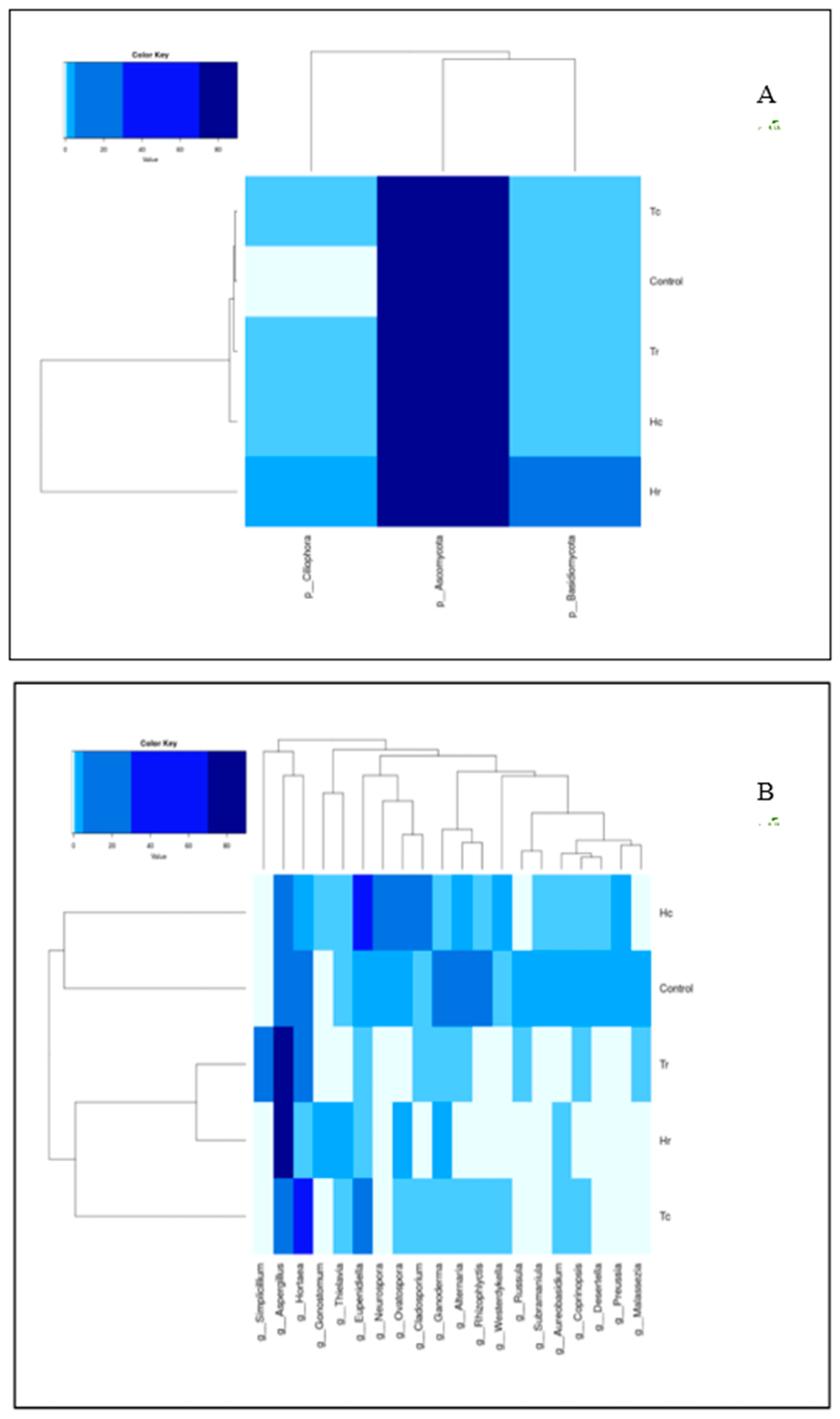
| Sample Name | Total Reads 1 | OTUs | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| Control-16S | 27204681 | 120 | 95.79 | 87.83 | 58 |
| Hc-16S | 67619892 | 858 | 93.51 | 85.88 | 58 |
| Hr-16S | 60276009 | 986 | 93.02 | 84.97 | 59 |
| Tc-16S | 61078118 | 1120 | 94.19 | 86.83 | 57 |
| Tr-16S | 56330547 | 1559 | 92.96 | 84.78 | 58 |
| Sample Name | Total Reads (Mb) | OTUs | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|
| Control—ITS | 34119109 | 102 | 97.31 | 92.28 | 48 |
| Hc—ITS | 39738492 | 162 | 97 | 92.65 | 50 |
| Hr—ITS | 45080296 | 87 | 95.71 | 90.08 | 49 |
| Tc—ITS | 47581365 | 184 | 96.56 | 91.76 | 54 |
| Tr—ITS | 48912745 | 130 | 96.01 | 90.18 | 53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baeshen, N.N.; Baz, L.; Shami, A.Y.; Ashy, R.A.; Jalal, R.S.; Abulfaraj, A.A.; Refai, M.; Majeed, M.A.; Abuzahrah, S.S.; Abdelkader, H.; et al. Composition, Abundance, and Diversity of the Soil Microbiome Associated with the Halophytic Plants Tamarix aphylla and Halopeplis perfoliata on Jeddah Seacoast, Saudi Arabia. Plants 2023, 12, 2176. https://doi.org/10.3390/plants12112176
Baeshen NN, Baz L, Shami AY, Ashy RA, Jalal RS, Abulfaraj AA, Refai M, Majeed MA, Abuzahrah SS, Abdelkader H, et al. Composition, Abundance, and Diversity of the Soil Microbiome Associated with the Halophytic Plants Tamarix aphylla and Halopeplis perfoliata on Jeddah Seacoast, Saudi Arabia. Plants. 2023; 12(11):2176. https://doi.org/10.3390/plants12112176
Chicago/Turabian StyleBaeshen, Naseebh N., Lina Baz, Ashwag Y. Shami, Ruba A. Ashy, Rewaa S. Jalal, Aala A. Abulfaraj, Mohammed Refai, Mazen A. Majeed, Samah S. Abuzahrah, Hayam Abdelkader, and et al. 2023. "Composition, Abundance, and Diversity of the Soil Microbiome Associated with the Halophytic Plants Tamarix aphylla and Halopeplis perfoliata on Jeddah Seacoast, Saudi Arabia" Plants 12, no. 11: 2176. https://doi.org/10.3390/plants12112176
APA StyleBaeshen, N. N., Baz, L., Shami, A. Y., Ashy, R. A., Jalal, R. S., Abulfaraj, A. A., Refai, M., Majeed, M. A., Abuzahrah, S. S., Abdelkader, H., Baeshen, N. A., & Baeshen, M. N. (2023). Composition, Abundance, and Diversity of the Soil Microbiome Associated with the Halophytic Plants Tamarix aphylla and Halopeplis perfoliata on Jeddah Seacoast, Saudi Arabia. Plants, 12(11), 2176. https://doi.org/10.3390/plants12112176








