The Role of Chromatin Modifications in the Evolution of Giant Plant Genomes
Abstract
1. Introduction
2. Results
2.1. The Repeat Profile of Fritillaria imperialis Is Characterized by a Large Fraction of Ty3/Gypsy Retroelements
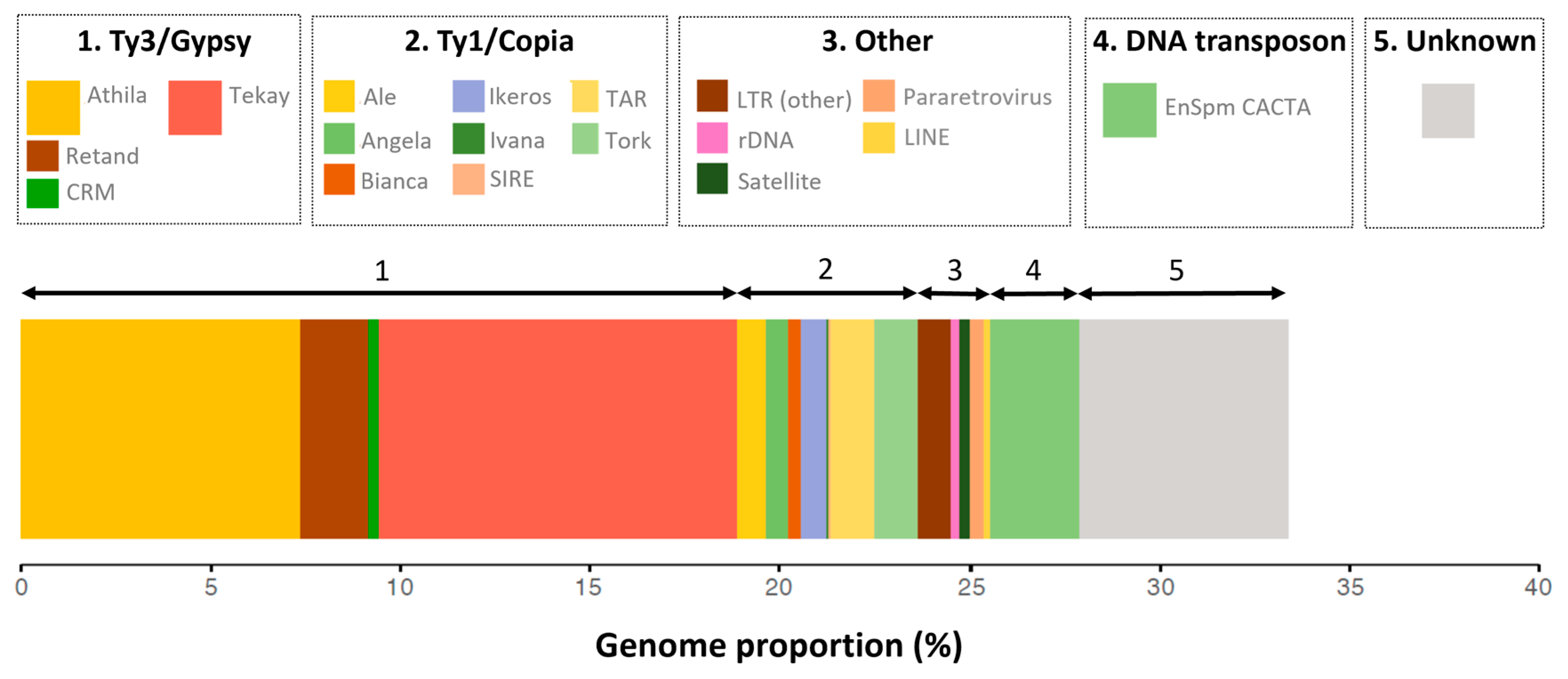
2.2. The Global Chromatin Landscape of Fritillaria imperialis Compared with Arabidopsis thaliana
2.2.1. Labelling of Arabidopsis and Fritillaria Nuclei to Detect Histones H3K4me1 and H3K4me2
2.2.2. Labelling of Histones H3K9me2, H3K9me3, H3K27me2, and H3K27me3
2.2.3. Labelling of Histones H3K9me1 and H3K27me1
3. Discussion
3.1. Global Distribution of Post-Translational Histone Modification
3.1.1. H3K4 Methylation Marks Genes and Associated Regulatory Regions
3.1.2. H3K9me3 and H3K27me3 Are Associated with the ‘Dark Matter’ of the Genome
3.1.3. H3K9me1 and H3K27me1 Associated with Repeats
3.1.4. H3K9me2 and H3K27me2 Associated with Semi-Degraded Repeats
3.1.5. Predictions for Future Proposed Research
3.2. The Genomic Organization of Plant Genomes Revealed by Histone Marks
4. Materials and Methods
4.1. RepeatExplorer2 Analysis of Repeat Content
4.2. Immunolabelling
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome size diversity and its impact on the evolution of land plants. Genes 2018, 9, 88. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. New record holders for maximum genome size in eudicots and monocots. J. Bot. 2010, 2010, 527357. [Google Scholar] [CrossRef]
- Schubert, I.; Vu, G.T.H. Genome stability and evolution: Attempting a holistic view. Trends Plant Sci. 2016, 21, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Morton, J.; Pellicer, J.; Leitch, I.J.; Leitch, A.R. Genome downsizing after polyploidy: Mechanisms, rates and selection pressures. Plant J. 2021, 107, 1003–1015. [Google Scholar] [CrossRef] [PubMed]
- Kirik, A.; Salomon, S.; Puchta, H. Species-specific double-strand break repair and genome evolution in plants. EMBO J. 2000, 19, 5562–5566. [Google Scholar] [CrossRef] [PubMed]
- Vu, G.T.H.; Cao, H.X.; Reiss, B.; Schubert, I. Deletion-bias in DNA double-strand break repair differentially contributes to plant genome shrinkage. New Phytol. 2017, 214, 1712–1721. [Google Scholar] [CrossRef]
- Cossu, R.M.; Casola, C.; Giacomello, S.; Vidalis, A.; Scofield, D.G.; Zuccolo, A. LTR retrotransposons show low levels of unequal recombination and high rates of intraelement gene conversion in large plant genomes. Genome Biol. Evol. 2017, 9, 3449–3462. [Google Scholar] [CrossRef]
- Fedoroff, N.V. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Kelly, L.J.; Renny-Byfield, S.; Pellicer, J.; Macas, J.; Novak, P.; Neumann, P.; Lysak, M.A.; Day, P.D.; Berger, M.; Fay, M.F.; et al. Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 2015, 208, 596–607. [Google Scholar] [CrossRef]
- Novák, P.; Guignard, M.S.; Neumann, P.; Kelly, L.J.; Mlinarec, J.; Koblížková, A.; Dodsworth, S.; Kovařík, A.; Pellicer, J.; Wang, W.; et al. Repeat-sequence turnover shifts fundamentally in species with large genomes. Nat. Plants 2020, 6, 1325–1329. [Google Scholar] [CrossRef]
- Maumus, F.; Quesneville, H. Impact and insights from ancient repetitive elements in plant genomes. Curr. Opin. Plant Biol. 2016, 30, 41–46. [Google Scholar] [CrossRef]
- Ma, J.X.; Bennetzen, J.L. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 2006, 103, 383–388. [Google Scholar] [CrossRef]
- Ma, J.X.; Devos, K.M.; Bennetzen, J.L. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004, 14, 860–869. [Google Scholar] [CrossRef]
- Becher, H.; Ma, L.; Kelly, L.J.; Kovarik, A.; Leitch, I.J.; Leitch, A.R. Endogenous pararetrovirus sequences associated with 24 nt small RNAs at the centromeres of Fritillaria imperialis L. (Liliaceae), a species with a giant genome. Plant J. 2014, 80, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Hatlen, A.; Kelly, L.J.; Becher, H.; Wang, W.; Kovarik, A.; Leitch, I.J.; Leitch, A.R. Angiosperms are unique amongst land plants for the occurrence of key genes in the RNA-Directed DNA methylation (RdDM) pathway. Genome Biol. Evol. 2015, 7, 2648–2662. [Google Scholar] [CrossRef] [PubMed]
- Houben, A.; Demidov, D.; Gernand, D.; Meister, A.; Leach, C.R.; Schubert, I. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 2003, 33, 967–973. [Google Scholar] [CrossRef]
- Fuchs, J.; Demidov, D.; Houben, A.; Schubert, I. Chromosomal histone modification patterns—From conservation to diversity. Trends Plant Sci. 2006, 11, 199–208. [Google Scholar] [CrossRef]
- Fuchs, J.; Jovtchev, G.; Schubert, I. The chromosomal distribution of histone methylation marks in gymnosperms differs from that of angiosperms. Chromosome Res. 2008, 16, 891–898. [Google Scholar] [CrossRef]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Fuchs, J.; Schubert, I. Chromosomal distribution and functional interpretation of epigenetic histone marks in plants. In Plant Cytogenetics: Genome Structure and Chromosome Function; Bass, H.W., Birchler, J.A., Eds.; Springer: New York, NY, USA, 2012; pp. 231–253. [Google Scholar]
- Maumus, F.; Quesneville, H. Deep investigation of Arabidopsis thaliana junk DNA reveals a continuum between repetitive elements and genomic dark matter. PLoS ONE 2014, 9, e94101. [Google Scholar] [CrossRef] [PubMed]
- Naish, M.; Alonge, M.; Wlodzimierz, P.; Tock, A.J.; Abramson, B.W.; Schmücker, A.; Mandáková, T.; Jamge, B.; Lambing, C.; Kuo, P.; et al. The genetic and epigenetic landscape of the Arabidopsis centromeres. Science 2021, 374, eabi7489. [Google Scholar] [CrossRef]
- Baud, A.; Wan, M.; Nouaud, D.; Francillonne, N.; Anxolabehere, D.; Quesneville, H. Traces of transposable elements in genome dark matter co-opted by flowering gene regulation networks. Peer Community J. 2022, 2, e14. [Google Scholar] [CrossRef]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef]
- Di Stefano, M.; Nützmann, H.-W.; Marti-Renom, M.A.; Jost, D. Polymer modelling unveils the roles of heterochromatin and nucleolar organizing regions in shaping 3D genome organization in Arabidopsis thaliana. Nucleic Acids Res. 2021, 49, 1840–1858. [Google Scholar] [CrossRef] [PubMed]
- Rabl, C. Über Zelltheilung. Morphol. Jahrb. 1885, 10, 214–330. [Google Scholar]
- Leitch, A.R. Higher levels of organization in the interphase nucleus of cycling and differentiated cells. Microbiol. Mol. Biol. Rev. 2000, 64, 138–150. [Google Scholar] [CrossRef]
- Mei, W.; Stetter Markus, G.; Gates Daniel, J.; Stitzer Michelle, C.; Ross-Ibarra, J. Adaptation in plant genomes: Bigger is different. Am. J. Bot. 2018, 105, 16–19. [Google Scholar] [CrossRef]
- Novak, P.; Neumann, P.; Macas, J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinform. 2010, 11, 378. [Google Scholar] [CrossRef]
- Lysak, M.; Fransz, P.; Schubert, I. Cytogenetic analyses of Arabidopsis. In Arabidopsis Protocols; Salinas, J., Sanchez-Serrano, J.J., Eds.; Humana Press: Totowa, NJ, USA, 2006; pp. 173–186. [Google Scholar]
- Doležel, J.; Binarova, P.; Lucretti, S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant. 1989, 31, 113–120. [Google Scholar] [CrossRef]
- Marques, A.; Fuchs, J.; Ma, L.; Heckmann, S.; Guerra, M.; Houben, A. Characterization of eu- and heterochromatin of Citrus with a focus on the condensation behavior of 45S rDNA chromatin. Cytogenet. Genome Res. 2011, 134, 72–82. [Google Scholar] [CrossRef] [PubMed]


| Repeat Type | Genome Proportion |
|---|---|
| (% of the Genome) | |
| LTR retrotransposons | |
| Ty1/Copia-like | 4.72 |
| Ty3/Gypsy-like | 18.91 |
| LTR (other) | 0.90 |
| Non-LTR retrotransposons | |
| LINEs | 0.19 |
| DNA transposons | |
| EnSpm/CACTA-like | 2.31 |
| Other | |
| Pararetrovirus | 0.38 |
| Ribosomal DNA | 0.22 |
| Satellite repeats | 0.25 |
| Unknown | 5.50 |
| TOTAL | 33.4 |
| Histone Mark Detected | Antibody Used | Dilution Used | Catalogue Number in Upstate® |
|---|---|---|---|
| H3K4me1 | Rabbit anti-H3K4me1 | 1:200 | 07-436 |
| H3K4me2 | Rabbit anti-H3K4me2 | 1:300 | 07-030 |
| H3K9me1 | Rabbit anti-H3K9me1 | 1:200 | 07-395 |
| H3K9me2 | Rabbit anti-H3K9me2 | 1:300 | 07-441 |
| H3K9me3 | Rabbit anti-H3K9me3 | 1:300 | 07-473 |
| H3K27me1 | Rabbit anti-H3K27me1 | 1:100 | 07-448 |
| H3K27me2 | Rabbit anti-H3K27me2 | 1:50 | 07-452 |
| H3K27me3 | Rabbit anti-H3K27me3 | 1:100 | 07-449 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leitch, A.R.; Ma, L.; Dodsworth, S.; Fuchs, J.; Houben, A.; Leitch, I.J. The Role of Chromatin Modifications in the Evolution of Giant Plant Genomes. Plants 2023, 12, 2159. https://doi.org/10.3390/plants12112159
Leitch AR, Ma L, Dodsworth S, Fuchs J, Houben A, Leitch IJ. The Role of Chromatin Modifications in the Evolution of Giant Plant Genomes. Plants. 2023; 12(11):2159. https://doi.org/10.3390/plants12112159
Chicago/Turabian StyleLeitch, Andrew R., Lu Ma, Steven Dodsworth, Jörg Fuchs, Andreas Houben, and Ilia J. Leitch. 2023. "The Role of Chromatin Modifications in the Evolution of Giant Plant Genomes" Plants 12, no. 11: 2159. https://doi.org/10.3390/plants12112159
APA StyleLeitch, A. R., Ma, L., Dodsworth, S., Fuchs, J., Houben, A., & Leitch, I. J. (2023). The Role of Chromatin Modifications in the Evolution of Giant Plant Genomes. Plants, 12(11), 2159. https://doi.org/10.3390/plants12112159













