Abstract
Fruit nutritional value, plant growth, and yield can be compromised by deficient copper (Cu) bioavailability, which often appears in arable lands. This condition causes low Cu content and modifications in the ripening-associated processes in tomato fruit. This research studies the transcriptomic changes that occur in red ripe tomato fruit grown under suboptimal Cu conditions to shed light on the molecular mechanisms underlying this stress. Comparative RNA-sequencing and functional analyses revealed that Cu deficiency during cultivation activates signals for metal ion transport, cellular redox homeostasis, pyridoxal phosphate binding, and amino acid metabolism while repressing the response to phosphate starvation in harvested fruit. Transcriptomic analyses highlighted a number of novel Cu stress-responsive genes of unknown function and indicated that Cu homeostasis regulation in tomato fruit may involve additional components than those described in model plants. It also studied the regulation of high-affinity Cu transporters and a number of well-known Cu stress-responsive genes during tomato fruit ripening depending on Cu availability, which allowed potential candidates to be targeted for biotechnological improvements in reproductive tissues. We provide the first study characterizing the molecular responses of fruit to Cu deficiency stress for any fruit crop.
1. Introduction
Given their sessile nature, plants have developed a wide variety of responses to environmental cues, including those that focus on mitigating both biotic and abiotic stresses. Among abiotic factors, environmental nutrient availability is especially relevant because plants are at the basis of trophic chains and their nutritional deficiencies are often transferred to consumers [1]. In this context, plant resilience depends on metabolic, physiological, and developmental responses to adapt to variations in mineral nutrient availability. The most common plant responses to cope with nutrient scarcity involve the induction and reallocation of specific high-affinity transporters and the accumulation and/or secretion of enzymes and compounds that facilitate nutrient remobilization from soil [2]. Much less is known, however, about the changes that occur in fruits in response to exposure to deficient mineral nutrient stress during growth.
A high percentage of European arable lands are defined as deficient in bioavailable copper (Cu) because of soil alkalization and/or high organic matter contents in their composition [3]. Although plants act as effective miners when Cu is scarce, with severe restrictions they can show diverse deficiency symptoms, which mostly affect fruit production and cause important economic losses in agriculture [1,4,5,6,7]. This is aggravated by the fact that one climate change effect is growing atmospheric CO2 levels, which result in greater carbohydrate synthesis in plants and a general loss of their nutritional quality. Indeed, some meta-analyses indicate that Cu and other metal deficiencies would be exacerbated in forthcoming years due to a rise in CO2, which would increase problems of obesity and the ‘hidden hunger’ in the world [8,9]. Overall, the integration between Cu homeostasis and the nutritional value of food is needed and will become increasingly relevant in the near future, according to predicted climate change effects, because environmental stress conditions could strongly impact food production and quality. To the best of our knowledge, only Lafuente et al. [10] have recently described how exposing plants to Cu deficiency during cultivation influences the nutritional status and antioxidant capacity of harvested fruit when focusing on tomatoes. Still, there is no information about the components of the Cu homeostasis network in tomatoes or the molecular mechanisms driven by Cu deficiency stress in this fruit.
Plant cells require Cu to maintain essential processes such as photosynthesis, respiration, scavenging ROS, cell wall lignification, response to pathogens, hormone perception, and metabolism [11,12,13]. The dual nature of Cu as an essential micronutrient, but a toxic one in excess, makes a complex homeostasis network necessary whereby Cu content is tightly, but dynamically, regulated [14,15,16]. Cu is soluble in the cupric (Cu2+) form but is relatively inert in the cuprous (Cu+) state. When available in growth media, Cu2+ enters plant root cells through promiscuous divalent cation transporters (YSL, ZIP) [11,17,18]. However, when scarce, plants use a Cu+-specific mobilization system based on the acidification of the external medium by H+-ATPases, plasma membrane NADPH-dependent cupric reductases (FRO4 and FRO5) [13,19], and the participation of high-affinity Cu+ transporters (COPTs) that incorporate Cu+ inside cells from the apoplast [12,20,21,22]. In line with this, there is a prevailing dogma of protein-protein interactions that mediate Cu+ delivery by which COPT members facilitate Cu+ entry across the membrane and modulate its transfer to specific metallochaperones for targeted distribution to different locations and organelles [23,24]. Cu delivery to cytosolic metalloproteins, such as Cu/Zinc (Zn) superoxide dismutases (Cu/ZnSOD) CSD1 and CSD2, is mediated by the Cu chaperone for superoxide dismutase (CCS) [25]. In chloroplasts, the major consumers of Cu in plants, CCS cuprochaperone also delivers Cu to chloroplastic Cu/ZnSOD, Cu+ P-type ATPase PAA2, and plastocyanin, among other proteins [26,27,28]. Likewise, CCH and ATOX1 are cuprochaperones that deliver Cu+ to the Cu+-ATPases located at the endoplasmic reticulum and are more related to ethylene signaling, while COX17 mediates Cu delivery to the cytochrome c oxidase (COX) of the respiratory chain in mitochondria [29,30,31,32,33]. By doing so, cuprochaperones minimize the immediate toxicity caused by highly reactive Cu+ traffic, which may affect the integrity of membranes and Ca2+/K+ channels and trigger Ca2+- and reactive oxygen species (ROS)-mediated downstream signals [26,34,35].
Therefore, to conciliate dynamic changes in Cu supply and demand and to adapt to environmental metal restrictions or excesses, plants have evolved sophisticated mechanisms that involve a complex homeostatic network to control Cu uptake, delivery to target proteins, and detoxification. In response to Cu scarcity, in a nutshell, plants activate Cu uptake and mobilization and minimize Cu utilization in dispensable pathways by prioritizing Cu for essential processes. In this context, the SQUAMOSA PROMOTER-BINDING-LIKE 7 (SPL7) transcription factor is a key player in promoting Cu uptake under Cu limitation by inducing not only the COPT members located at the plasma membrane [36], but also the expression of iron (Fe) superoxide dismutase FSD1, the CCS chaperone, and several genes that encode laccases and plastocyanin. In addition, SPL7 modulates posttranscriptional regulation events by inducing microRNAs, such as miR398. This, in turn, mediates the mRNA down-regulation of CSD1 and CSD2 [27,36,37]. The substitution of CSD1 and CSD2 for their Fe counterparts, FSD1, when Cu is shortened is considered a hallmark strategy for replacing dispensable Cu proteins with other metalloproteins by performing a similar task. It is surprising that, in spite of its relevance for triggering the Cu-deficiency stress signal and orchestrating the subsequent response, the expression of the SPL7 transcription factor is not induced by Cu limitation. Instead, the SPL7 function is regulated by Cu content throughout conformational changes in its structure, which activate/inactivate its ability to induce the Cu deficiency response [38].
Most of our current understanding of Cu homeostasis regulation comes from research performed on Arabidopsis model plants. In horticultural crops, however, research into this topic started a few years ago by identifying and characterizing COPT family members in different species [39,40,41,42,43,44,45,46]. Nevertheless, studies on their regulation under stress conditions and information about other Cu homeostasis network components are still lacking.
Encouraged by our previous data, which demonstrated that exposing tomato plants to low Cu availability during cultivation leads to deficient Cu levels in the peel of harvested fruit [10] and might potentially be transferred to consumers, in the present work we aimed to uncover the molecular mechanisms underlying the Cu deficiency response in tomato fruit by a transcriptomic approach and to focus on a commercial red ripe (RR) mature stage. We found that well-known Cu homeostasis network components, together with a number of proteins of unknown function, were regulated in RR fruit in response to this stress. The results also revealed that adaptation of fruit to low Cu availability might involve the regulation of other nutrients, such as phosphate. In addition, the study of the transcriptional changes of the relevant genes involved in the Cu-stress response, i.e., all the COPT family members, two promiscuous divalent transporters (ZIP and YSL), two cuprochaperones (CCH and ATOX1), and two superoxide dismutases (CSD and FeSOD), in three different ripening stages indicated that Cu homeostasis is modulated during the tomato fruit maturation process, and a Cu-deficient environment during cultivation provokes changes in the regulation of this gene network from the most immature stages.
2. Results
2.1. Fruit Transcriptomic Changes in Response to Cu-Deficient Growing Conditions
Given the lower Cu levels found in the pericarp of fruit harvested from plants cultivated under Cu deficiency compared to Cu sufficiency (control) conditions [10], the transcriptome of this tissue was studied in the RR fruit harvested from both environments (Figure 1). In all, 284 differentially expressed genes (DEGs, Padj < 0.05) were identified, of which 119 and 165 were up- and down-regulated, respectively, in response to Cu deficiency.
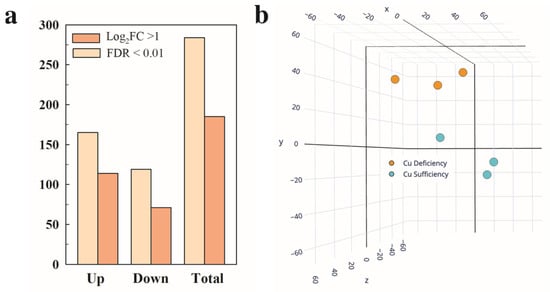
Figure 1.
Differential expression and multivariate analysis of tomato fruit in response to Cu deficiency growing conditions. (a) Number of differentially expressed genes (DEGs, Padj < 0.05) up- and down-regulated in the red ripe fruit harvested from the Cu deficiency conditions in relation to Cu sufficiency. Those DEGs that met an additional cutoff of 2 in the fold change (FC) expression levels (Log2 FC ≥ 1) are also indicated. (b) Principal component analysis (PCA) based on the total number of DEGs. The three axes in the PCA account for 80.1% of the total variance between the Cu sufficiency and Cu deficiency conditions (X = 36.15%, Y = 28.33%, Z = 15.62%). Three biological replicates from each condition were used for all the analyses.
When a cutoff of log2FC > 1 (2 in the fold change (FC)) induction/repression levels was set, those numbers were lowered to 71 and 114, respectively (Figure 1a). All the DEGs were used for the principal component analysis (PCA). Two different groups of samples were found that corresponded to the Cu sufficiency and Cu deficiency conditions. The three axes in the PCA accounted for 80.1% of the total variance among conditions (X = 36.15%, Y = 28.33%, and Z = 15.62%). The three biological replicates among each condition were also closely grouped, showing good reproducibility of the transcriptomic results (Figure 1b). It should be noted that in both the Cu sufficiency and Cu deficiency groups of samples, one of the three replicates showed more variability, but despite this, the samples grouped correctly in the PCA, and the statistical analysis (Padj < 0.05) provided a reasonable number of DEGs (Figure 1), which supports the robustness of the data.
The full list of DEGs (284 genes) between the RR fruit harvested from the Cu sufficiency and Cu deficiency conditions is shown in Table S1. Of them, the list of the top 10 induced/repressed genes by Cu deficiency is provided in Table 1.

Table 1.
List of the top 10 induced and repressed genes in tomato fruit in response to Cu deficiency stress growing conditions.
The most induced genes were related to mono- and divalent cation transport and translocation. Thus, the highest induction (~6-fold) was for the high-affinity copper transporter SlCOPT2, followed by the cation/H+ exchanger SlCHX18 (~5-fold) and a gene encoding a calcium-translocating P-type ATPase (~4.8-fold). Then a set of genes with diverse functions, including a WRKY transcription factor, a glycosyl hydrolase, a cytochrome P450, and a chitinase, were induced more than 4.5-fold in this fruit compared to those harvested from the control sufficiency growing conditions (Table 1). It should be noted that the genes encoding for cuprochaperones CCH (Solyc11g007200) and ATOX1 (Solyc05g055310) were also induced by the Cu stress condition, albeit at lower FC levels (1.9 and 1.8, respectively, Table S1). It is noticeable that two genes of unknown function (Solyc03g111725 and Solyc09g097770) with no orthologous gene within the Arabidopsis genome were more than 4-fold induced (Table 1), and 26 more non-annotated genes were induced by Cu deficiency (Table S1). This means that almost 17% (28 out of 165) of the genes induced by this stress were unknown proteins in the tomato genome. Interestingly, the most repressed (~7-fold) gene (Solyc10g049533) and three additional genes showing fold change-repression values of about 3.8 (Solyc01g109100, Solyc01g109090, and Solyc07g053055) were also described as unknown proteins (Table 1), and 30 more non annotated genes were repressed by low Cu availability (Table S1). Therefore, more than 28% (34 out of 119) of the repressed genes had an unknown function. On the list of the top 10 repressed genes (Table 1), an inflorescence meristem receptor-like kinase (IMK2), a Ser/Thr protein kinase, and diverse genes encoding peptidases, proteases, and hydrolases also appeared. Last, a gene encoding a SPX domain protein (SPX3), related to the regulation of phosphate transport and sensing, was more than 3-fold repressed by Cu deficiency.
To narrow down the molecular responses driven by Cu deficiency stress under the growing conditions on the RR tomato fruit, a gene ontology analysis was performed on the total list of DEGs obtained from the transcriptomic comparison. Many biological processes (BPs) were induced by Cu deficiency (Figure S1), of which the most specific are shown in Figure 2. In contrast, the only overrepresented BP in the set of down-regulated genes was ‘cellular response to phosphate starvation’, which contained three SPX genes ranging from 1.7- to 3.4-fold change repressions (Figure 2). The ‘pyridoxal phosphate binding’ molecular function (MF) was overrepresented in the set of induced genes. The DEGs included in this MF mostly encode pyridoxal-5′-phosphate (PLP)-dependent transferases and other enzymes that need PLP to function, such as ACC synthase and a tyrosine transaminase protein (Figure 2). Of the induced BPs, ‘metal ion transport’ was especially relevant in the context of this work. This BP included the genes encoding for the SlCOPT2 transporter, a calcium-translocating P-type ATPase, a heavy metal-associated (HMA) Cu transporter protein, and two Cu chaperones (SlCCH and SlATOX1). The ‘cell redox homeostasis’ BP was also overrepresented and composed of several thioredoxin/glutaredoxin family protein members (Figure 2).
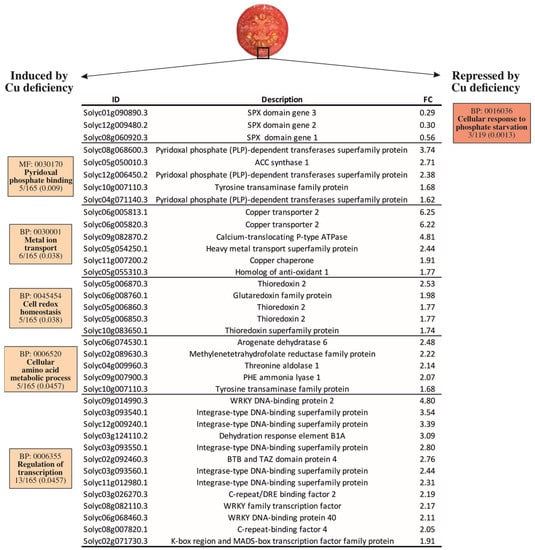
Figure 2.
Gene ontology analysis of the differentially expressed genes (DEGs) in the pericarp of the tomato fruit harvested from the plants grown under Cu sufficiency or Cu deficiency conditions. In boxes, the GO code is provided for each molecular function (MF) or biological process (BP), together with the term description, the number of DEGs belonging to that term on the total list of the induced (165) or repressed (119) DEGs, and the adjusted p-value (Padj < 0.05). ID: Gene identification number according to the iTAG3.2 database; description refers to the annotation of the best Arabidopsis thaliana hit for the selected gene. FC: Fold Change.
The ‘cellular amino acid metabolic process’ and the ‘regulation of transcription’ BPs were overrepresented in the up-regulated genes by Cu deficiency. Belonging to the ‘cellular amino acid metabolic process’ BP, plastid-localized arogenate dehydratase 6, involved in phenylalanine biosynthesis, was the most induced gene, while the phenylalanine ammonia lyase (SlPAL), at the entry point of the phenylpropanoids metabolism, was also ~2-fold induced by Cu deficiency compared to the control fruit. In line with this, a threonine aldolase that catalyzes the condensation between glycine and acetaldehyde and a tyrosine transaminase involved in L-phenylalanine degradation were also induced. A gene encoding a methylenetetrahydrofolate reductase (MTHFR) family protein and involved in methionine metabolism was also included in this BP (Figure 2). Last, several DEGs were grouped in the ‘regulation of transcription’ BP. These genes were mostly integrase-type, WRKY, and C-repeat/DRE DNA-binding transcription factors and ranged from 1.9- to 4.8-fold inductions by Cu stress versus the control fruit (Figure 2).
2.2. Changes Caused by Low Cu Availability in the Regulation of High-Affinity Cu Transporters (SlCOPTs) during Tomato Fruit Ripening
High-affinity Cu transporter family (SlCOPTs) members have been recently identified and characterized in the Solanum lycopersicum genome [39]. As SlCOPT2 was the most induced gene in the pericarp of tomato fruit in response to Cu deficiency growing conditions, we decided to extend this transcriptional study to all the members of this family by also considering different ripening stages (Figure 3).
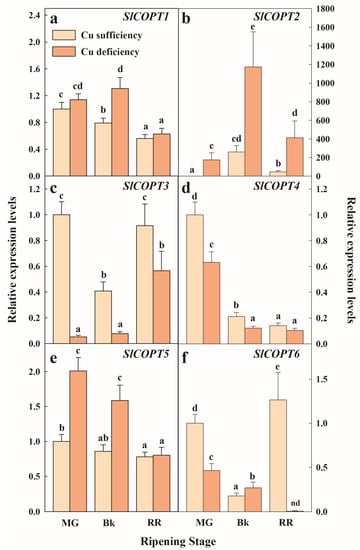
Figure 3.
Effect of Cu availability during growth on SlCOPTs gene expression in the fruit pericarp tissue at three ripening stages. Bars represent the mean values of three biological replicates ± standard error. Expression levels are relative to those obtained for the MG fruit harvested from the plants grown under Cu sufficiency conditions for SlCOPT1 (a), SlCOPT2 (b), SlCOPT3 (c), SlCOPT4 (d), SlCOPT5 (e) and SlCOPT6 (f), separately. In each panel, different letters indicate significant differences between growing conditions and the fruit ripening stages, according to a two-way ANOVA analysis. SlCOPTs are named after their identification within the Solanum lycopersicum genome, according to Romero et al. [39]. MG: Mature Green; Bk: Breaker; RR: Red Ripe; nd: not detected expression.
Under the Cu sufficiency control conditions, SlCOPT2 expression transiently increased with maturation and peaked in the breaker (Bk) stage (Figure 3b). Conversely, the transcript levels of SlCOPT3 and SlCOPT6 showed a transitory decrease in Bk to achieve similar levels to the most immature stage (mature green, MG) in the RR fruit (Figure 3c,f). SlCOPT1, SlCOPT4, and SlCOPT5, however, continuously decreased with fruit maturation under the control conditions (Figure 3a,d,e). Similarly, the genes encoding SlCOPT1, SlCOPT4, SlCOPT5, and SlCOPT6 were repressed with ripening in the fruit grown under Cu deficiency conditions. Contrarily, SlCOPT3 gene expression increased at the end of the ripening process, and SlCOPT2 transcripts accumulated with ripening and sharply peaked in the Bk stage (Figure 3).
The expression profile of SlCOPT2 in the Cu-deficient fruit was parallel to that described under the control conditions but was around 5- to 6-fold higher in any maturity stage. It is also worth noticing in the MG stage that the expression of this gene under Cu deficiency was already ~170-fold higher than under control conditions (Figure 3b). Low Cu availability transiently induced the expression of SlCOPT1 in Bk (Figure 3a), and that of SlCOPT5 in the MG and Bk stages (Figure 3e). However, the Cu deficiency condition did not always cause the induction of SlCOPTs compared to the control fruit. In fact, the SlCOPT3 expression levels were consistently lower in the Cu-deficient fruit during ripening (Figure 3c), and the SlCOPT4 transcript decreased with Cu scarcity in the MG and Bk stages compared to the control fruit (Figure 3d). Similarly, SlCOPT6 gene expression was mostly repressed by Cu deficiency during ripening compared to the Cu sufficiency conditions, except for a slight induction in Bk (Figure 3f).
2.3. Regulation of Cu Stress-Responsive Genes during Tomato Fruit Ripening
In order to better understand the regulation of Cu homeostasis components in tomato, a set of well-known Cu stress-responsive genes was selected, and their expression profiles were studied in tomato fruit in three ripening stages (Figure 4). Because they were included in the ‘metal ion transport’ BP that resulted from the GO analysis (Figure 2), the genes encoding the SlCCH and SlATOX1 cuprochaperones were selected. In addition, two superoxide dismutases (SlCSD and SlFeSOD2) were included in these analyses because they have been reported to be good molecular markers of Cu stress. Last, two additional unspecific divalent transporters (SlZIP11 and SlYSL2) were also studied.
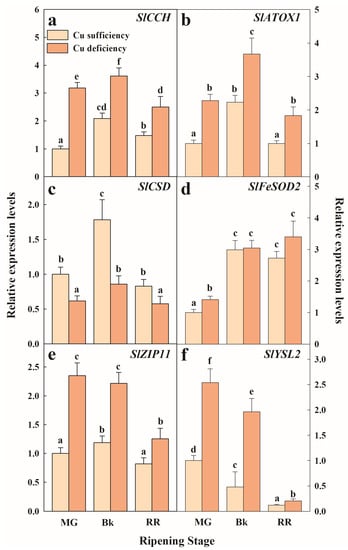
Figure 4.
Effect of Cu availability during growth on Cu-homeostasis-related gene expression in the fruit pericarp tissue at three ripening stages. Bars represent the mean values of three biological replicates ± standard error. Expression levels are relative to those obtained for the MG fruit harvested from the plants grown under Cu sufficiency conditions for SlCCH (a), SlATOX1 (b), SlCSD (c), SlFeSOD2 (d), SlZIP11 (e) and SlYSL2 (f), separately. In each panel, different letters indicate significant differences between growing conditions and fruit ripening stages, according to a two-way ANOVA analysis. MG: Mature Green; Bk: Breaker; RR: Red Ripe.
Our results generally indicated that under either Cu sufficiency control conditions or Cu deficiency stress, the genes related to Cu homeostasis were regulated in tomato fruit during the ripening process. A general expression pattern among the studied genes, except for SlFeSOD2 and SlYSL2, was found in the fruit that developed under Cu sufficiency, which consisted of similar relative expression levels in the most immature (MG) and mature (RR) fruit with a transitory peak in the accumulation of transcripts in the Bk stage (Figure 4). This pattern was also observed when examining the expression of SICCH, SlATOX1, and SlCSD in the Cu-deficiency-grown fruit (Figure 4). Statistical differences between conditions were still found in all the genes. Hence, the transcript levels of both Cu chaperones SlCCH and SlATOX1 were about 2-fold higher under Cu deficiency in all the ripening stages (Figure 4a,b). Conversely, the gene expression levels of Cu/Zn superoxide dismutase (SlCSD) were lower during maturation in the fruit exposed to Cu stress compared to the control fruit (Figure 4c). The expression levels of SlFeSOD2 were, however, the lowest in the MG stage and increased thereafter in the Bk stage, but remained unchanged until the end of the ripening process independently of the Cu condition. The expression levels of the Cu-stressed fruit were higher than in the control fruit, but only in the MG stage (Figure 4d). The transcript accumulation of divalent transporters SlZIP11 and SlYSL2 was also influenced by Cu deficiency during ripening. Thus, in spite of SlZIP11 expression transiently peaking in Bk in the control fruit and remaining steady until the Bk stage to thereafter decrease in the RR fruit under Cu-deficient conditions, transcript levels were always higher in the Cu-stressed fruit (Figure 4e). In contrast, the SlYSL2 pattern continuously decreased while ripening progressed under both conditions, although expression levels were about 2-fold higher in the Cu-deficient fruit at any maturity stage (Figure 4f).
3. Discussion
It is known that low Cu bioavailability leads to reduced growth, distortion, and chlorosis of young leaves and decreases fertility and seed formation in plants and cereal crops [1,17,47,48,49,50,51]. However, much less has been investigated in fruit species. Among the few that have been studied, the effect of Cu limitation during cultivation on the tomato plant phenotype was described decades ago [4,5,6,7], and only recently have the effects of this stress on fruit nutritional value and quality been described [10]. In this work, we identified the genome-wide molecular responses driven by this stress in tomato fruit in the commercial RR stage. To our knowledge, this is the first characterization of the mechanistic basis of Cu deficiency stress in any fruit species by focusing on fruit tissue.
3.1. Cu Deficiency Stress Regulates Cu Intake, Mobilisation and Economization during Tomato Fruit Ripening
A key point to controlling Cu homeostasis lies at the level of Cu intake from the apoplast. The comparative transcriptomic analyses between the RR fruit harvested from the plants grown under Cu sufficiency and Cu deficiency conditions revealed that SlCOPT2, which is one of the six COPT family members in Solanum lycopersicum [39], was the most induced gene in response to Cu stress in the whole tomato genome (Table 1). This gene belongs to the ‘metal ion transport’ BP induced in the RR fruit by Cu-limited conditions (Figure 2). A deeper transcriptional study further demonstrated that all the SlCOPT members were regulated throughout the fruit ripening process independently of exposure to Cu stress (Figure 3). Of them, only SlCOPT1, SlCOPT2, SlCOPT5, and SlCOPT6 were, at some point, induced by Cu deficiency stress compared to the control conditions. In fact, only the SlCOPT2 transcript levels were consistently higher under Cu-deficient conditions for the whole maturation process, including the RR stage (Figure 3b). This agrees with findings by Romero et al. [39], who showed that SlCOPT1 and SlCOPT2 were fully able to efficiently transport Cu through the plasma membrane, and both genes were induced by Cu shortage in tomato leaf and root tissues. SlCOPT5 and SlCOPT6 showed much more deficient functionality in spite of their encoding genes also being induced by Cu deficiency in roots and leaves [39]. The present results reveal that, as SlCOPT5 and SlCOPT6 are also up-regulated in fruit (Figure 3e,f), their participation in Cu distribution through plant tissues cannot be ruled out. Together, in accordance with studies in model plants [12,14,15,24,52,53,54,55,56], the results presented herein suggest that the induction of Cu uptake by high-affinity Cu transporters is a key response to low Cu bioavailability in tomato, and SlCOPT2 is a good molecular marker of Cu deficiency stress in this fruit. In this context, it is important to mention that the expression levels of two promiscuous divalent cation transporters, SlZIP11 and SlYSL2, which have been related to Cu and Fe transport and to micronutrients’ stress responses in other species [11,13,17], were also regulated during fruit maturation and induced by Cu deficiency independently of the ripening stage (Figure 4e,f). In addition, the cation/H+ antiporter CHX18, which has been linked with metal and other abiotic stress responses [57,58], was the second most induced gene by Cu scarcity (Table 1). Likewise, a heavy metal transporter (Solyc05g054250) and a P-type ATPase (Solyc09g082870), both of which belong to the ‘metal ion transport’ BP, were induced by Cu deficiency in the RR fruit (Figure 2). It is important to note that these genes are involved in the acidification and depolarization of the apoplast, which allows Cu2+ to reduce into Cu+ and be incorporated by COPTs into the cell [11,12,13,53]. These results suggest that Cu shortage during plant growth, which results in low Cu levels in the fruit pericarp [10], would trigger the induction of Cu-specific and general divalent transporters and energy-consuming components of Cu homeostasis to cope with the consequences of this stress condition in fruit.
Besides inducing Cu uptake, exposing tomatoes to limiting Cu availability during plant growth provoked the induction of cuprochaperones in fruit. Indeed, the expression patterns of SlCCH and SlATOX1 under Cu deficiency paralleled those of the control conditions throughout the maturation process, but with expression levels around 2-fold higher (Figure 2 and Figure 4a,b). This is a strategy that probably aims to avoid, to some extent, the oxidative damage caused by Cu+ entrance throughout COPT transporters, as reported in other plant species [11,12,13,53]. Further support for this idea comes from the induction of the ‘cell redox homeostasis’ BP in the Cu-deficient fruit (Figure 2), which involves a number of thioredoxin/glutaredoxin family protein members whose functions include ROS scavenging, antioxidation, and Fe-S cluster metabolism [59].
Another important point for controlling Cu homeostasis under metal scarcity is the minimization of Cu utilization in dispensable pathways. Substituting Cu/Zn superoxide dismutase (CSD) for its Fe counterpart (FeSOD) is a hallmark of Cu deficiency stress [12,26,37,60]. Accordingly, the results presented herein indicate that SlCSD and SlFeSOD2 expression patterns vary differently during ripening and in response to Cu stress in tomato fruit. Thus, while SlCSD transcript levels remained almost steady and always lower in the Cu-deficient fruit than in the control fruit, SlFeSOD2 gene expression increased with ripening and was higher under Cu scarcity than under the control conditions in the MG stage (Figure 4c,d), when Fe content is higher in the Cu-deficient fruit [10]. Therefore, these data suggest that tomato fruit economizes Cu availability during ripening through Cu substitution in non-essential metalloproteins by another metal, such as Fe, to perform analogous functions.
3.2. Cu Stress Response during Tomato Fruit Ripening Iunvolves the Regulation of Phosphate Homeostasis
Regarding the interaction of Cu homeostasis with other micronutrients [11,56,61,62,63,64], a Cu-Fe interdependence where phosphate (Pi) metabolism might also be involved has been suggested in model plants [55]. Perea-García et al. demonstrated that AtCOPT2 is pivotal for the response to simultaneous Cu and Fe scarcity and plays a part in the antagonistic response of these metals’ deficiency and Pi starvation [55]. Indeed, authors report that the expression of the genes encoding SPX domain proteins, which are molecular markers of Pi-starved plants and participate in Pi homeostasis as phosphate level sensors [65,66,67,68], is induced in copt2 mutant plants [55]. In agreement with this, our results indicated that the ‘cellular response to Pi starvation’ BP, which included three SPX-coding genes, was repressed in the Cu-deficient fruit (Figure 2). In line with Pi homeostasis, it is also interesting to note the induction of the ‘pyridoxal phosphate binding’ BP by Cu deficiency stress in tomato fruit (Figure 2). Pyridoxal 5’-phosphate (PLP) is considered the active form of vitamin B6, an essential cofactor in amino acid biosynthetic pathways and for a variety of enzymatic reactions like those related to heme/chlorophyll and ethylene synthesis [69]. Of the genes included in this BP, a number of PLP-dependent enzymes were found (Figure 2), such as the ACC synthase involved in ethylene formation. Similarly, several PLP-dependent enzymes belonging to the ‘cellular amino acid metabolic process’ BP were also overrepresented in the set of up-regulated genes by Cu deficiency stress. These enzymes are related to methionine (MTHFR), but mostly to the phenylalanine metabolism. In the last one, it is worth noting the ~2-fold induction by the Cu deficiency of the gene coding for phenylalanine ammonia lyase (SlPAL) because PAL was at the entry point of the phenylpropanoids metabolism, which may lead to metabolites performing an important antioxidant function [70]. Altogether, we interpret these results as a strategy of fruit to inhibit the response to Pi starvation under limited Cu availability. As this might be due to increased Pi contents in fruit as a consequence of low Cu accumulation, the aim is to avoid any possible cross-reactions that could trigger cell damage. Moreover, but not excluding them, these responses might be geared to increase the antioxidant capacity of fruit by means of the induction of SPX activity and the accumulation of phenylpropanoids and other secondary metabolites that derive from those amino acids.
Last, the ‘regulation of transcription’ BP, including a large number of transcription factors among which the WRKY and integrase-type DNA-binding proteins predominate, was also induced in the RR fruit in response to Cu deficiency (Figure 2). There is no information about the specific targets of these proteins to allow us to understand their participation in the Cu stress response. However, the marked inductions observed in this set of genes suggest a multitarget transcriptional response by which tomato fruit may attempt to adapt to low Cu levels.
4. Materials and Methods
4.1. Fruit Material and Growing Conditions
Tomato (S. lycopersicum L. cv. Moneymaker) seeds were sterilized and thereafter stratified before growing them in perlite in a greenhouse with 16 h of light at 22 °C and 19 °C at night, as described in [10]. Thirty plants were grouped into two sets of fifteen plants each, and then sorted into three biological replicates of five plants per group. The fifteen plants in the first set were irrigated with a home-made 0.5X Hoagland solution lacking any source of Cu [10] and prepared in demineralized water to produce Cu deficiency. The second set of plants was used as the control (control, +Cu). According to previous reports for this tomato cultivar [39], the watering solution used in the control plants was supplemented with 5 μM CuSO4⋅5 H2O (Sigma-Aldrich, Madrid, Spain) to generate Cu sufficiency growing conditions. Plants were allowed to grow, and fruit were harvested from each biological replicate in the MG (when fruit had reached their full size, and pulp and peel were entirely green), Bk (when fruit started losing their green color), and RR (when the fruit surface is completely red and displays the expected commercial appearance) ripening stages. The harvested fruit was immediately delivered to the laboratory for further analysis. The pericarp of three fruit from each biological replicate was frozen, homogenized, and stored at −80 °C for further analysis.
4.2. RNA Extraction and Gene Expression Analyses
Total RNA was extracted from the pericarp tissue of the fruit harvested in the different above-indicated ripening stages. RNA was extracted with Trizol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s directions, as described in [71]. RNA concentration was determined by a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA), and its integrity was assessed by agarose gel staining. cDNA was synthesized by following the manufacturer’s instructions in the Maxima H Minus First Strand cDNA Synthesis Kit (ThermoFisher Scientific, Waltham, MA, USA), as in [72]. Real-time quantitative PCRs were carried out as described in [73]. Gene-specific primer pairs (Table S1), 25 ng of cDNA, and SYBR Green qPCR MasterMix (Roche Diagnostics Ltd., Vienna, Austria) were used to generate relative gene expression data in a LightCycler480 System (Roche) instrument. Genes SlACT, SlCAC, SlSAND, and SlRPL2 were used to normalize the expression levels of target genes [74] using the Relative Expression Software Tool software (REST-MC, rest.gene-quantification.info). Values are the means of three biological replicates with two technical replicates ± SD.
4.3. RNA-Sequencing, Data Processing and Normalization
Total RNA integrity was assessed in a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) by the RNA 6000 Nano Kit (Agilent). Sequencing libraries were constructed with the TruSeq Stranded mRNA Library Prep Kit® with PolyA selection for ribo depletion (Illumina Inc., San Diego, CA, USA) following the manufacturer’s recommendations and three biological replicates of 2 μg of RNA per RR fruit pericarp sample for both the Cu sufficiency and Cu deficiency conditions. Libraries were sequenced on an Illumina NextSeq 500 platform, and 75-bp single-end reads were generated by the Genome Facility of the SCSIE-UV (Valencia, Spain). Data processing and normalization were performed as in [75]. Briefly, raw sequence reads were checked for quality by the FastP and FastQC software. Clean data were obtained by filtering sequence reads by at least a mean Q28 and mapped to the reference Solanum lycopersicum ITAG3.2 genome sequence (Phytozome genome ID: 514) by using the default settings in the TopHat2 v2.1.0 [76] software with the strand-specific alignment type. Only the mapped reads over the protein-coding genes with the RNA-seq quantification pipeline were considered for the raw read counts by assuming opposing strand specificity. The differential expression analysis between the two samples of our experimental design was performed by the edgeR R/Bioconductor package (v3.20.9) [77,78] in the R (v3.4.4) environment [79] based on the negative binomial distribution. Genes were considered DEGs when the significance of the comparison resulted in an adjusted [80] value of p ≤ 0.05. The Log2 RPM method was followed to estimate the unique gene expression levels. The number of DEGs that met a cutoff of Log2 fold change (FC) > 1 was also assessed. A PCA was conducted with the Log2 RPM values and was 3D-plotted by plot.ly. The total number of DEGs was used for gene ontology (GO) categorization purposes. The biological process enrichment analysis of DEGs was implemented by the TopGO (v2.30.0) package [81] with the default “weight01” algorithm. A GO term was considered significantly enriched if three DEGs or more were annotated for that term with a classic Fisher value of p < 0.05.
4.4. Statistical Analyses
The statistical analyses were performed using the STATGRAPHICS software.3. The gene expression data were subjected to analyses of variance (ANOVA), and the significance of the differences was determined by Tukey’s test (p < 0.05) on the mean values for all the ripening stages and both growing conditions.
5. Conclusions
One important conclusion from this study is that the fruit molecular mechanisms underlying the response to Cu deficiency mostly mimic those described in plants to adapt to this stress condition by focusing on the activation of Cu uptake, the regulation of the redox machinery to avoid oxidative damage, and the minimization of Cu utilization in dispensable pathways by prioritizing Cu utilization for essential processes. Interestingly, the results from the present research work into tomato fruit have revealed for the first time that responses to Cu deficiency may be commonly regulated in fruit, cereal crops, and model plants. This study also unravels a number of genes with unknown function putatively involved in Cu homeostasis, or at least highly responsive to Cu stress, that are uniquely involved in the tomato fruit response. Importantly, this is the case for 17% and 28% of the genes induced and repressed, respectively, in fruit by exposure to Cu scarcity during growth (Table 1 and Table S1). These facts further support the notion that Cu homeostasis in fruits cannot be explained by mere extrapolation from model plants, and new players/responsive genes are yet to be characterized. Further research is still needed to understand the role of these genes in the adaptive mechanisms of fruit to low Cu levels and to assess whether these responses are also found in the tomato plant’s molecular mechanisms driven by this stress.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12102062/s1, Figure S1: Complete list of the biological processes (BP) overrepresented on the list of up-regulated genes in response to the Cu deficiency conditions in the pericarp of tomato fruit in the RR ripening stage. In the yellow boxes, the GO code is provided for each BP, together with the term description, the number of DEGs belonging to that term on the total list of up-regulated genes, and the adjusted p-value (Padj < 0.05); Table S1: Total list of the induced (165) and repressed (119) genes in tomato fruit in response to Cu deficiency stress under growing conditions; Table S2: Forward (F) and reverse (R) primers used in this study.
Author Contributions
Conceptualization, P.R.; Methodology, P.R.; Formal analysis, P.R. and M.T.L.; Investigation, P.R. and M.T.L.; Writing—original draft preparation, P.R.; Project administration, P.R.; Funding acquisition, P.R. and M.T.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the TOMACOP Project (H2020-MSCA-IF-799712), within the framework of the Marie Skłodowska-Curie Actions and the European Horizon 2020 program, and by the ABACuS Project (CIDEXG/2022/58) within the Plan Generation Talent (Plan GenT) from the Conselleria de Innovación, Universidades, Ciencia y Sociedad Digital of the Generalitat Valenciana (GVA).
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The technical assistance of R. Sampedro is gratefully acknowledged. We also acknowledge the technical support of C. Navajo at the Plant Production Facility of SCSIE-UV, and the instruments and bioinformatic assistance of V. Sentandreu at the Genome Facility of SCSIE-UV.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Marschner, P. Marschner’s Mineral Nutrition of Higher Plants; Elsevier: Amsterdam, The Netherlands, 2012; ISBN 9780123849052. [Google Scholar]
- Schachtman, D.P.; Shin, R. Nutrient Sensing and Signaling: NPKS. Annu. Rev. Plant Biol. 2007, 58, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, C.; Panagos, P.; Lugato, E.; Huang, J.-H.; Orgiazzi, A.; Jones, A.; Fernández-Ugalde, O.; Borrelli, P.; Montanarella, L. Copper Distribution in European Topsoils: An Assessment Based on LUCAS Soil Survey. Sci. Total. Environ. 2018, 636, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.; Graves, C.J.; Winsor, G.W. Effects of Copper Deficiency and Liming on the Yield, Quality and Copper Status of Tomatoes, Lettuce and Cucumbers Grown in Peat. Sci. Hortic. 1978, 9, 199–205. [Google Scholar] [CrossRef]
- Arnon, D.I.; Stout, P.R. The Essentiality of Certain Elements in Minute Quantity for Plants with Special Reference to Copper. Plant Physiol. 1939, 14, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Piper, C.S. Investigations on Copper Deficiency in Plants. J. Agric. Sci. 1942, 32, 143–178. [Google Scholar] [CrossRef]
- Bailey, L.F.; McHargue, J.S. Copper Deficiency in Tomatoes. Am. J. Bot. 1943, 30, 558. [Google Scholar] [CrossRef]
- Weigel, H.J. Plant Quality Declines as CO2 Levels Rise. Elife 2014, 3, e03233. [Google Scholar] [CrossRef]
- Loladze, I. Hidden Shift of the Ionome of Plants Exposed to Elevated CO2 Depletes Minerals at the Base of Human Nutrition. Elife 2014, 3, e02245. [Google Scholar] [CrossRef]
- Lafuente, M.T.; Sampedro, R.; Vélez, D.; Romero, P. Deficient Copper Availability on Organoleptic and Nutritional Quality of Tomato Fruit. Plant Sci. 2023, 326, 111537. [Google Scholar] [CrossRef]
- Puig, S.; Andrés-Colás, N.; García-Molina, A.; Peñarrubia, L. Copper and Iron Homeostasis in Arabidopsis: Responses to Metal Deficiencies, Interactions and Biotechnological Applications. Plant Cell Environ. 2007, 30, 271–290. [Google Scholar] [CrossRef]
- Burkhead, J.L.; Gogolin Reynolds, K.A.; Abdel-Ghany, S.E.; Cohu, C.M.; Pilon, M. Copper Homeostasis. New Phytol. 2009, 182, 799–816. [Google Scholar] [CrossRef] [PubMed]
- Peñarrubia, L.; Romero, P.; Carrió-Seguí, A.; Andrés-Bordería, A.; Moreno, J.; Sanz, A. Temporal Aspects of Copper Homeostasis and Its Crosstalk with Hormones. Front. Plant Sci. 2015, 6, 255. [Google Scholar] [CrossRef] [PubMed]
- Migocka, M.; Malas, K. Plant Responses to Copper: Molecular and Regulatory Mechanisms of Copper Uptake, Distribution and Accumulation in Plants. In Plant Micronutrient Use Efficiency: Molecular and Genomic Perspectives in Crop Plants; Kamiya, T., Burritt, D.J., Tran, L.-S.P., Fujiwara, T.B.T.-P.M.U.E., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 71–86. ISBN 978-0-12-812104-7. [Google Scholar]
- Krämer, U.; Clemens, S. Functions and Homeostasis of Zinc, Copper, and Nickel in Plants. Top. Curr. Genet. 2006, 14, 216–271. [Google Scholar]
- Schulten, A.; Krämer, U. Interactions between Copper Homeostasis and Metabolism in Plants; Springer: Berlin/Heidelberg, Germany, 2017; pp. 111–146. [Google Scholar]
- Sheng, H.; Jiang, Y.; Ishka, M.R.; Chia, J.-C.; Dokuchayeva, T.; Kavulych, Y.; Zavodna, T.-O.; Mendoza, P.; Huang, R.; Smieshka, L.; et al. YSL3-Mediated Copper Distribution Is Required for Fertility, Grain Yield, and Size in Brachypodium. bioRxiv 2019, 13. [Google Scholar] [CrossRef]
- Waters, B.M.; Chu, H.H.; DiDonato, R.J.; Roberts, L.A.; Eisley, R.B.; Lahner, B.; Salt, D.E.; Walker, E.L. Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 Reveal Their Roles in Metal Ion Homeostasis and Loading of Metal Ions in Seeds. Plant Physiol. 2006, 141, 1446–1458. [Google Scholar] [CrossRef] [PubMed]
- Bernal, M.; Casero, D.; Singh, V.; Wilson, G.T.; Grande, A.; Yang, H.; Dodani, S.C.; Pellegrini, M.; Huijser, P.; Connolly, E.L.; et al. Transcriptome Sequencing Identifies SPL7-Regulated Copper Acquisition Genes FRO4 / FRO5 and the Copper Dependence of Iron Homeostasis in Arabidopsis. Plant Cell 2012, 24, 738–761. [Google Scholar] [CrossRef]
- Pilon, M. Moving Copper in Plants. New Phytol. 2011, 192, 305–307. [Google Scholar] [CrossRef]
- Sancenón, V.; Puig, S.; Mira, H.; Thiele, D.J.; Peñarrubia, L. Identification of a Copper Transporter Family in Arabidopsis Thaliana. Plant Mol. Biol. 2003, 51, 577–587. [Google Scholar] [CrossRef]
- Puig, S. Function and Regulation of the Plant COPT Family of High-Affinity Copper Transport Proteins. Adv. Bot. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Puig, S.; Lee, J.; Lau, M.; Thiele, D.J. Biochemical and Genetic Analyses of Yeast and Human High Affinity Copper Transporters Suggest a Conserved Mechanism for Copper Uptake. J. Biol. Chem. 2002, 277, 26021–26030. [Google Scholar] [CrossRef]
- Pope, C.R.; Flores, A.G.; Kaplan, J.H.; Unger, V.M. Structure and Function of Copper Uptake Transporters. Curr. Top. Membr. 2012, 69, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.J.; Winge, D.R. Copper Metallochaperones. Annu. Rev. Biochem. 2010, 79, 537–562. [Google Scholar] [CrossRef] [PubMed]
- Ravet, K.; Pilon, M. Copper and Iron Homeostasis in Plants: The Challenges of Oxidative Stress. Antioxid Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghany, S.E.; Burkhead, J.L.; Gogolin, K.A.; Andrés-Colás, N.; Bodecker, J.R.; Puig, S.; Peñarrubia, L.; Pilon, M. AtCCS Is a Functional Homolog of the Yeast Copper Chaperone Ccs1/Lys7. FEBS Lett. 2005, 579, 2307–2312. [Google Scholar] [CrossRef]
- Blaby-Haas, C.E.; Merchant, S.S. Lysosome-Related Organelles as Mediators of Metal Homeostasis. J. Biol. Chem. 2014, 289, 28129–28136. [Google Scholar] [CrossRef]
- Mira, H.; Martínez-García, F.; Peñarrubia, L. Evidence for the Plant-Specific Intercellular Transport of the Arabidopsis Copper Chaperone CCH. Plant J. 2001, 25, 521–528. [Google Scholar] [CrossRef]
- Garcia, L.; Welchen, E.; Gonzalez, D.H. Mitochondria and Copper Homeostasis in Plants. Mitochondrion 2014, 19, 269–274. [Google Scholar] [CrossRef]
- Balandin, T.; Castresana, C. AtCOX17, an Arabidopsis Homolog of the Yeast Copper Chaperone COX17. Plant Physiol. 2002, 129, 1852–1857. [Google Scholar] [CrossRef]
- Attallah, C.V.; Welchen, E.; Pujol, C.; Bonnard, G.; Gonzalez, D.H. Characterization of Arabidopsis Thaliana Genes Encoding Functional Homologues of the Yeast Metal Chaperone Cox19p, Involved in Cytochrome c Oxidase Biogenesis. Plant Mol. Biol. 2007, 65, 343–355. [Google Scholar] [CrossRef]
- Company, P.; González-Bosch, C. Identification of a Copper Chaperone from Tomato Fruits Infected with Botrytis Cinerea by Differential Display. Biochem. Biophys. Res. Commun. 2003, 304, 825–830. [Google Scholar] [CrossRef]
- Rodrigo-Moreno, A.; Andrés-Colás, N.; Poschenrieder, C.; Gunsé, B.; Peñarrubia, L.; Shabala, S. Calcium- and Potassium-Permeable Plasma Membrane Transporters Are Activated by Copper in Arabidopsis Root Tips: Linking Copper Transport with Cytosolic Hydroxyl Radical Production. Plant Cell Env. 2013, 36, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A Tidal Wave of Signals: Calcium and ROS at the Forefront of Rapid Systemic Signaling. Trends Plant Sci 2014, 19, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of Copper Homeostasis by Micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Molina, A.; Xing, S.; Huijser, P. Functional Characterisation of Arabidopsis SPL7 Conserved Protein Domains Suggests Novel Regulatory Mechanisms in the Cu Deficiency Response. BMC Plant Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Gabrielli, A.; Sampedro, R.; Perea-García, A.; Puig, S.; Lafuente, M.T. Identification and Molecular Characterization of the High-Affinity Copper Transporters Family in Solanum Lycopersicum. Int. J. Biol. Macromol. 2021, 192, 600–610. [Google Scholar] [CrossRef]
- Yuan, M.; Li, X.; Xiao, J.; Wang, S. Molecular and Functional Analyses of COPT/Ctr-Type Copper Transporter-like Gene Family in Rice. BMC Plant Biol. 2011, 11, 69. [Google Scholar] [CrossRef]
- Wang, Q.; Wei, N.; Jin, X.; Min, X.; Ma, Y.; Liu, W. Molecular Characterization of the COPT/Ctr-Type Copper Transporter Family under Heavy Metal Stress in Alfalfa. Int. J. Biol. Macromol. 2021, 181, 644–652. [Google Scholar] [CrossRef]
- Wang, H.; Du, H.; Li, H.; Huang, Y.; Ding, J.; Liu, C.; Wang, N.; Lan, H.; Zhang, S. Identification and Functional Characterization of the ZmCOPT Copper Transporter Family in Maize. PLoS ONE 2018, 13, e0199081. [Google Scholar] [CrossRef]
- Martins, V.; Bassil, E.; Hanana, M.; Blumwald, E.; Gerós, H. Copper Homeostasis in Grapevine: Functional Characterization of the Vitis Vinifera Copper Transporter 1. Planta 2014, 240, 91–101. [Google Scholar] [CrossRef]
- Jung, H.; Gayomba, S.R.; Yan, J.; Vatamaniuk, O.K. Brachypodium Distachyon as a Model System for Studies of Copper Transport in Cereal Crops. Front. Plant Sci. 2014, 5, 236. [Google Scholar] [CrossRef]
- Senovilla, M.; Abreu, I.; Escudero, V.; Cano, C.; Bago, A.; Imperial, J.; González-Guerrero, M. MtCOPT2 Is a Cu+ Transporter Specifically Expressed in Medicago Truncatula Mycorrhizal Roots. Mycorrhiza 2020, 30, 781–788. [Google Scholar] [CrossRef] [PubMed]
- Escaray, F.J.; Antonelli, C.J.; Copello, G.J.; Puig, S.; Peñarrubia, L.; Ruiz, O.A.; Perea-García, A. Characterization of the Copper Transporters from Lotus Spp. and Their Involvement under Flooding Conditions. Int. J. Mol. Sci. 2019, 20, 3136. [Google Scholar] [CrossRef]
- Vatamaniuk, O.K. Plant Movement and LAC of It: How Copper Facilitates Explosive Seed Dispersal. Proc. Natl. Acad. Sci. USA 2022, 119, e2208331119. [Google Scholar] [CrossRef] [PubMed]
- Rahmati Ishka, M.; Vatamaniuk, O.K. Copper Deficiency Alters Shoot Architecture and Reduces Fertility of Both Gynoecium and Androecium in Arabidopsis Thaliana. Plant Direct 2020, 4, e00288. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chia, J.C.; Sheng, H.; Jung, H.-I.; Zavodna, T.O.; Zhang, L.; Huang, R.; Jiao, C.; Craft, E.J.; Fei, Z.; et al. Arabidopsis Pollen Fertility Requires the Transcription Factors CITF1 and SPL7 That Regulate Copper Delivery to Anthers and Jasmonic Acid Synthesis. Plant Cell 2017, 29, 3012–3029. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Colás, N.; Perea-García, A.; Puig, S.; Peñarrubia, L. Deregulated Copper Transport Affects Arabidopsis Development Especially in the Absence of Environmental Cycles. Plant Physiol. 2010, 153, 170–184. [Google Scholar] [CrossRef]
- Sancenón, V.; Puig, S.; Mateu-Andrés, I.; Dorcey, E.; Thiele, D.J.; Peñarrubia, L. The Arabidopsis Copper Transporter COPT1 Functions in Root Elongation and Pollen Development. J. Biol. Chem. 2004, 279, 15348–15355. [Google Scholar] [CrossRef]
- Peñarrubia, L.; Andrés-Colás, N.; Moreno, J.; Puig, S. Regulation of Copper Transport in Arabidopsis Thaliana: A Biochemical Oscillator? J. Biol. Inorg. Chem. 2010, 15, 29–36. [Google Scholar] [CrossRef]
- Ishka, M.R.; Chia, J.C.; Vatamaniuk, O.K. Advances in Understanding of Copper Function and Transport in Plants. Cation Transp. Plants 2022, 205–226. [Google Scholar] [CrossRef]
- Castro, P.H.; Lilay, G.H.; Assunção, A.G.L. Chapter 1-Regulation of Micronutrient Homeostasis and Deficiency Response in Plants A2-Hossain, Mohammad Anwar; Kamiya, T., Burritt, D.J., Tran, L.-S.P., Fujiwara, T.B.T.-P.M.U.E., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 1–15. ISBN 978-0-12-812104-7. [Google Scholar]
- Perea-García, A.; Garcia-Molina, A.; Andrés-Colás, N.; Vera-Sirera, F.; Pérez-Amador, M.A.; Puig, S.; Peñarrubia, L. Arabidopsis Copper Transport Protein COPT2 Participates in the Cross Talk between Iron Deficiency Responses and Low-Phosphate Signaling. Plant Physiol. 2013, 162, 180–194. [Google Scholar] [CrossRef]
- Yruela, I. Copper in Plants: Acquisition, Transport and Interactions. Funct. Plant Biol. 2009, 36, 409–430. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Liao, Q.; Fu, H.; Ye, Z.; Mao, Y.; Luo, J.; Wang, Y.; Yuan, H.; Xin, J. Effect of Potassium Intake on Cadmium Transporters and Root Cell Wall Biosynthesis in Sweet Potato. Ecotoxicol. Environ. Saf. 2023, 250, 114501. [Google Scholar] [CrossRef] [PubMed]
- Talke, I.N.; Hanikenne, M.; Krämer, U. Zinc-Dependent Global Transcriptional Control, Transcriptional Deregulation, and Higher Gene Copy Number for Genes in Metal Homeostasis of the Hyperaccumulator Arabidopsis Halleri. Plant Physiol. 2006, 142, 148–167. [Google Scholar] [CrossRef] [PubMed]
- Meyer, Y.; Siala, W.; Bashandy, T.; Riondet, C.; Vignols, F.; Reichheld, J.P. Glutaredoxins and Thioredoxins in Plants. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2008, 1783, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Li, J.; Han, H.; Du, R.; Wang, X. Physiological and Molecular Mechanisms of Plant Responses to Copper Stress. Int. J. Mol. Sci. 2022, 23, 12950. [Google Scholar] [CrossRef]
- Srivastava, S. Plant-Metal Interactions; Srivastava, S., Srivastava, A.K., Suprasanna, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 3-030-20732-3. [Google Scholar]
- Abou Seeda, M.A.; Abou El-Nour, E.A.A.; Yassen, A.A.; Gad Mervat, M.; Sahar, M.Z. Interaction of Copper, Zinc, and Their Importance in Plant Physiology: Review, Acquisition and Transport. Middle East J. Appl. Sci. 2020, 10, 407–434. [Google Scholar] [CrossRef]
- Ghasemi-Fasaei, R.; Ronaghi, A. Interaction of Iron with Copper, Zinc, and Manganese in Wheat as Affected by Iron and Manganese in a Calcareous Soil. J. Plant Nutr. 2008, 31, 839–848. [Google Scholar] [CrossRef]
- Gayomba, S.R.; Zhai, Z.; Jung, H.-I.; Vatamaniuk, O.K. Local and Systemic Signaling of Iron Status and Its Interactions with Homeostasis of Other Essential Elements. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; González, E.; Bustos, R.; Linhares, F.; Leyva, A.; Paz-Ares, J. The Transcriptional Control of Plant Responses to Phosphate Limitation. J. Exp. Bot. 2004, 55, 285–293. [Google Scholar] [CrossRef]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a Sub-Family of Arabidopsis Genes with the SPX Domain Reveals Their Diverse Functions in Plant Tolerance to Phosphorus Starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Chien, P.-S.; Chiang, C.-P.; Leong, S.J.; Chiou, T.-J. Sensing and Signaling of Phosphate Starvation–from Local to Long Distance. Plant Cell Physiol. 2018, 59, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Shang, W.; Li, C.; Jia, L.; Wang, X.; Xing, G.; Zheng, W.M. Evolution of the SPX Gene Family in Plants and Its Role in the Response Mechanism to Phosphorus Stress. Open Biol. 2018, 8, 170231. [Google Scholar] [CrossRef]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal Phosphate Enzymes: Mechanistic, Structural, and Evolutionary Considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef] [PubMed]
- Howles, P.A.; Sewalt, V.J.H.; Paiva, N.L.; Elkind, Y.; Bate, N.J.; Lamb, C.; Dixon, R.A. Overexpression of L-Phenylalanine Ammonia-Lyase in Transgenic Tobacco Plants Reveals Control Points for Flux into Phenylpropanoid Biosynthesis. Plant Physiol. 1996, 112, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Rose, J.K.C. A Relationship between Tomato Fruit Softening, Cuticle Properties and Water Availability. Food Chem. 2019, 295, 300–310. [Google Scholar] [CrossRef]
- Romero, P.; Lafuente, M.T. Relative Humidity Regimes Modify Epicuticular Wax Metabolism and Fruit Properties during Navelate Orange Conservation in an ABA-Dependent Manner. Food Chem. 2022, 369, 130946. [Google Scholar] [CrossRef]
- Romero, P.; Alférez, F.; Lafuente, M.T. Involvement of Phospholipases and Sucrose in Carbon Starvation-Induced Non-Chilling Peel Pitting in Citrus Fruit. Postharvest Biol. Technol. 2020, 169, 111295. [Google Scholar] [CrossRef]
- Martin, L.B.B.; Romero, P.; Fich, E.A.; Domozych, D.S.; Rose, J.K.C. Cuticle Biosynthesis in Tomato Leaves Is Developmentally Regulated by Abscisic Acid. Plant Physiol. 2017, 174, 1384–1398. [Google Scholar] [CrossRef]
- Romero, P.; Lafuente, M.T. Abscisic Acid Deficiency Alters Epicuticular Wax Metabolism and Morphology That Leads to Increased Cuticle Permeability During Sweet Orange (Citrus sinensis) Fruit Ripening. Front. Plant Sci. 2020, 11, 1914. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate Alignment of Transcriptomes in the Presence of Insertions, Deletions and Gene Fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- R Core Team, R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 24 April 2023).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved Scoring of Functional Groups from Gene Expression Data by Decorrelating GO Graph Structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).