Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants
Abstract
:1. Introduction
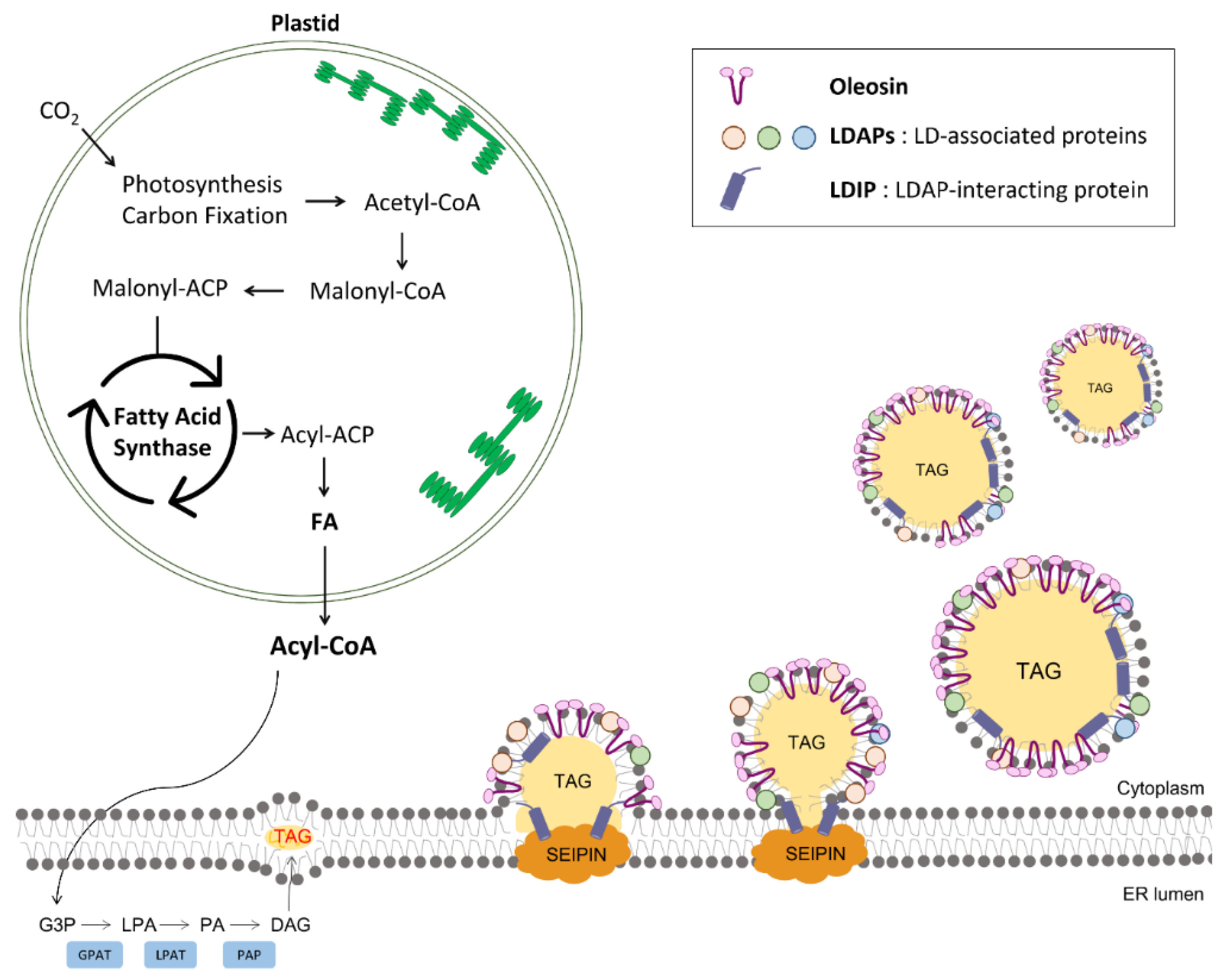
2. Biogenesis of LDs in Plant Cells
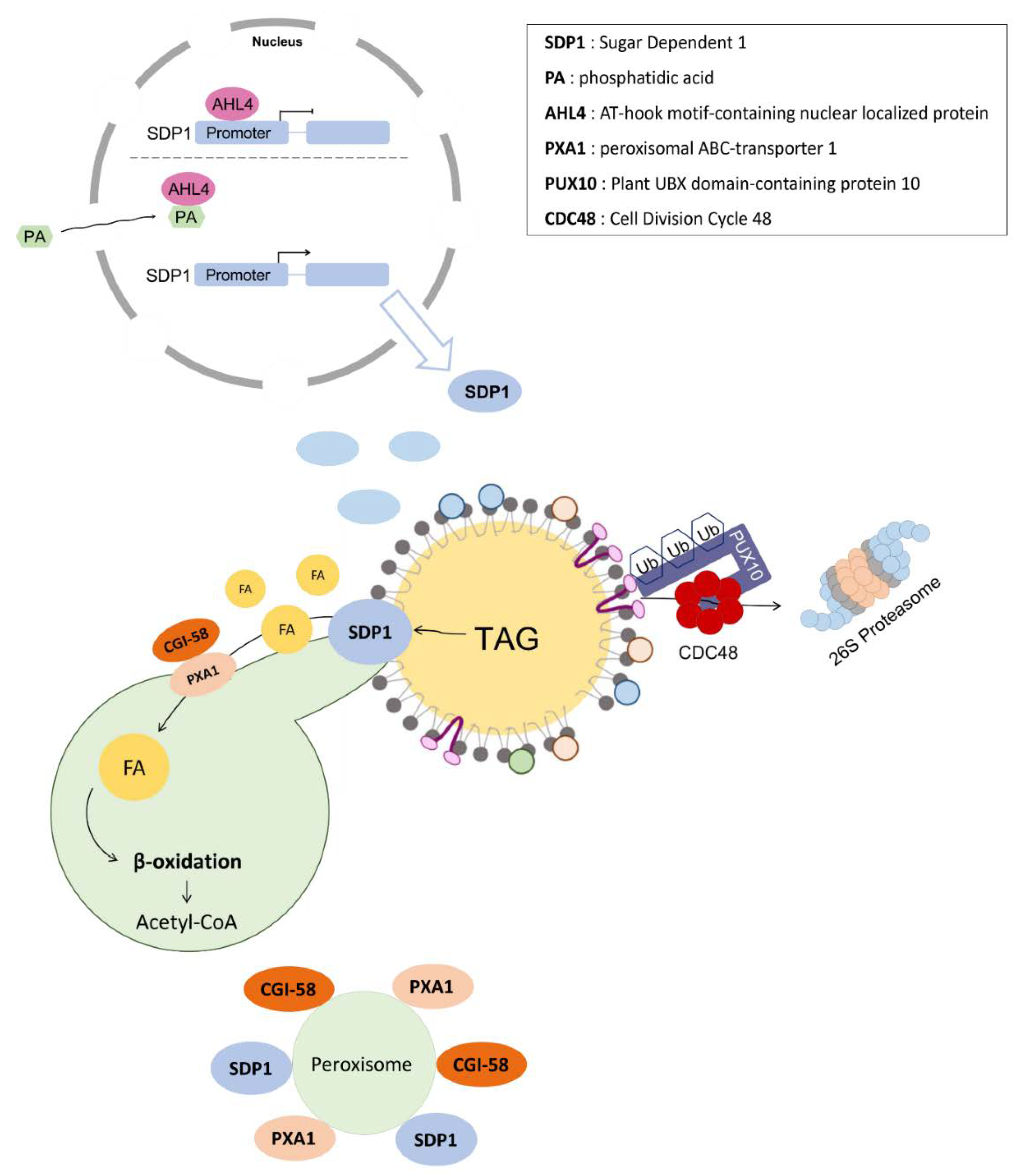
3. Mobilization of LDs in Plants: Pivotal Roles of LD-Associated Lipases
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lundquist, P.K.; Shivaiah, K.K.; Espinoza-Corral, R. Lipid droplets throughout the evolutionary tree. Prog. Lipid Res. 2020, 78, 101029. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cordes, K.R.; Farese, R.V.; Walther, T.C. Lipid droplets at a glance. J. Cell Sci. 2009, 122, 749–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onal, G.; Kutlu, O.; Gozuacik, D.; Dokmeci Emre, S. Lipid Droplets in Health and Disease. Lipids Health Dis. 2017, 16, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Gross, D.A.; Silver, D.L. Cytosolic lipid droplets: From mechanisms of fat storage to disease. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 304–326. [Google Scholar] [CrossRef]
- Cermelli, S.; Guo, Y.; Gross, S.P.; Welte, M.A. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 2006, 16, 1783–1795. [Google Scholar] [CrossRef] [Green Version]
- Herker, E.; Ott, M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrin. Met. 2011, 22, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Brink, J.T.R.; Fourie, R.; Sebolai, O.; Albertyn, J.; Pohl, C.H. The role of lipid droplets in microbial pathogenesis. J. Med. Microbiol. 2021, 70, 001383. [Google Scholar] [CrossRef]
- Bosch, M.; Sweet, M.J.; Parton, R.G.; Pol, A. Lipid droplets and the host-pathogen dynamic: FATal attraction? J. Cell Biol. 2021, 220, e202104005. [Google Scholar] [CrossRef]
- Murphy, D.J. The dynamic roles of intracellular lipid droplets: From archaea to mammals. Protoplasma 2012, 249, 541–585. [Google Scholar] [CrossRef]
- de Vries, J.; Ischebeck, T. Ties between Stress and Lipid Droplets Pre-date Seeds. Trends Plant Sci. 2020, 25, 1203–1214. [Google Scholar] [CrossRef]
- Romani, F.; Banic, E.; Florent, S.N.; Kanazawa, T.; Goodger, J.Q.D.; Mentink, R.A.; Dierschke, T.; Zachgo, S.; Ueda, T.; Bowman, J.L.; et al. Oil Body Formation in Marchantia polymorpha Is Controlled by MpC1HDZ and Serves as a Defense against Arthropod Herbivores. Curr. Biol. 2020, 30, 2815–2828. [Google Scholar] [CrossRef]
- Velazquez, A.P.; Tatsuta, T.; Ghillebert, R.; Drescher, I.; Graef, M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 2016, 212, 621–631. [Google Scholar] [CrossRef]
- Chitraju, C.; Mejhert, N.; Haas, J.T.; Diaz-Ramirez, L.G.; Grueter, C.A.; Imbriglio, J.E.; Pinto, S.; Koliwad, S.K.; Walther, T.C.; Farese, R.V., Jr. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab. 2017, 26, 407–418.e403. [Google Scholar] [CrossRef]
- Unger, R.H.; Orci, L. Lipotoxic diseases of nonadipose tissues in obesity. Int. J. Obes. 2000, 24 (Suppl. 4), S28–S32. [Google Scholar] [CrossRef] [Green Version]
- Bailey, A.P.; Koster, G.; Guillermier, C.; Hirst, E.M.; MacRae, J.I.; Lechene, C.P.; Postle, A.D.; Gould, A.P. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell 2015, 163, 340–353. [Google Scholar] [CrossRef] [Green Version]
- Dejgaard, S.Y.; Presley, J.F. Interactions of Lipid Droplets with the Intracellular Transport Machinery. Int. J. Mol. Sci. 2021, 22, 2776. [Google Scholar] [CrossRef]
- Henne, M. And three’s a party: Lysosomes, lipid droplets, and the ER in lipid trafficking and cell homeostasis. Curr. Opin. Cell Biol. 2019, 59, 40–49. [Google Scholar] [CrossRef]
- van Wijk, K.J.; Kessler, F. Plastoglobuli: Plastid Microcompartments with Integrated Functions in Metabolism, Plastid Developmental Transitions, and Environmental Adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef]
- Suzuki, M. Regulation of lipid metabolism via a connection between the endoplasmic reticulum and lipid droplets. Anat. Sci. Int. 2017, 92, 50–54. [Google Scholar] [CrossRef]
- Rottet, S.; Besagni, C.; Kessler, F. The role of plastoglobules in thylakoid lipid remodeling during plant development. Biochim. Biophys. Acta 2015, 1847, 889–899. [Google Scholar] [CrossRef] [Green Version]
- Holzl, G.; Dormann, P. Chloroplast Lipids and Their Biosynthesis. Annu. Rev. Plant Biol. 2019, 70, 51–81. [Google Scholar] [CrossRef]
- Jarvis, P.; Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef]
- Sadali, N.M.; Sowden, R.G.; Ling, Q.; Jarvis, R.P. Differentiation of chromoplasts and other plastids in plants. Plant Cell Rep. 2019, 38, 803–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.; Yi, T.; Ha, S.H. Diversity of Plastid Types and Their Interconversions. Front. Plant Sci. 2021, 12, 692024. [Google Scholar] [CrossRef] [PubMed]
- Espinoza-Corral, R.; Heinz, S.; Klingl, A.; Jahns, P.; Lehmann, M.; Meurer, J.; Nickelsen, J.; Soll, J.; Schwenkert, S. Plastoglobular protein 18 is involved in chloroplast function and thylakoid formation. J. Exp. Bot. 2019, 70, 3981–3993. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Friso, G.; Rowland, E.; Majsec, K.; van Wijk, K.J. The Plastoglobule-Localized Metallopeptidase PGM48 Is a Positive Regulator of Senescence in Arabidopsis thaliana. Plant Cell 2016, 28, 3020–3037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundquist, P.K.; Poliakov, A.; Giacomelli, L.; Friso, G.; Appel, M.; McQuinn, R.P.; Krasnoff, S.B.; Rowland, E.; Ponnala, L.; Sun, Q.; et al. Loss of plastoglobule kinases ABC1K1 and ABC1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 2013, 25, 1818–1839. [Google Scholar] [CrossRef] [Green Version]
- Youssef, A.; Laizet, Y.; Block, M.A.; Marechal, E.; Alcaraz, J.P.; Larson, T.R.; Pontier, D.; Gaffe, J.; Kuntz, M. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 2010, 61, 436–445. [Google Scholar] [CrossRef]
- Piller, L.E.; Abraham, M.; Dormann, P.; Kessler, F.; Besagni, C. Plastid lipid droplets at the crossroads of prenylquinone metabolism. J. Exp. Bot. 2012, 63, 1609–1618. [Google Scholar] [CrossRef]
- Chapman, K.D.; Aziz, M.; Dyer, J.M.; Mullen, R.T. Mechanisms of lipid droplet biogenesis. Biochem. J. 2019, 476, 1929–1942. [Google Scholar] [CrossRef]
- Zienkiewicz, K.; Zienkiewicz, A. Degradation of Lipid Droplets in Plants and Algae-Right Time, Many Paths, One Goal. Front. Plant Sci. 2020, 11, 579019. [Google Scholar] [CrossRef]
- Smith, S.; Witkowski, A.; Joshi, A.K. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 2003, 42, 289–317. [Google Scholar] [CrossRef]
- Cronan, J.E., Jr.; Waldrop, G.L. Multi-subunit acetyl-CoA carboxylases. Prog. Lipid Res. 2002, 41, 407–435. [Google Scholar] [CrossRef]
- Rawsthorne, S. Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 2002, 41, 182–196. [Google Scholar] [CrossRef]
- Wang, Z.; Benning, C. Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc. Trans. 2012, 40, 457–463. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Gugel, I.L.; Giavalisco, P.; Zeisler, V.; Schreiber, L.; Soll, J.; Philippar, K. FAX1, a novel membrane protein mediating plastid fatty acid export. PLoS Biol. 2015, 13, e1002053. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Zhang, Y.; Meng, H.; Li, S.; Wang, S.; Xiao, Z.; Chang, P.; Zhang, X.; Li, Q.; Guo, L.; et al. Characterization of Fatty Acid Exporters involved in fatty acid transport for oil accumulation in the green alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 2019, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Schnurr, J.A.; Shockey, J.M.; de Boer, G.J.; Browse, J.A. Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 2002, 129, 1700–1709. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.; Li, H.Y.; Zhang, J.P.; Chan, S.W.; Chye, M.L. Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Mol. Biol. 2008, 68, 571–583. [Google Scholar] [CrossRef] [Green Version]
- Leung, K.C.; Li, H.Y.; Mishra, G.; Chye, M.L. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Mol. Biol. 2004, 55, 297–309. [Google Scholar] [CrossRef]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef] [Green Version]
- Dahlqvist, A.; Stahl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef] [Green Version]
- Fei, W.; Du, X.; Yang, H. Seipin, adipogenesis and lipid droplets. Trends Endocrinol. Metab. 2011, 22, 204–210. [Google Scholar] [CrossRef]
- Cai, Y.; Goodman, J.M.; Pyc, M.; Mullen, R.T.; Dyer, J.M.; Chapman, K.D. Arabidopsis SEIPIN Proteins Modulate Triacylglycerol Accumulation and Influence Lipid Droplet Proliferation. Plant Cell 2015, 27, 2616–2636. [Google Scholar] [CrossRef] [Green Version]
- Pyc, M.; Cai, Y.; Greer, M.S.; Yurchenko, O.; Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Turning Over a New Leaf in Lipid Droplet Biology. Trends Plant Sci. 2017, 22, 596–609. [Google Scholar] [CrossRef]
- Wang, H.J.; Becuwe, M.; Housden, B.E.; Chitraju, C.; Porras, A.J.; Graham, M.M.; Liu, X.R.N.; Thiam, A.R.; Savage, D.B.; Agarwal, A.K.; et al. Seipin is required for converting nascent to mature lipid droplets. eLife 2016, 5, e16582. [Google Scholar] [CrossRef] [Green Version]
- Cartwright, B.R.; Binns, D.D.; Hilton, C.L.; Han, S.; Gao, Q.; Goodman, J.M. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell 2015, 26, 726–739. [Google Scholar] [CrossRef]
- Salo, V.T.; Belevich, I.; Li, S.; Karhinen, L.; Vihinen, H.; Vigouroux, C.; Magre, J.; Thiele, C.; Holtta-Vuori, M.; Jokitalo, E.; et al. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 2016, 35, 2699–2716. [Google Scholar] [CrossRef]
- Greer, M.S.; Cai, Y.; Gidda, S.K.; Esnay, N.; Kretzschmar, F.K.; Seay, D.; McClinchie, E.; Ischebeck, T.; Mullen, R.T.; Dyer, J.M.; et al. SEIPIN Isoforms Interact with the Membrane-Tethering Protein VAP27-1 for Lipid Droplet Formation. Plant Cell 2020, 32, 2932–2950. [Google Scholar] [CrossRef]
- Siao, W.; Wang, P.; Voigt, B.; Hussey, P.J.; Baluska, F. Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum-plasma membrane contact sites. J. Exp. Bot. 2016, 67, 6161–6171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taurino, M.; Costantini, S.; De Domenico, S.; Stefanelli, F.; Ruano, G.; Delgadillo, M.O.; Sanchez-Serrano, J.J.; Sanmartin, M.; Santino, A.; Rojo, E. SEIPIN Proteins Mediate Lipid Droplet Biogenesis to Promote Pollen Transmission and Reduce Seed Dormancy. Plant Physiol. 2018, 176, 1531–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.Y.; Park, K.Y.; Seo, Y.S.; Kim, W.T. Arabidopsis Small Rubber Particle Protein Homolog SRPs Play Dual Roles as Positive Factors for Tissue Growth and Development and in Drought Stress Responses. Plant Physiol. 2016, 170, 2494–2510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyc, M.; Gidda, S.K.; Seay, D.; Esnay, N.; Kretzschmar, F.K.; Cai, Y.; Doner, N.M.; Greer, M.S.; Hull, J.J.; Coulon, D.; et al. LDIP cooperates with SEIPIN and LDAP to facilitate lipid droplet biogenesis in Arabidopsis. Plant Cell 2021, 33, 3076–3103. [Google Scholar] [CrossRef]
- Pyc, M.; Cai, Y.Q.; Gidda, S.K.; Yurchenko, O.; Park, S.; Kretzschmar, F.K.; Ischebeck, T.; Valerius, O.; Braus, G.H.; Chapman, K.D.; et al. Arabidopsis lipid droplet-associated protein (LDAP) interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 2017, 92, 1182–1201. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.H.C. Plant Lipid Droplets and Their Associated Proteins: Potential for Rapid Advances. Plant Physiol. 2018, 176, 1894–1918. [Google Scholar] [CrossRef] [Green Version]
- Murphy, D.J. Structure, function and biogenesis of storage lipid bodies and oleosins in plants. Prog. Lipid Res. 1993, 32, 247–280. [Google Scholar] [CrossRef]
- Huang, C.Y.; Huang, A.H.C. Unique Motifs and Length of Hairpin in Oleosin Target the Cytosolic Side of Endoplasmic Reticulum and Budding Lipid Droplet. Plant Physiol. 2017, 174, 2248–2260. [Google Scholar] [CrossRef] [Green Version]
- Siloto, R.M.; Findlay, K.; Lopez-Villalobos, A.; Yeung, E.C.; Nykiforuk, C.L.; Moloney, M.M. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 2006, 18, 1961–1974. [Google Scholar] [CrossRef] [Green Version]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and functions of lipid droplets in plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: From Yeast to Man. J. Lipid Res. 2012, 53, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Mosblech, A.; Feussner, I.; Heilmann, I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 2009, 47, 511–517. [Google Scholar] [CrossRef]
- Lin, L.J.; Tai, S.S.; Peng, C.C.; Tzen, J.T. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 2002, 128, 1200–1211. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Cheng, Z.J.; Gan, L.; Zhang, H.; Wu, F.Q.; Lin, Q.B.; Wang, J.L.; Wang, J.; Guo, X.P.; Zhang, X.; et al. OsHSD1, a hydroxysteroid dehydrogenase, is involved in cuticle formation and lipid homeostasis in rice. Plant Sci. 2016, 249, 35–45. [Google Scholar] [CrossRef]
- Peramuna, A.; Bae, H.; Quinonero Lopez, C.; Fromberg, A.; Petersen, B.; Simonsen, H.T. Connecting moss lipid droplets to patchoulol biosynthesis. PLoS ONE 2020, 15, e0243620. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Graham, I.A. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 2001, 6, 72–78. [Google Scholar] [CrossRef]
- Deruyffelaere, C.; Purkrtova, Z.; Bouchez, I.; Collet, B.; Cacas, J.L.; Chardot, T.; Gallois, J.L.; D’Andrea, S. PUX10 Is a CDC48A Adaptor Protein That Regulates the Extraction of Ubiquitinated Oleosins from Seed Lipid Droplets in Arabidopsis. Plant Cell 2018, 30, 2116–2136. [Google Scholar] [CrossRef] [Green Version]
- Kretzschmar, F.K.; Mengel, L.A.; Muller, A.O.; Schmitt, K.; Blersch, K.F.; Valerius, O.; Braus, G.H.; Ischebeck, T. PUX10 Is a Lipid Droplet-Localized Scaffold Protein That Interacts with CELL DIVISION CYCLE48 and Is Involved in the Degradation of Lipid Droplet Proteins. Plant Cell 2018, 30, 2137–2160. [Google Scholar] [CrossRef] [Green Version]
- Eastmond, P.J. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 2006, 18, 665–675. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.A.; Quettier, A.L.; Shaw, E.; Eastmond, P.J. Seed Storage Oil Mobilization Is Important But Not Essential for Germination or Seedling Establishment in Arabidopsis. Plant Physiol. 2011, 157, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Fan, J.; Yan, C.; Roston, R.; Shanklin, J.; Xu, C. Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward beta-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 2014, 26, 4119–4134. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.A.; van Erp, H.; Quettier, A.L.; Shaw, E.; Menard, G.; Kurup, S.; Eastmond, P.J. The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 2013, 162, 1282–1289. [Google Scholar] [CrossRef] [Green Version]
- Thazar-Poulot, N.; Miquel, M.; Fobis-Loisy, I.; Gaude, T. Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. USA 2015, 112, 4158–4163. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Hayashi, Y.; Otomo, M.; Mano, S.; Oikawa, K.; Hayashi, M.; Nishimura, M. Sucrose Production Mediated by Lipid Metabolism Suppresses the Physical Interaction of Peroxisomes and Oil Bodies during Germination of Arabidopsis thaliana. J. Biol. Chem. 2016, 291, 19734–19745. [Google Scholar] [CrossRef] [Green Version]
- Chapman, K.D.; Trelease, R.N. Acquisition of membrane lipids by differentiating glyoxysomes: Role of lipid bodies. J. Cell Biol. 1991, 115, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- Zolman, B.K.; Silva, I.D.; Bartel, B. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 2001, 127, 1266–1278. [Google Scholar] [CrossRef]
- Park, S.; Gidda, S.K.; James, C.N.; Horn, P.J.; Khuu, N.; Seay, D.C.; Keereetaweep, J.; Chapman, K.D.; Mullen, R.T.; Dyer, J.M. The alpha/beta hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell 2013, 25, 1726–1739. [Google Scholar] [CrossRef] [Green Version]
- Cai, G.; Kim, S.C.; Li, J.; Zhou, Y.; Wang, X. Transcriptional Regulation of Lipid Catabolism during Seedling Establishment. Mol. Plant 2020, 13, 984–1000. [Google Scholar] [CrossRef]
- Kelly, A.A.; Shaw, E.; Powers, S.J.; Kurup, S.; Eastmond, P.J. Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol. J. 2013, 11, 355–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.J.; Yang, S.W.; Mao, H.Z.; Veena, S.P.; Yin, J.L.; Chua, N.H. Gene silencing of Sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol. Biofuels 2014, 7, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trites, M.J.; Clugston, R.D. The role of adipose triglyceride lipase in lipid and glucose homeostasis: Lessons from transgenic mice. Lipids Health Dis. 2019, 18, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweiger, M.; Lass, A.; Zimmermann, R.; Eichmann, T.O.; Zechner, R. Neutral lipid storage disease: Genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E289–E296. [Google Scholar] [CrossRef] [Green Version]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Strauss, J.G.; Gorkiewicz, G.; Zechner, R. Adipose triglyceridelipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Nur, B.G.; Gencpinar, P.; Yuzbasioglu, A.; Emre, S.D.; Mihci, E. Chanarin-Dorfman syndrome: Genotype-Phenotype Correlation. Eur. J. Med. Genet. 2015, 58, 238–242. [Google Scholar] [CrossRef]
- James, C.N.; Horn, P.J.; Case, C.R.; Gidda, S.K.; Zhang, D.; Mullen, R.T.; Dyer, J.M.; Anderson, R.G.; Chapman, K.D. Disruption of the Arabidopsis CGI-58 homologue produces Chanarin-Dorfman-like lipid droplet accumulation in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 17833–17838. [Google Scholar] [CrossRef] [Green Version]
- Muller, A.O.; Ischebeck, T. Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol. 2018, 217, 1062–1076. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.J.; Zaikova, K.; Yeom, S.-J.; Kim, Y.-S.; Lee, D.W. Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants. Plants 2022, 11, 1243. https://doi.org/10.3390/plants11091243
Choi YJ, Zaikova K, Yeom S-J, Kim Y-S, Lee DW. Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants. Plants. 2022; 11(9):1243. https://doi.org/10.3390/plants11091243
Chicago/Turabian StyleChoi, Yun Ju, Kseniia Zaikova, Soo-Jin Yeom, Yeong-Su Kim, and Dong Wook Lee. 2022. "Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants" Plants 11, no. 9: 1243. https://doi.org/10.3390/plants11091243
APA StyleChoi, Y. J., Zaikova, K., Yeom, S.-J., Kim, Y.-S., & Lee, D. W. (2022). Biogenesis and Lipase-Mediated Mobilization of Lipid Droplets in Plants. Plants, 11(9), 1243. https://doi.org/10.3390/plants11091243





