Effects of Harpin and Flg22 on Growth Enhancement and Pathogen Defense in Cannabis sativa Seedlings
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant and Microbe Materials
2.2. Pathogenicity Test
2.3. Gene Expression Analysis
2.4. Growth Test
3. Results
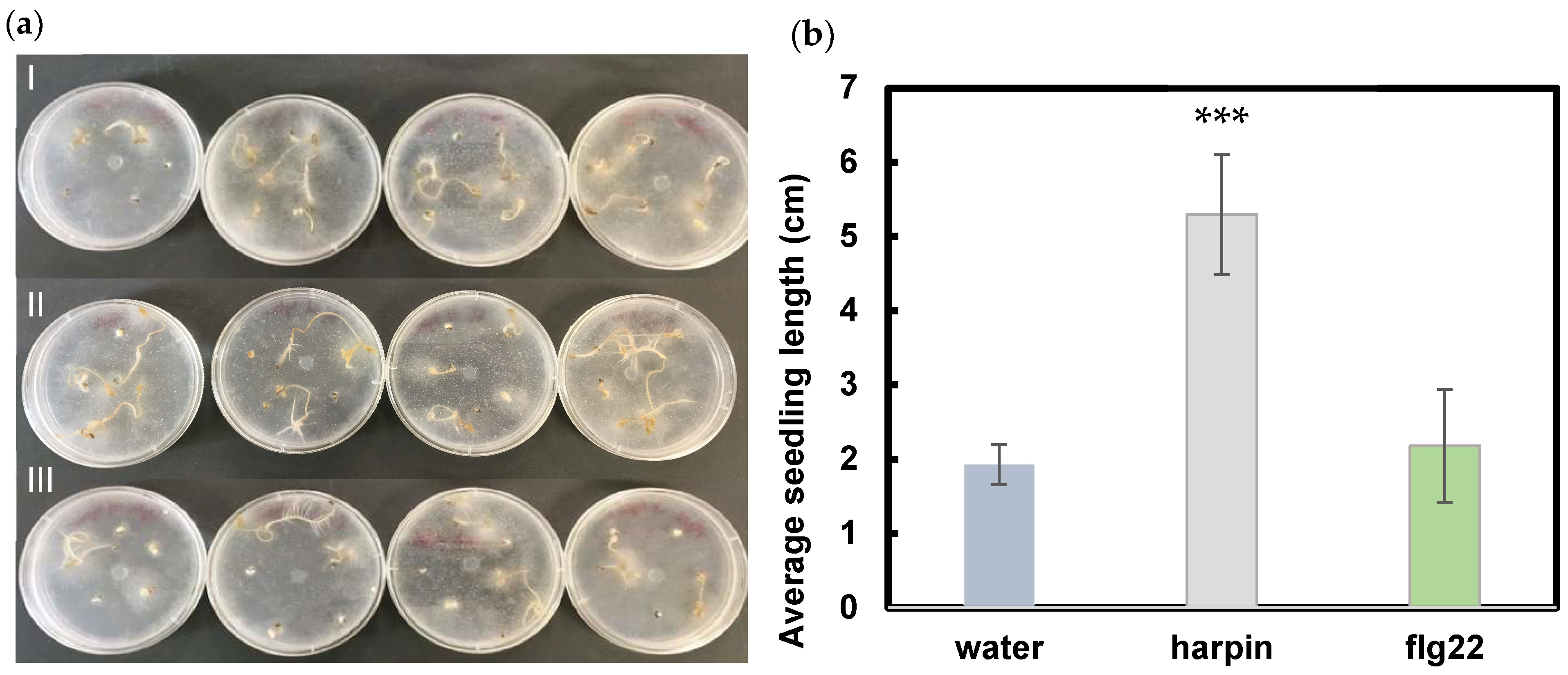
3.1. Harpin Induces Resistance to P. aphanidermatum in Hemp Seedlings
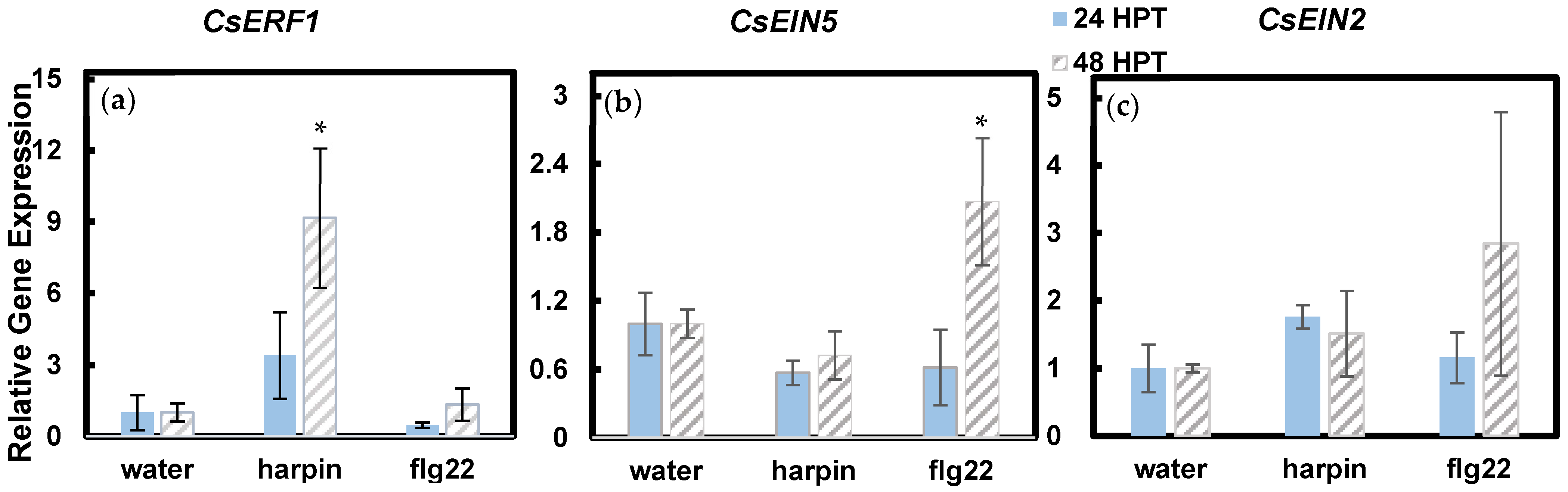
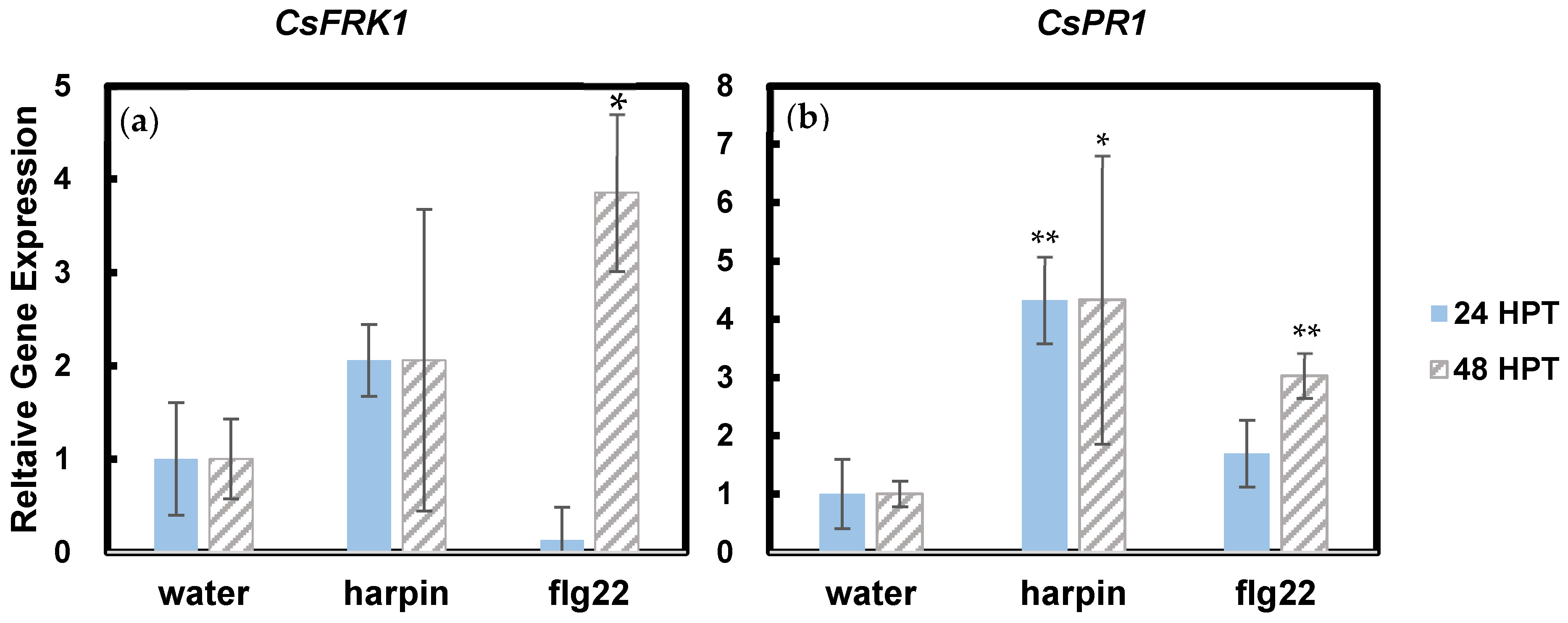
3.2. Harpin and Flg22 Upregulate Certain Expression of Ethylene-Responsive Genes and Defense Genes
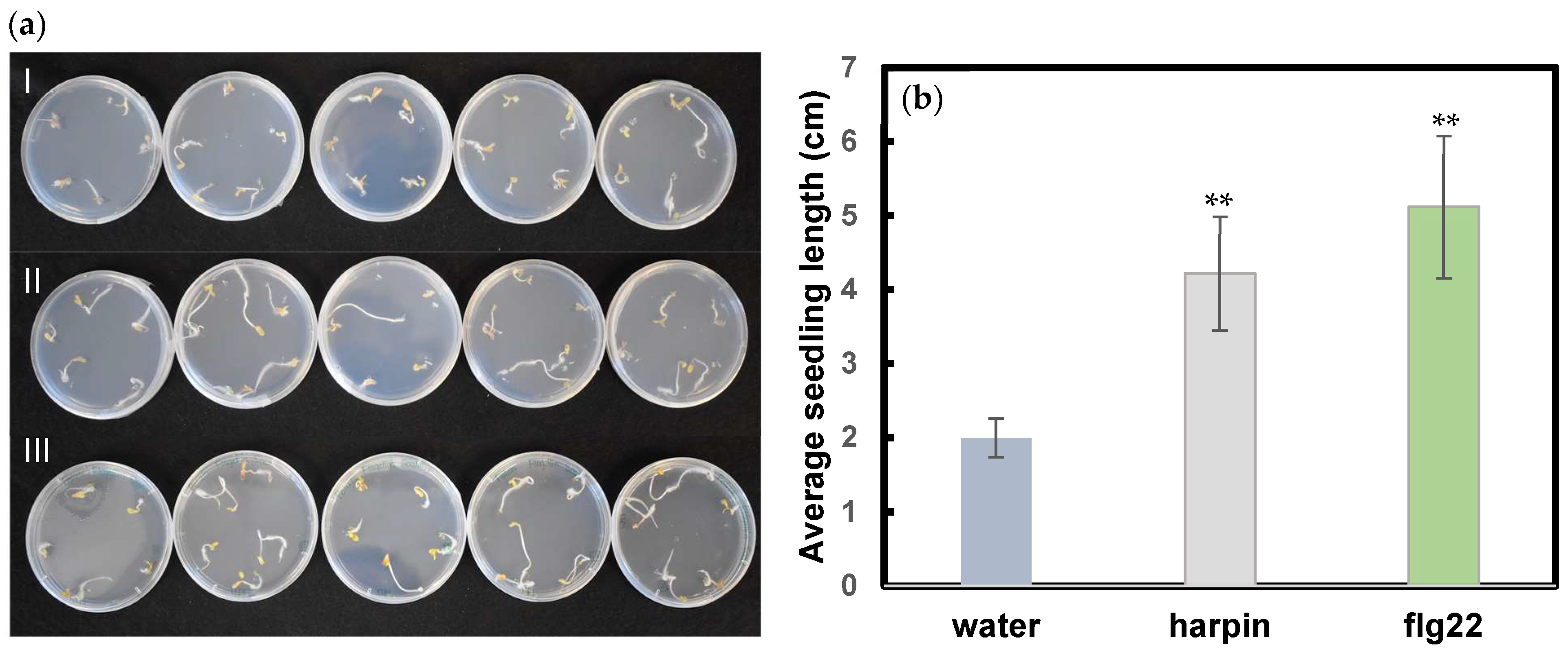
3.3. Both Harpin and Flg22 Promote Hemp Seedling Growth
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PAMP | Pathogen-Associated Molecular Pattern |
| PRR | Pattern Recognition Receptors |
| FLG22 | Flagellin 22 |
| PTI | PAMP-Triggered Immunity |
| ERF1 | Ethylene Response Factor 1 |
| EIN5 | Ethylene Insensitive Protein 5 |
| EIN2 | Ethylene Insensitive Protein 2 |
| FRK1 | Flagellin22-induced Receptor-like Kinase 1 |
| PR1 | Pathogenesis-Related Protein 1 |
| ETI | Effector-Triggered Immunity |
| JA | Jasmonic acid |
| SA | Salicylic Acid |
References
- Livaja, M.; Zeidler, D.; von Rad, U.; Durner, J. Transcriptional Responses of Arabidopsis thaliana to the Bacteria-Derived PAMPs Harpin and Lipopolysaccharide. Immunobiology 2008, 213, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.P.; Peng, J.; Bao, Z.; Meng, X.; Bonasera, J.M.; Chen, G.; Beer, S.V.; Dong, H. Downstream Divergence of the Ethylene Signaling Pathway for Harpin-Stimulated Arabidopsis Growth and Insect Defense. Plant Physiol. 2004, 136, 3628–3638. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Han, B.; Xu, M.; Han, L.; Zhao, Y.; Liu, Z.; Dong, H.; Zhang, C. Plant Growth Enhancement and Associated Physiological Responses Are Coregulated by Ethylene and Gibberellin in Response to Harpin Protein Hpa1. Planta 2014, 239, 831–846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mersmann, S.; Bourdais, G.; Rietz, S.; Robatzek, S. Ethylene Signaling Regulates Accumulation of the FLS2 Receptor and Is Required for the Oxidative Burst Contributing to Plant Immunity. Plant Physiol. 2010, 154, 391–400. [Google Scholar] [CrossRef]
- Chang, X.; Nick, P. Defence Signalling Triggered by Flg22 and Harpin Is Integrated into a Different Stilbene Output in Vitis Cells. PLoS ONE 2012, 7, e40446. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.; Harnrak, A.; Chen, Y.C.; Hsu, C.M. A Harpin-Induced Ethylene-Responsive Factor Regulates Plant Growth and Responses to Biotic and Abiotic Stresses. Biochem. Biophys. Res. Commun. 2010, 402, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.-I.; Yasuda, S. Pattern Recognition Receptors and Signaling in Plant–Microbe Interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Zipfel, C. Pattern-Recognition Receptors in Plant Innate Immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef]
- Jelenska, J.; Davern, S.M.; Standaert, R.F.; Mirzadeh, S.; Greenberg, J.T. Flagellin Peptide Flg22 Gains Access to Long-Distance Trafficking in Arabidopsis via Its Receptor, FLS2. J. Exp. Bot. 2017, 68, 1769–1783. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Felix, G.; Boller, T. A Single Locus Determines Sensitivity to Bacterial Flagellin in Arabidopsis thaliana. Plant J. 1999, 18, 277–284. [Google Scholar] [CrossRef]
- Reboutier, D.; Frankart, C.; Briand, J.; Biligui, B.; Sandrine, L.; Rona, J.-P.; Barny, M.-A.; Bouteau, F. The HrpNea Harpin from Erwinia amylovora Triggers Differential Responses on the Nonhost Arabidopsis thaliana Cells and on the Host Apple Cells. Mol. Plant-Microbe Interact. 2007, 20, 94–100. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.-F.; Etheridge, N.; Schaller, G.E. 2004 Ethylene Signaling Pathway. Ann. Bot. 2005, 95, 901–915. [Google Scholar] [CrossRef] [PubMed]
- El Ghaouth, A.; Arul, J.; Grenier, J.; Benhamou, N.; Asselin, A.; Belanger, R. Effect of Chitosan on Cucumber Plants: Suppression of Pythium aphanidermatum and Induction of Defense Reactions. Am. Phytophatological Soc. 1993, 84, 313–320. [Google Scholar] [CrossRef]
- Broders, K.D.; Lipps, P.E.; Paul, P.A.; Dorrance, A.E. Characterization of Pythium Spp. Associated with Corn and Soybean Seed and Seedling Disease in Ohio. Plant Dis. 2007, 91, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Punja, Z.K.; Rodriguez, G. Fusarium and Pythium Species Infecting Roots of Hydroponically Grown Marijuana (Cannabis Sativa L.) plants. Can. J. Plant Pathol. 2018, 40, 498–513. [Google Scholar] [CrossRef]
- Mao, J.L.; Miao, Z.Q.; Wang, Z.; Yu, L.H.; Cai, X.T.; Xiang, C.B. Arabidopsis ERF1 Mediates Cross-Talk between Ethylene and Auxin Biosynthesis during Primary Root Elongation by Regulating ASA1 Expression. PLoS Genet. 2016, 12, e1006076. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 Integrates Signals from Ethylene and Jasmonate Pathways in Plant Defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jia, Z.; Lü, B.; Shi, H.; Shao, W.; Dong, H. Transcription Factor AtMYB44 Regulates Induced Expression of the ETHYLENE INSENSITIVE2 Gene in Arabidopsis Responding to a Harpin Protein. Mol. Plant-Microbe Interact. 2011, 24, 377–389. [Google Scholar] [CrossRef]
- Janda, M.; Lamparová, L.; Zubíková, A.; Burketová, L.; Martinec, J.; Krčková, Z. Temporary Heat Stress Suppresses PAMP-Triggered Immunity and Resistance to Bacteria in Arabidopsis thaliana. Mol. Plant Pathol. 2019, 20, 1005–1012. [Google Scholar] [CrossRef]
- Pečenková, T.; Pleskot, R.; Žárský, V. Subcellular Localization of Arabidopsis Pathogenesis-Related 1 (PR1) Protein. Int. J. Mol. Sci. 2017, 18, 825. [Google Scholar] [CrossRef]
- Göhre, V.; Jones, A.M.E.; Sklenář, J.; Robatzek, S.; Weber, A.P.M. Molecular Crosstalk between PAMP-Triggered Immunity and Photosynthesis. Mol. Plant-Microbe Interact. 2012, 25, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Booth, J.K.; Page, J.E.; Bohlmann, J. Terpene Synthases from Cannabis sativa. PLoS ONE 2017, 12, e0173911. [Google Scholar] [CrossRef]
- Small, E.; Brookes, B. Temperature and Moisture Content for Storage Maintenance of Germination Capacity of Seeds of Industrial Hemp, Marijuana, and Ditchweed Forms of Cannabis sativa. J. Nat. Fibers 2012, 9, 240–255. [Google Scholar] [CrossRef]
- Oyebanji, O.B.; Nweke, O.; Odebunmi, O.; Galadima, N.B.; Nnodi, U.N.; Afolabi, A.S.; Ogbadu, G.H. Simple, Effective and Economical Explant-Surface Sterilization Protocol for Cowpea, Rice and Sorghum Seeds. Afr. J. Biotechnol. 2009, 8, 5395–5399. [Google Scholar]
- Luecke, N.C.; Crawford, K.M.; Koike, S.T.; Stanghellini, H.; Burkhardt, A. First Report of Root Rot Caused by Pythium dissotocum on Hydroponically Grown Collard Greens (Brassica oleracea Var. acephala). Plant Dis. 2021, 106, 1075. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Dunand, C. Waving and Skewing: How Gravity and the Surface of Growth Media Affect Root Development in Arabidopsis. New Phytol. 2007, 176, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, T.; Long, J.; Yin, Q.; Zhang, Y.; Chen, L.; Liu, R.; Gao, T.; Dong, H. Productivity and Biochemical Properties of Green Tea in Response to Full-Length and Functional Fragments of HpaG Xooc, a Harpin Protein from the Bacterial Rice Leaf Streak Pathogen Xanthomonas oryzae pv. oryzicola. J. Biosci. 2007, 32, 1119–1131. [Google Scholar] [CrossRef]
- Dong, Y.; Li, P.; Zhang, C. Harpin Hpa1 Promotes Flower Development in Impatiens and Parochetus Plants. Bot. Stud. 2016, 57, 22. [Google Scholar] [CrossRef][Green Version]
- Hu, J.; Masson, R. First Report of Crown and Root Rot Caused by Pythium aphanidermatum on Industrial Hemp (Cannabis sativa) in Arizona. Plant Dis. 2021, 101, 1038. [Google Scholar] [CrossRef]
- Chen, J.; Li, Z.; Cheng, Y.; Gao, C.; Guo, L.; Wang, T.; Xu, J. First Report of Pythium aphanidermatum Causing Crown and Root Rot on Industrial Hemp in China. Plant Dis. 2022, 106, 1076. [Google Scholar] [CrossRef]
- Chuang, H.W. The Dual Function of Harpin in Disease Resistance and Growth in Phalaenopsis Orchids. Orchid. Biotechnol. III 2017, 3, 309–329. [Google Scholar] [CrossRef]
- Akbudak, N.; Tezcan, H.; Akbudak, B.; Seniz, V. The Effect of Harpin Protein on Plant Growth Parameters, Leaf Chlorophyll, Leaf Colour and Percentage Rotten Fruit of Pepper Plants Inoculated with Botrytis cinerea. Sci. Hortic. 2006, 109, 107–112. [Google Scholar] [CrossRef]
- Chang, X.; Seo, M.; Takebayashi, Y.; Kamiya, Y.; Riemann, M.; Nick, P. Jasmonates Are Induced by the PAMP Flg22 but Not the Cell Death-Inducing Elicitor Harpin in Vitis Rupestris. Protoplasma 2017, 254, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Delaney, T.P.; Bauer, D.W.; Beer, S.V. Harpin Induces Disease Resistance in Arabidopsis through the Systemic Acquired Resistance Pathway Mediated by Salicylic Acid and the NIM1 Gene. Plant J. 1999, 20, 207–215. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, S.; Yang, D.; Fan, Z. Revealing Shared and Distinct Genes Responding to JA and SA Signaling in Arabidopsis by Meta-Analysis. Front. Plant Sci. 2020, 11, 908. [Google Scholar] [CrossRef]
- Choi, M.S.; Kim, W.; Lee, C.; Oh, C.S. Harpins, Multifunctional Proteins Secreted by Gram-Negative Plant-Pathogenic Bacteria. Mol. Plant-Microbe Interact. 2013, 26, 1115–1122. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Leon-Reyes, A.; van der Ent, S.; van Wees, S.C.M. Networking by Small-Molecule Hormones in Plant Immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Mishina, T.E.; Zeier, J. Pathogen-Associated Molecular Pattern Recognition Rather than Development of Tissue Necrosis Contributes to Bacterial Induction of Systemic Acquired Resistance in Arabidopsis. Plant J. 2007, 50, 500–513. [Google Scholar] [CrossRef]
- Tsuda, K.; Sato, M.; Glazebrook, J.; Cohen, J.D.; Katagiri, F. Interplay between MAMP-Triggered and SA-Mediated Defense Responses. Plant J. 2008, 53, 763–775. [Google Scholar] [CrossRef]
- Xu, M.-Y.; Zhou, T.; Zhao, Y.-Y.; Li, J.-B.; Xu, H.; Dong, H.-S.; Zhang, C.-L. Transgenic Expression of a Functional Fragment of Harpin Protein Hpa1 in Wheat Represses English Grain Aphid Infestation. J. Integr. Agric. 2014, 13, 2565–2576. [Google Scholar] [CrossRef]
- Fu, M.; Xu, M.; Zhou, T.; Wang, D.; Tian, S.; Han, L.; Dong, H.; Zhang, C. Transgenic Expression of a Functional Fragment of Harpin Protein Hpa1 in Wheat Induces the Phloem-Based Defence against English Grain Aphid. J. Exp. Bot. 2014, 65, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Punja, Z.K. Emerging Diseases of Cannabis sativa and Sustainable Management. Pest Manag. Sci. 2021, 77, 3857–3870. [Google Scholar] [CrossRef] [PubMed]
| Gene | Function | PAMP | Reference |
|---|---|---|---|
| Ethylene Response Factor 1 [ERF1] | Transcriptional activator which acts downstream of the ethylene signaling pathway. It mediates development and regulates pathogen defense genes [16]. | harpin and flg22 | [2,17] |
| Ethylene Insensitive Protein 5 [EIN5] | Required for plant growth enhancement [2]. Acts downstream of CTR1, a negative regulator of the pathway. If ethylene is present, CTR1 does not act, and the pathway can be activated [3]. | harpin | [3] |
| Ethylene Insensitive Protein 2 [EIN2] | Required for insect resistance in plants [2]. Protein located downstream of CTR1 in the ethylene pathway. If CTR1 is inhibited, then EIN2 may be activated. It is a Nramp-like protein, meaning it is a natural resistance-associated macrophage protein [18]. | harpin | [2,18] |
| Flg22-Induced Receptor Like Kinase 1 [FRK1] | Associated with early defense signaling as well as mediating senescence. Following flg22 exposure in an organism, FRK1 regulation likely increases accordingly [19]. | flg22 | [19] |
| Pathogenesis Related Protein 1 [PR1] | Gene expression increases following pathogen detection [20]. It is an indicator of systemic acquired resistance. | flg22 | [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sands, L.B.; Cheek, T.; Reynolds, J.; Ma, Y.; Berkowitz, G.A. Effects of Harpin and Flg22 on Growth Enhancement and Pathogen Defense in Cannabis sativa Seedlings. Plants 2022, 11, 1178. https://doi.org/10.3390/plants11091178
Sands LB, Cheek T, Reynolds J, Ma Y, Berkowitz GA. Effects of Harpin and Flg22 on Growth Enhancement and Pathogen Defense in Cannabis sativa Seedlings. Plants. 2022; 11(9):1178. https://doi.org/10.3390/plants11091178
Chicago/Turabian StyleSands, Lauren B., Taylor Cheek, Joseph Reynolds, Yi Ma, and Gerald A. Berkowitz. 2022. "Effects of Harpin and Flg22 on Growth Enhancement and Pathogen Defense in Cannabis sativa Seedlings" Plants 11, no. 9: 1178. https://doi.org/10.3390/plants11091178
APA StyleSands, L. B., Cheek, T., Reynolds, J., Ma, Y., & Berkowitz, G. A. (2022). Effects of Harpin and Flg22 on Growth Enhancement and Pathogen Defense in Cannabis sativa Seedlings. Plants, 11(9), 1178. https://doi.org/10.3390/plants11091178







