Non-Destructive Measurement of the Pumpkin Rootstock Root Phenotype Using AZURE KINECT
Abstract
:1. Introduction
2. Materials and Methods
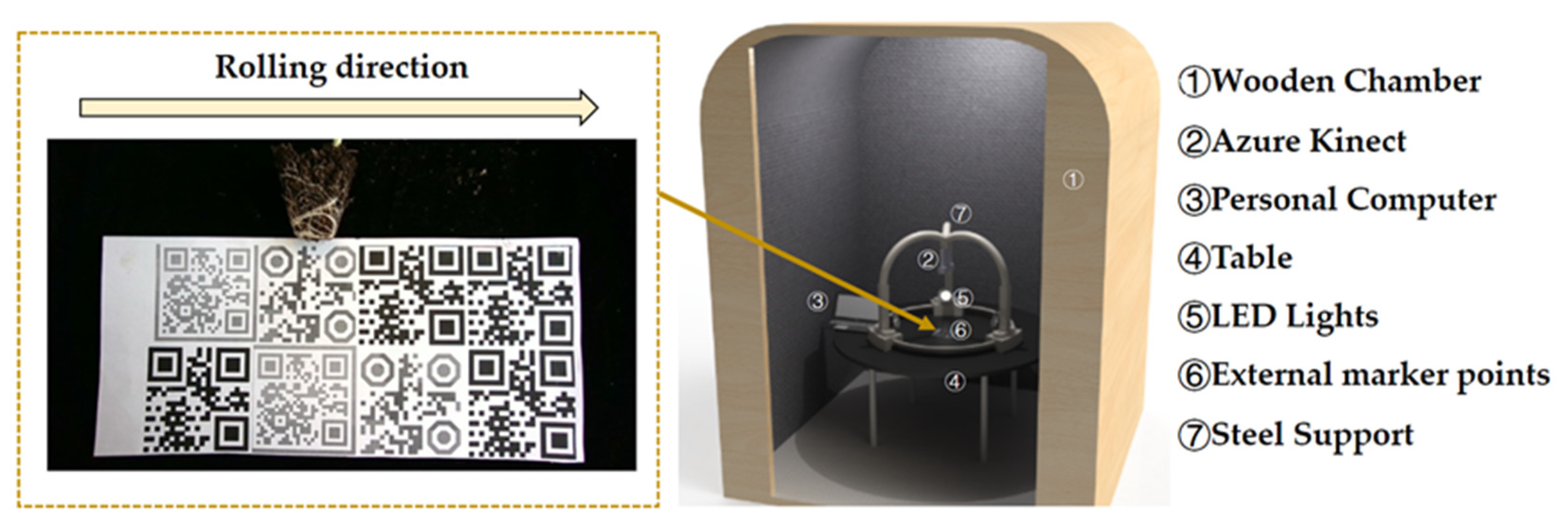
2.1. Experimental Location, Equipment, and Design
2.2. Growth Conditions and Plant Materials
2.3. Multi-View Color-Image and Depth-Image Mosaic Method for the Surface Root System of Root Ball of Pumpkin Rootstock Based on External Feature Points
2.4. Precision Segmentation Method for the Surface Root Image of Root Ball of Plug Seedling
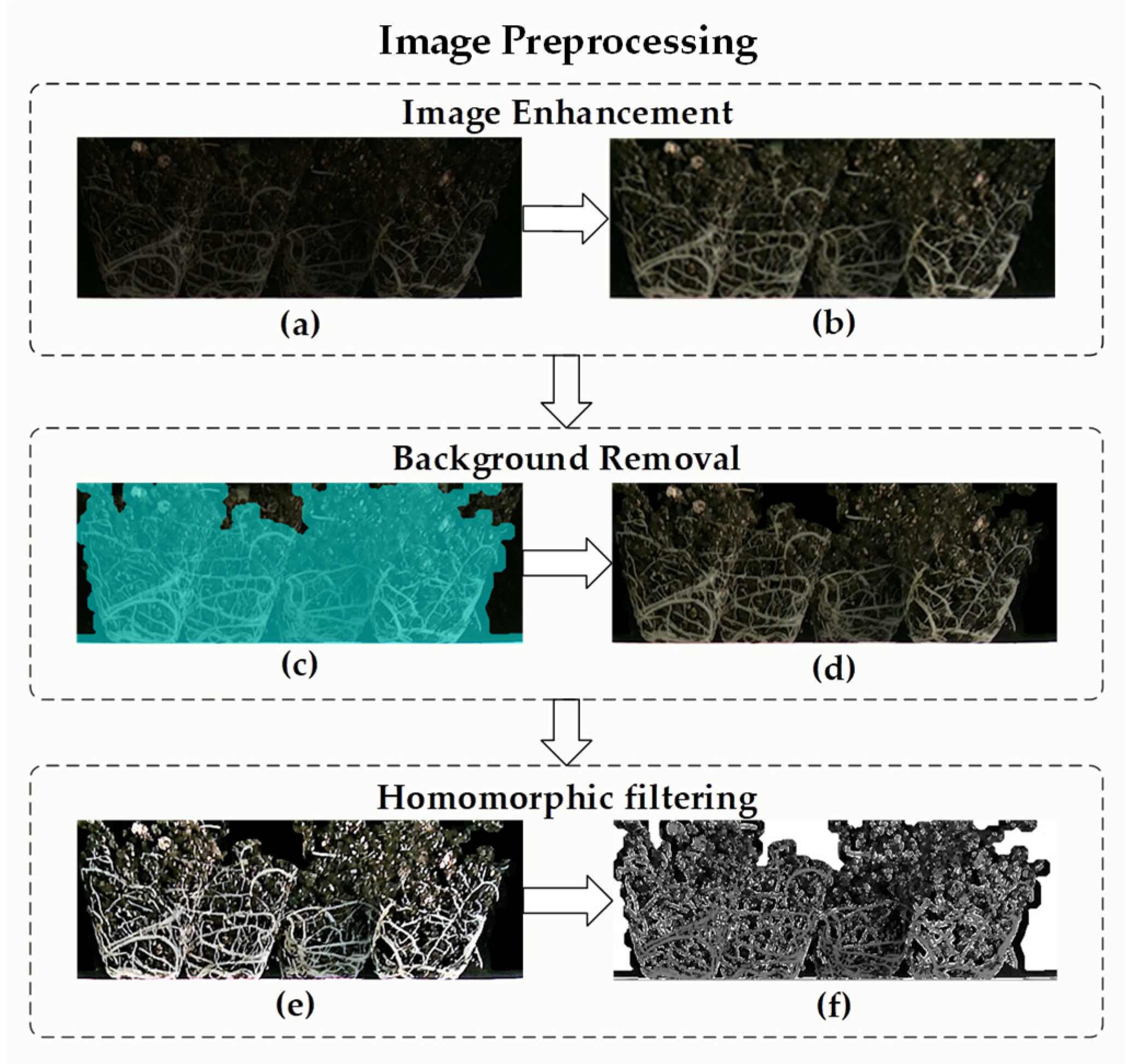
2.4.1. Surface Root Image Preprocessing for Root Ball of Plug Seedling
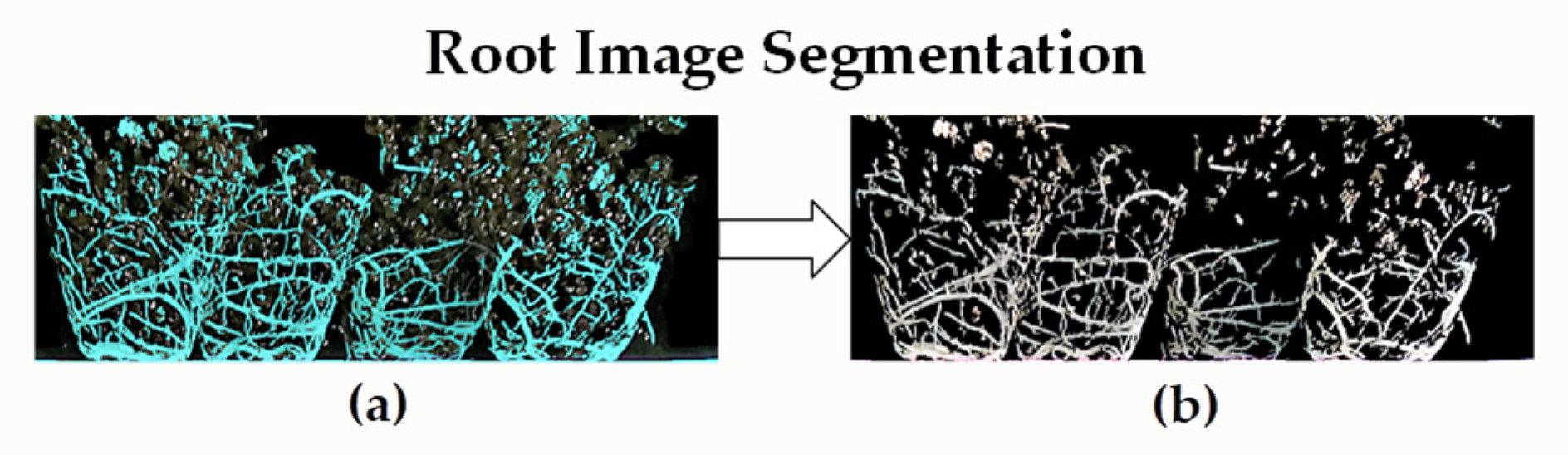
2.4.2. Surface Root Image Segmentation Algorithm
2.5. Phenotypic Data Mining, Non-Destructive Measurement, and Accurate Estimation
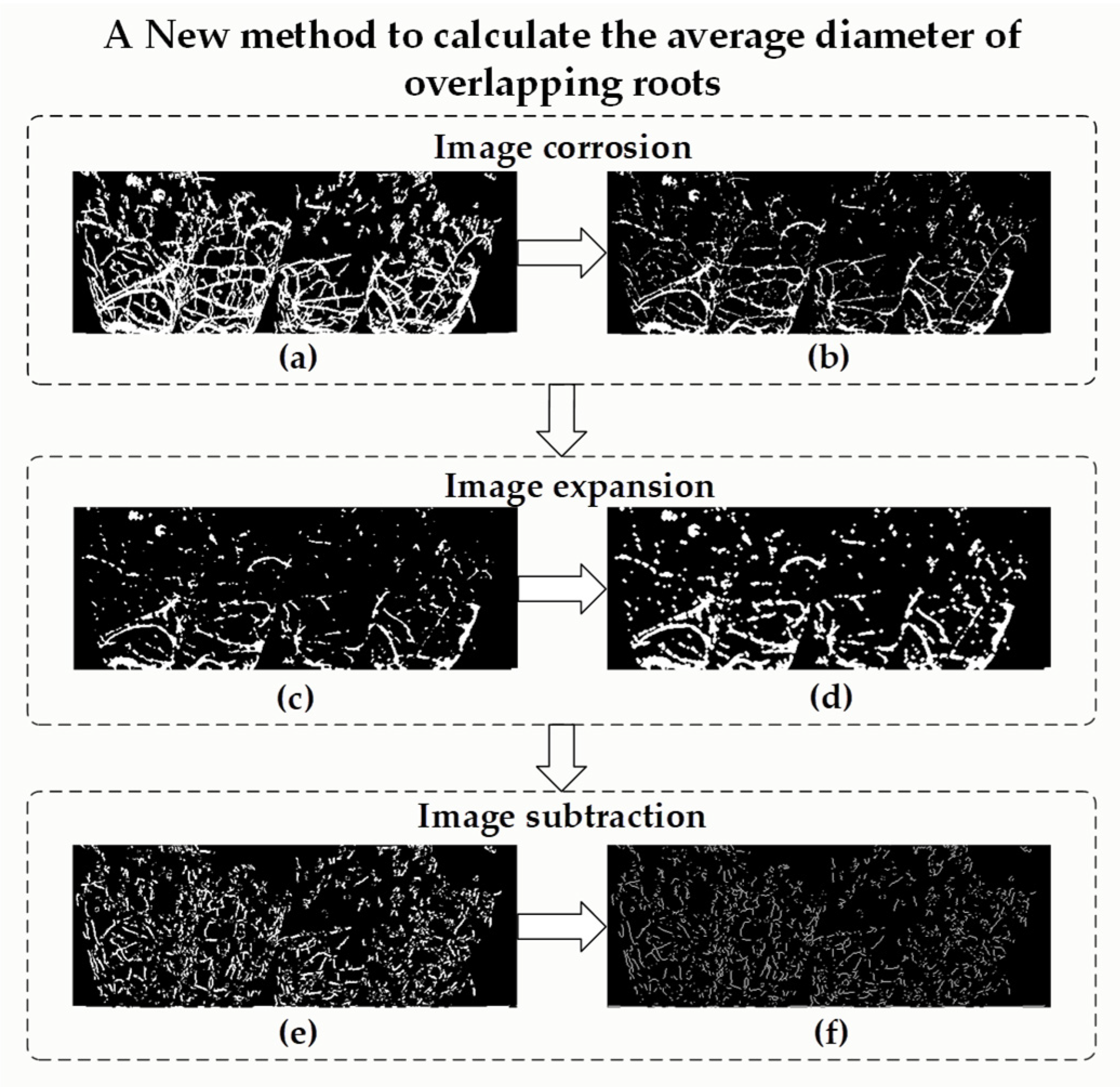
2.5.1. Non-Destructive Measuring Method for Average Diameter of Total Root
2.5.2. Calculation of Root Encapsulation (Density of Surface Root)
2.5.3. Prediction Model for the Surface Area and Length of Total Root
- 1.
- Estimating the Surface Area of Total Root (SATR)
- 2.
- Estimating the Length of Total Root (LTR)
- 3.
- Calculating Volume of Total Root (VTR)
3. Results
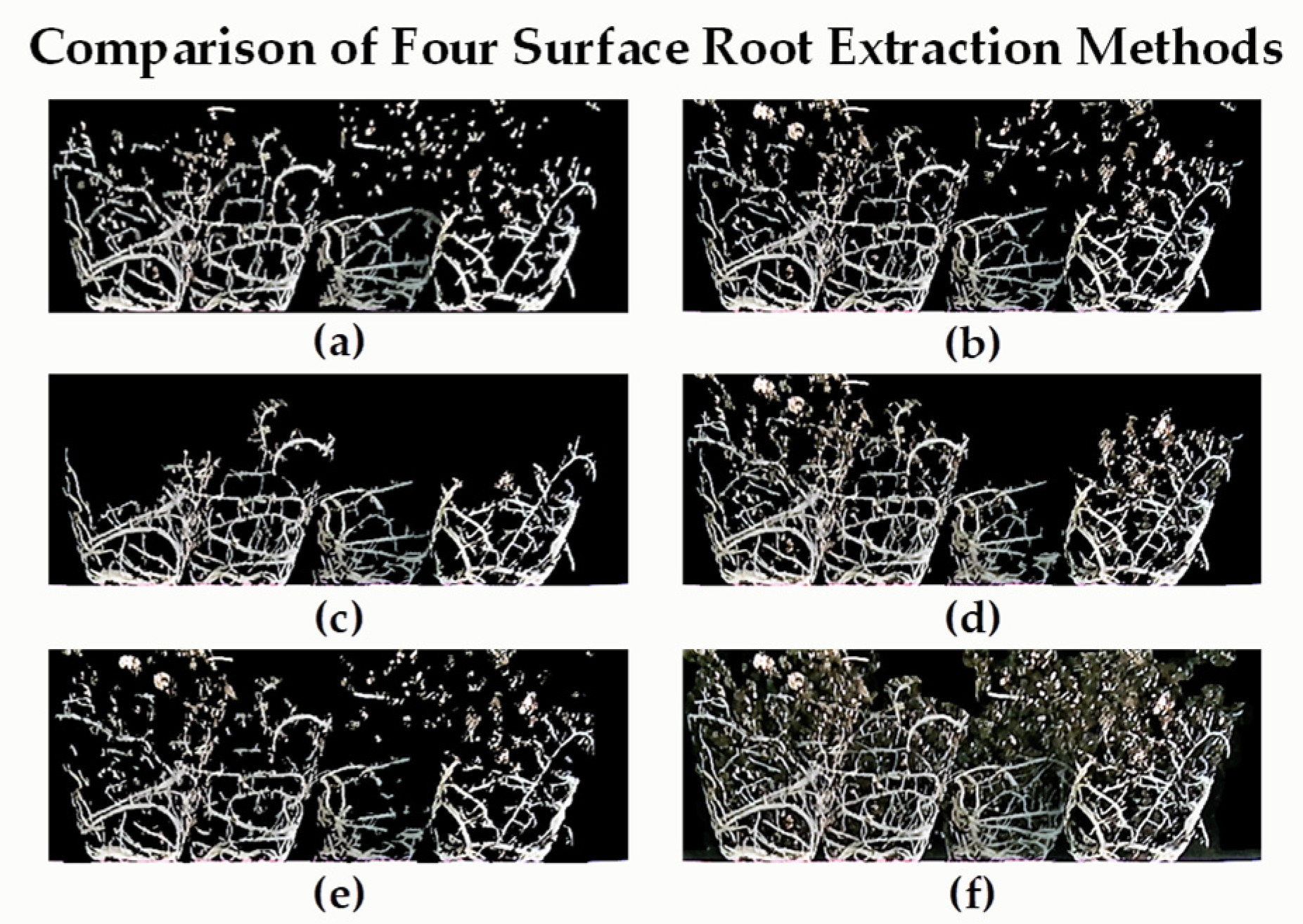
3.1. Performance Test of Images Using the Surface Root Segmentation Algorithms
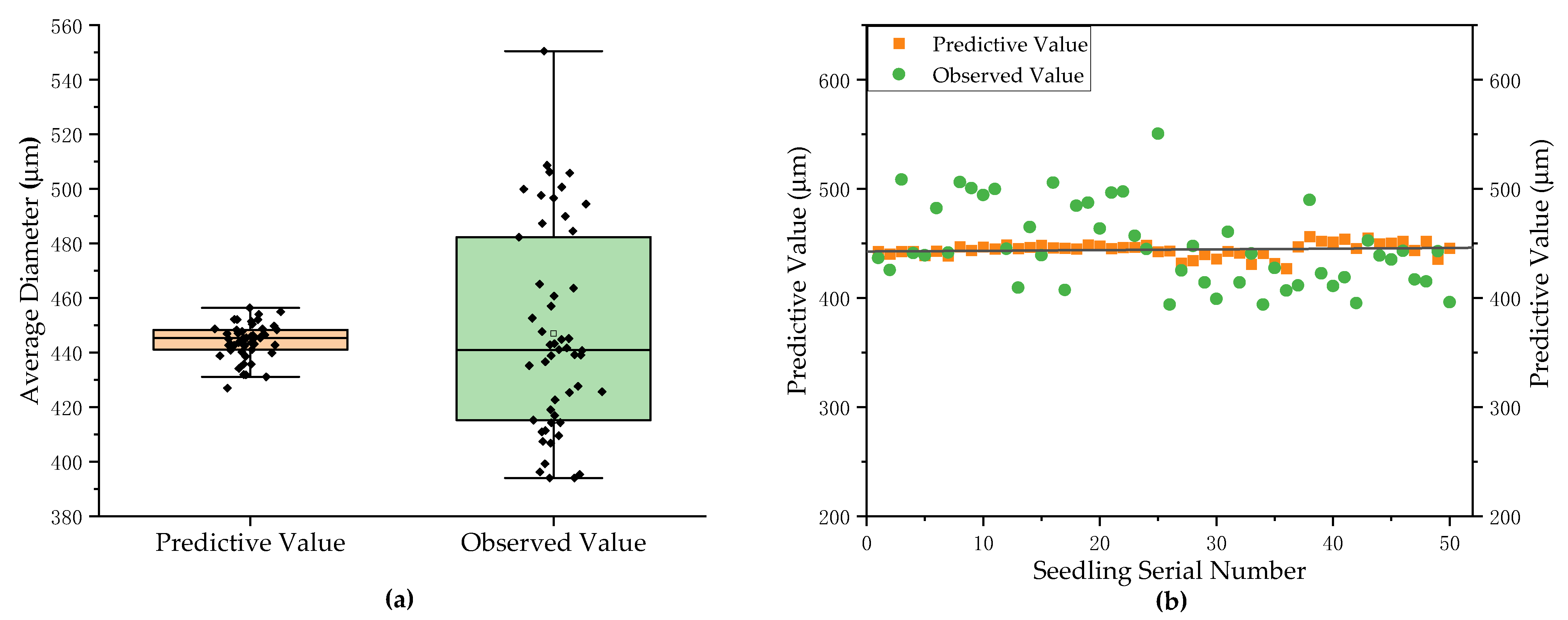
3.2. Measurement Results and Analysis of Average Diameter of Pumpkin Rootstock
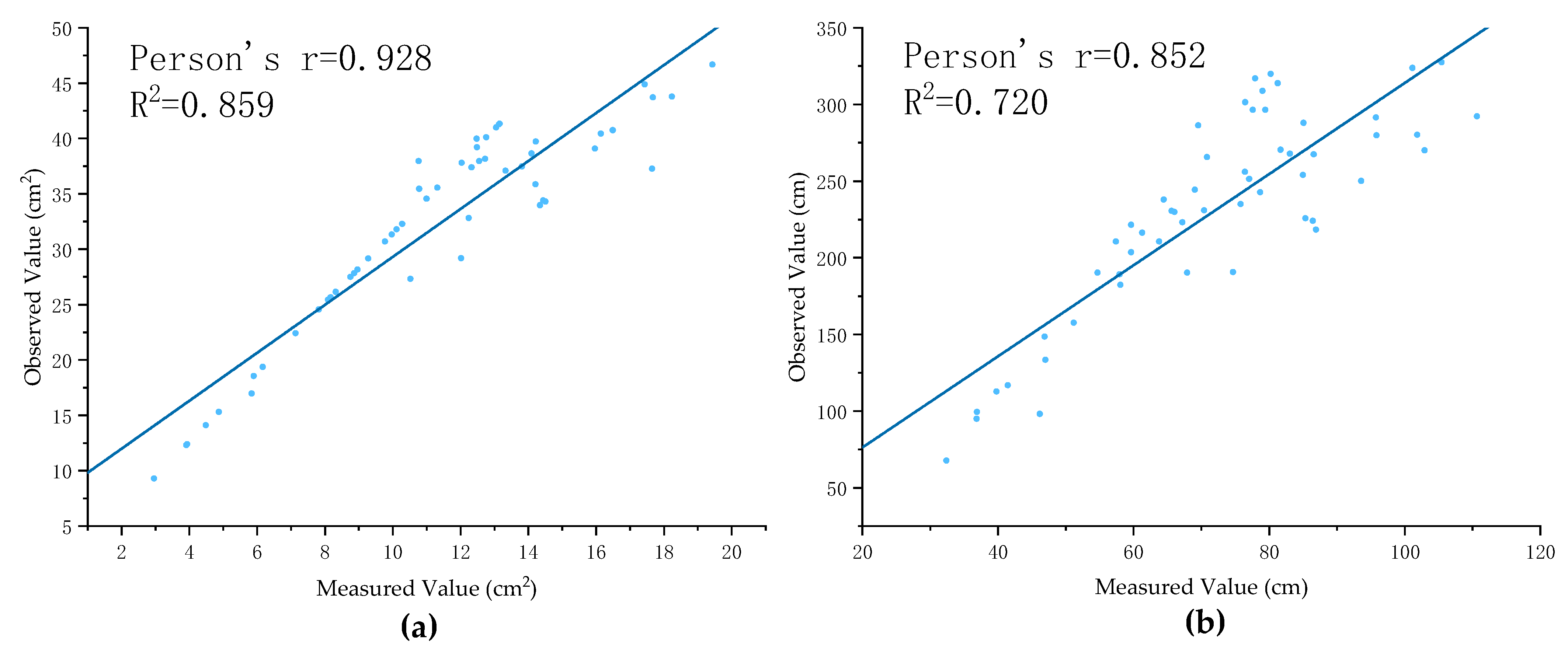
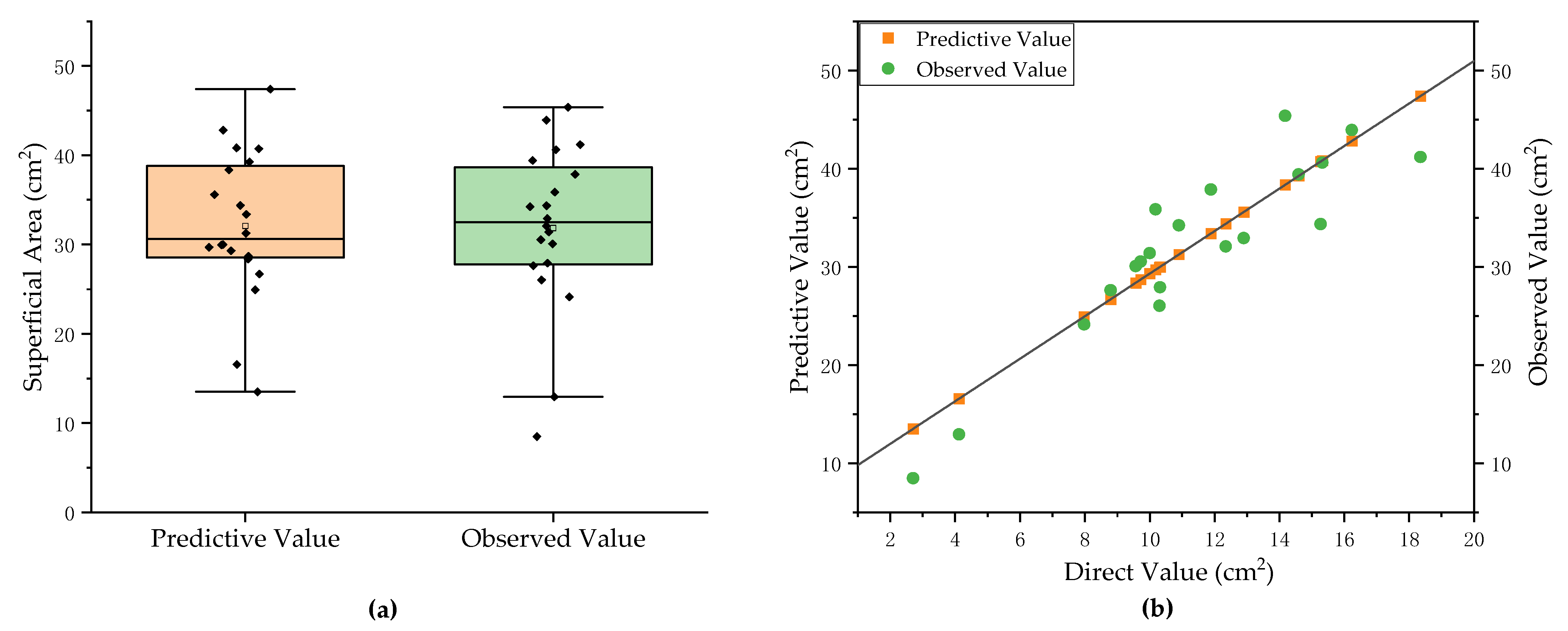
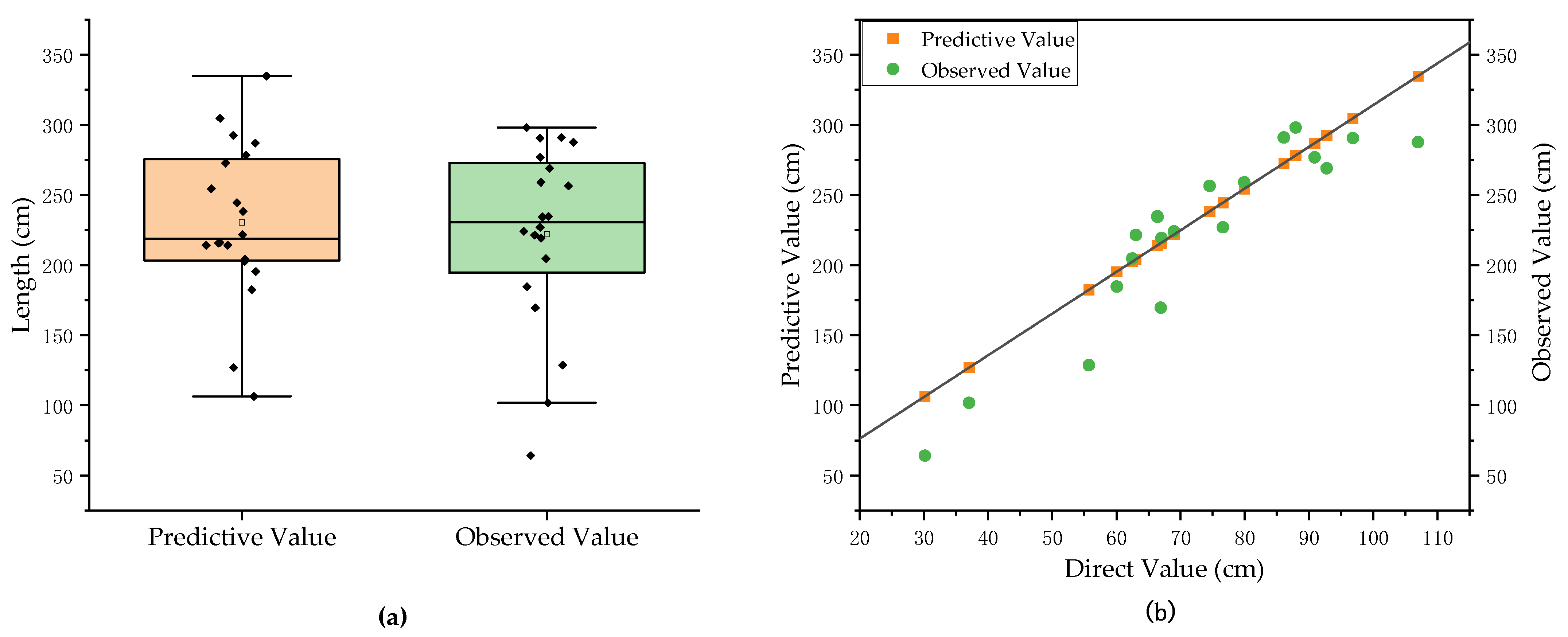
3.3. Prediction Results and Analysis of Total Root Phenotypes of Pumpkin Rootstocks
4. Discussion
4.1. Discussion of the Root Image Stitching Algorithm
4.2. Discussion of the Root Image Segmentation Algorithm
4.3. Discussion on the Calculation Method for the Root Phenotype
4.4. Discussion of Method Adaptability
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lee, S.G. Production of High Quality Vegetable Seedling Grafts. Acta Hortic. (ISHS) 2007, 759, 169–174. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Imtiaz, M.; Kong, Q.; Cheng, F.; Ahmed, W.; Huang, Y.; Bie, Z. Grafting: A Technique to Modify Ion Accumulation in Horticultural Crops. Front. Plant Sci. 2016, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suchoff, D.H.; Schultheis, J.R.; Kleinhenz, M.D.; Louws, F.; Gunter, C.C. Rootstock Improves High-tunnel Tomato Water Use Efficiency. HortTechnology 2018, 28, 344–353. [Google Scholar] [CrossRef] [Green Version]
- Ballesta, M.M.; Alcaraz-López, C.; Muries, B.; Mota-Cadenas, C.; Carvajal, M. Physiological aspects of rootstock-scion interactions. Sci. Hortic. 2010, 127, 112–118. [Google Scholar] [CrossRef]
- Bertucci, M.B.; Suchoff, D.; Jennings, K.; Monks, D.W.; Gunter, C.C.; Schultheis, J.R.; Louws, F. Comparison of Root System Morphology of Cucurbit Rootstocks for Use in Watermelon Grafting. HortTechnology 2018, 28, 625–636. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Pradal, C. Root phenotyping: Important and minimum information required for root modeling in crop plants. Breed. Sci. 2021, 71, 8. [Google Scholar] [CrossRef] [PubMed]
- Ghoncheh, Z.; Afkhami, F.; Fard, M.J.K.; Sorkhabi, R.E.; Aydin, U. Accuracy of digitally enhanced images compared with unprocessed digital images in the detection of external root resorption. Oral Radiol. 2017, 33, 133–139. [Google Scholar] [CrossRef]
- Teramoto, S.; Takayasu, S.; Kitomi, Y.; Arai-Sanoh, Y.; Tanabata, T.; Uga, Y. High-throughput three-dimensional visualization of root system architecture of rice using X-ray computed tomography. Plant Methods 2020, 16, 66. [Google Scholar] [CrossRef]
- van Dusschoten, D.; Metzner, R.; Kochs, J.; Postma, J.A.; Pflugfelder, D.; Bühler, J.; Schurr, U.; Jahnke, S. Quantitative 3D analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol. 2016, 170, 1176–1188. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Yan, H.; Zeng, L. Characterizing plant root parameters with deep learning-based heat pulse method. Geoderma 2022, 406, 115507. [Google Scholar] [CrossRef]
- Lu, W.; Wang, X.; Wang, F. Adaptive minirhizotron for pepper roots observation and its installation based on root system architecture traits. Plant Methods 2019, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dong, X.; Xue, Q.; Leskovar, D.I.; Jifon, J.; Butnor, J.R.; Marek, T. Ground penetrating radar (GPR) detects fine roots of agricultural crops in the field. Plant Soil 2018, 423, 517–531. [Google Scholar] [CrossRef]
- Alsalem, M.; Salehi, A.; Zhao, J.; Rewald, B.; Bodner, G. Combining image analyses tools for comprehensive characterization of root systems from soil-filled rhizobox phenotyping platforms. Int. Agrophysics 2021, 35, 257–268. [Google Scholar] [CrossRef]
- Chuanyu, W.; Xinyu, G.; Weiliang, W.; Sheng, W.; Shenghao, G. Phenotyping index acquisition method based on maize root images. Trans. Chin. Soc. Agric. Eng. 2021, 37, 169–176. [Google Scholar]
- Fang, H.; Ziwen, X.; Haigang, L.; Yan, G.; Baoguo, L.; Yunling, L.; Yuntao, M. Image-based root phenotyping for field-grown crops: An example under maize/soybean intercropping. J. Integr. Agric. 2021, 20, 2–15. [Google Scholar]
- Krzyzaniak, Y.; Cointault, F.; Loupiac, C.; Bernaud, E.; Ott, F.; Salon, C.; Laybros, A.; Han, S.; Héloir, M.C.; Adrian, M.; et al. In situ Phenotyping of Grapevine Root System Architecture by 2D or 3D Imaging: Advantages and Limits of Three Cultivation Methods. Front. Plant Sci. 2021, 12, 1278. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wang, M.; Yang, W.; Liu, Q.; Huang, C. Research on Non-destructive Measurement of Surface Root Traits of Rice Plants. J. Agric. Sci. Technol. 2017, 19, 66–77. [Google Scholar]
- Frangi, A.F.; Niessen, W.J.; Vincken, K.L.; Viergever, M.A. Multiscale vessel enhancement filtering. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 1998; Volume 1496, pp. 130–137. [Google Scholar]
- Yang, Z.L.; Guo, B.L. Image mosaic based on SIFT. In Proceedings of the 4th International Conference on Intelligent Information Hiding and Multimedia Signal Processing (IIH-MSP 2008), Harbin, China, 15–17 August 2008; pp. 1422–1425. [Google Scholar]
- Gao, T.; Zhu, F.; Paul, P.; Sandhu, J.; Doku, H.; Sun, J.; Pan, Y.; Staswick, P.; Walia, H.; Yu, H. Novel 3D Imaging Systems for High-Throughput Phenotyping of Plants. Remote Sens. 2021, 13, 2113. [Google Scholar] [CrossRef]
- Harris, C.G.; Stephens, M.J. A Combined Corner and Edge Detector. In Proceedings of the 4th Alvey Vision Conference, Manchester, UK, 31 August–2 September 1988; pp. 147–151. [Google Scholar]
- Brown, M.; Szeliski, R.; Winder, S. Multi-image matching using multi-scale oriented patches. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 510–517. [Google Scholar]
- Berg, A.; Malik, J. Geometric blur for template matching. In Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Kauai, HI, USA, 8–14 December 2001; Volume 1, pp. 607–614. [Google Scholar]
- Soille, P. Morphological Image Analysis: Principles and Applications. Sens. Rev. 1999, 28, 800–801. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Xu, S.; Huang, Y.; Bie, Z.; Notaguchi, M.; Zhou, J.; Wan, X.; Wang, Y.; Dong, W. Non-Destructive Measurement of the Pumpkin Rootstock Root Phenotype Using AZURE KINECT. Plants 2022, 11, 1144. https://doi.org/10.3390/plants11091144
Zhang M, Xu S, Huang Y, Bie Z, Notaguchi M, Zhou J, Wan X, Wang Y, Dong W. Non-Destructive Measurement of the Pumpkin Rootstock Root Phenotype Using AZURE KINECT. Plants. 2022; 11(9):1144. https://doi.org/10.3390/plants11091144
Chicago/Turabian StyleZhang, Moran, Shengyong Xu, Yuan Huang, Zhilong Bie, Michitaka Notaguchi, Jingyi Zhou, Xin Wan, Yuchen Wang, and Wanjing Dong. 2022. "Non-Destructive Measurement of the Pumpkin Rootstock Root Phenotype Using AZURE KINECT" Plants 11, no. 9: 1144. https://doi.org/10.3390/plants11091144
APA StyleZhang, M., Xu, S., Huang, Y., Bie, Z., Notaguchi, M., Zhou, J., Wan, X., Wang, Y., & Dong, W. (2022). Non-Destructive Measurement of the Pumpkin Rootstock Root Phenotype Using AZURE KINECT. Plants, 11(9), 1144. https://doi.org/10.3390/plants11091144







