Cultivable Fungal Endophytes in Roots, Rhizomes and Leaves of Posidonia oceanica (L.) Delile along the Coast of Sicily, Italy
Abstract
:1. Introduction
2. Results
2.1. Fungal Isolation and Identification
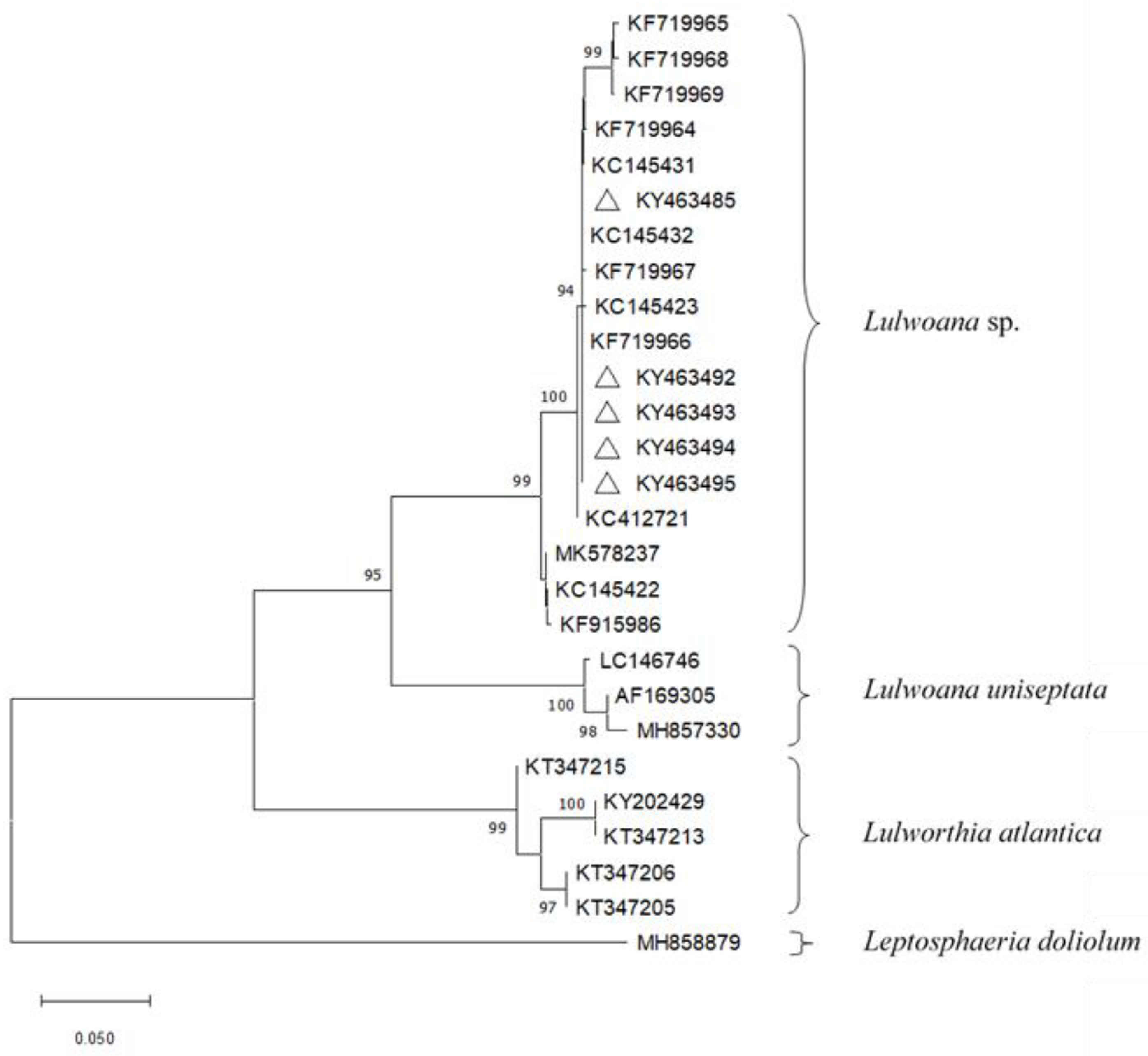
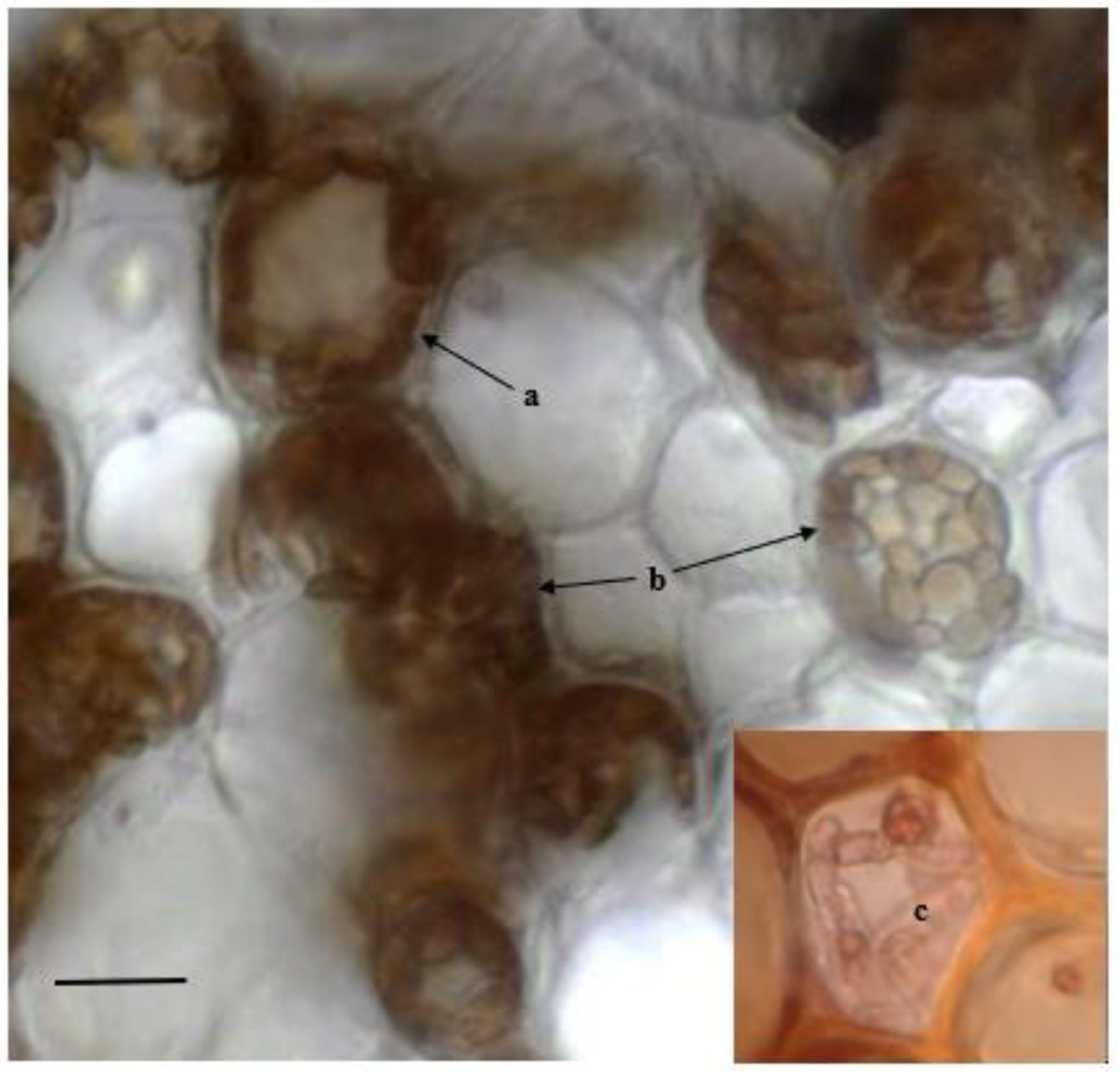
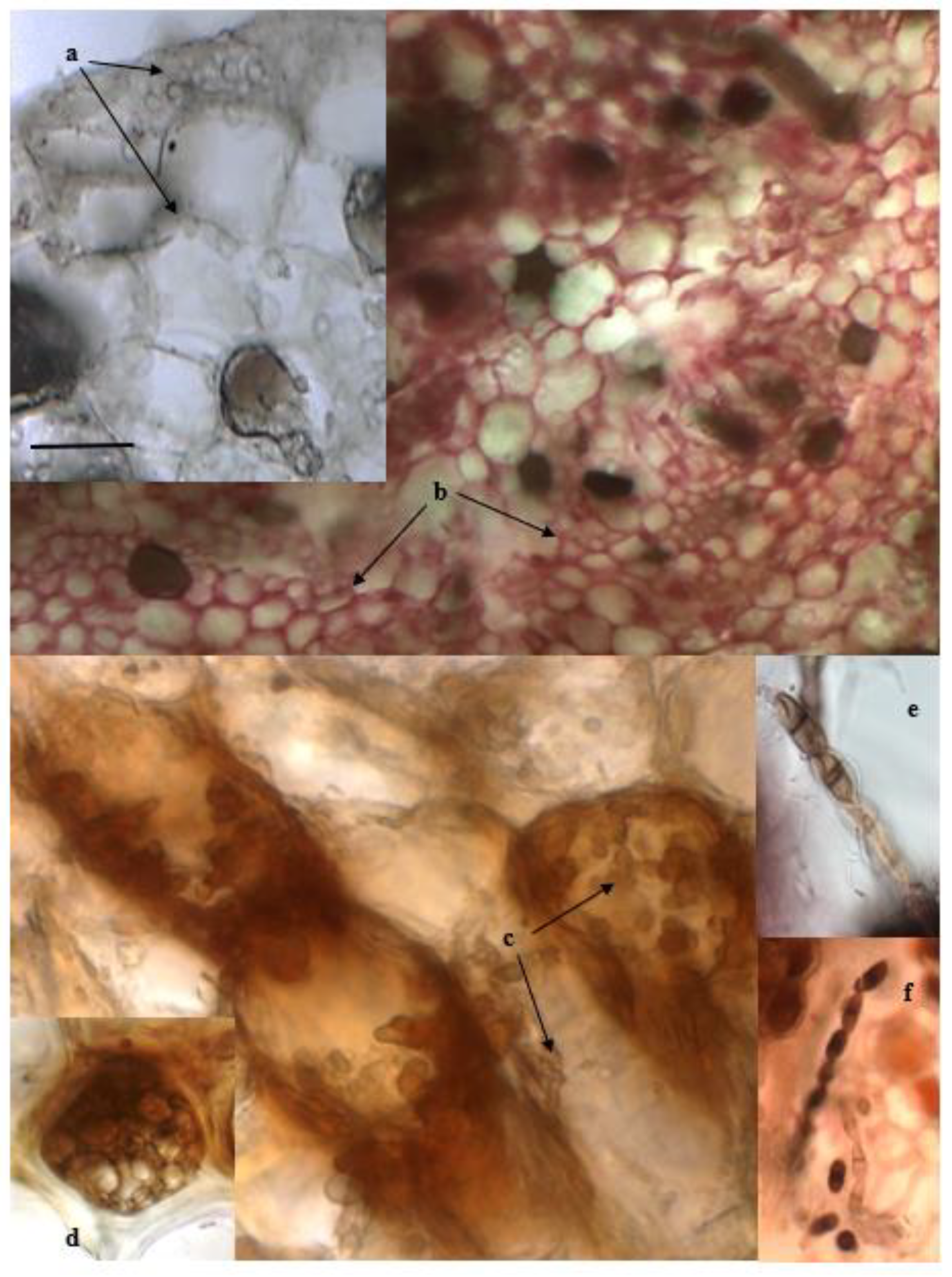
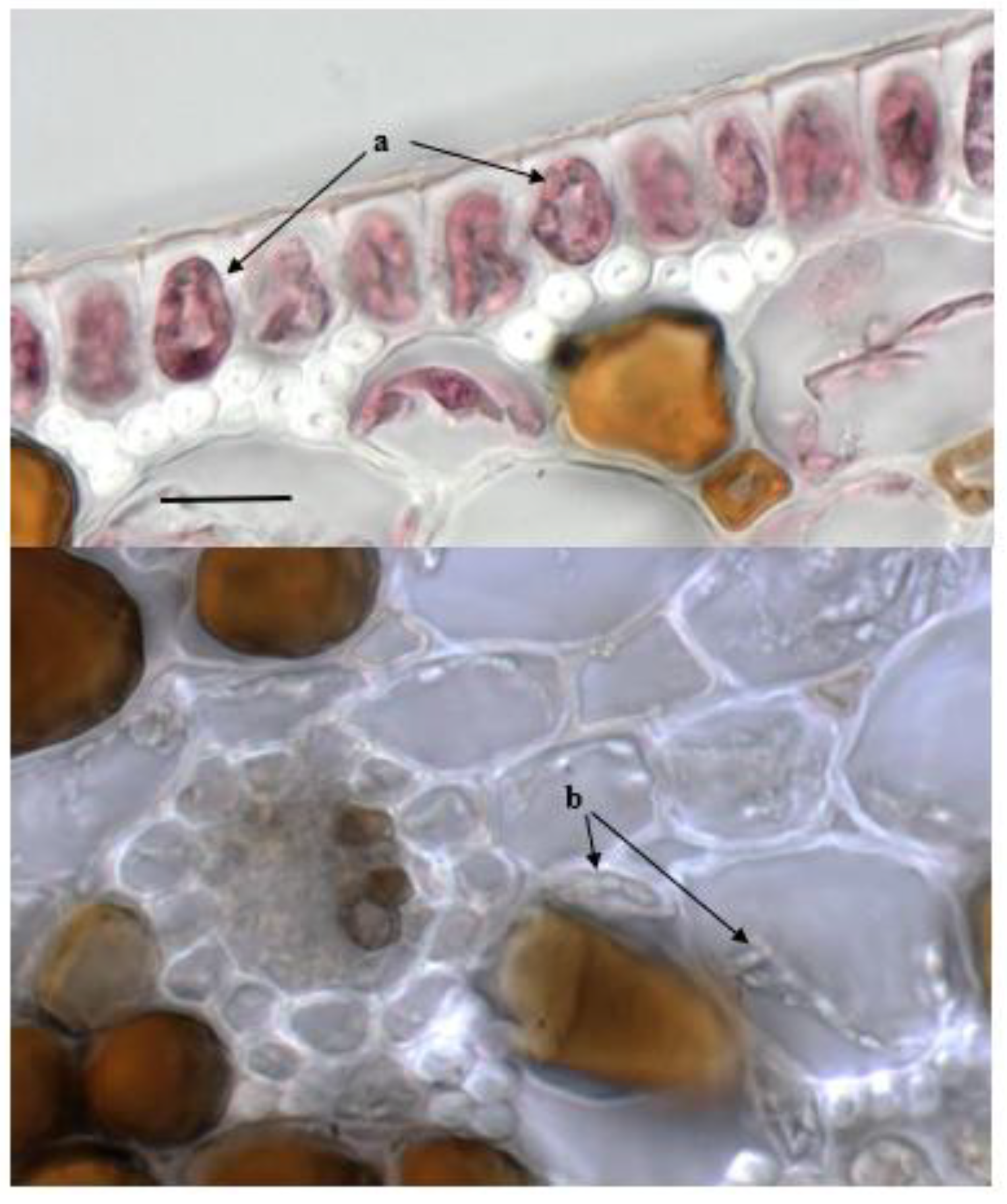
2.2. Detection of Fungal Endophytes in P. oceanica Organs
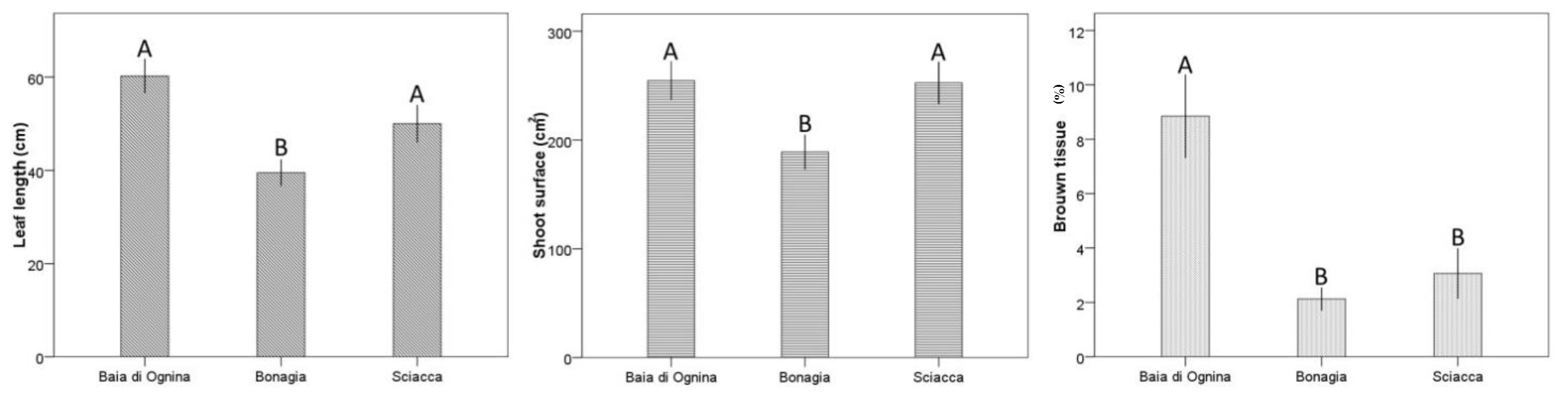
2.3. Leaf Biometry
3. Discussion
4. Materials and Methods
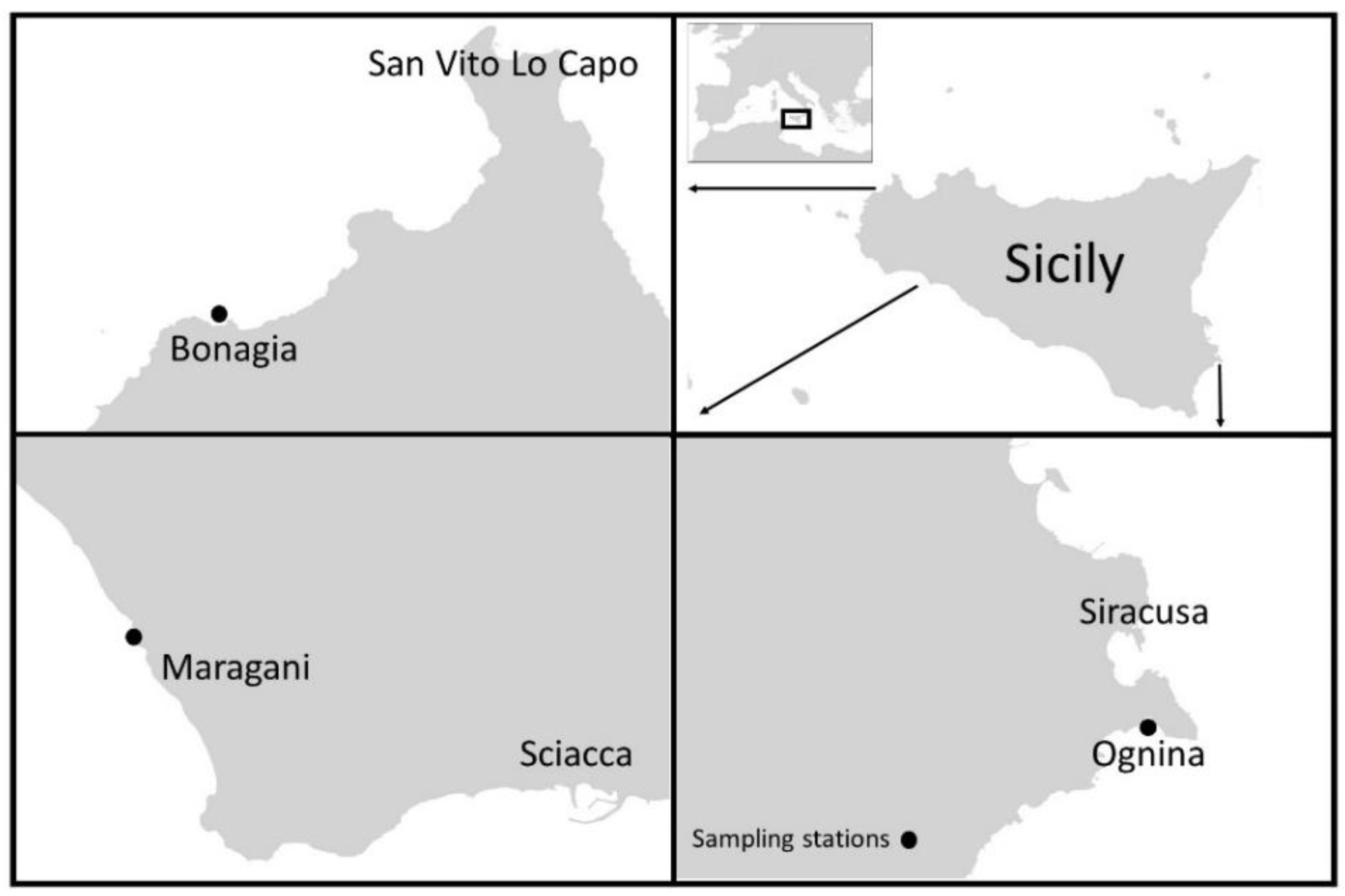
4.1. Study and Sampling Areas
4.2. Isolation of Fungal Endophytes from P. oceanica Organs
4.3. Molecular Identification and Phylogenetic Analyses
4.4. Microscopic Observations
4.5. Leaf Biometry of P. oceanica Samples
4.6. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Procaccini, G.; Buia, M.C.; Gambi, M.C.; Perez, M.; Pergent, G.; Pergent-Martini, C.; Romero, J. Seagrass status and extent along the Mediterranean coasts of Italy, France and Spain. In World Atlas of Seagrasses; Green, E.P., Short, F.T., Eds.; University of California Press: Berkeley, CA, USA, 2003; pp. 48–58. [Google Scholar]
- Hemminga, M.A.; Duarte, C.M. Seagrass Ecology; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Diaz, A.E. Patch dynamics of the Mediterranean seagrass Posidonia oceanica: Implications for recolonisation process. Aquat. Bot. 2008, 89, 397–403. [Google Scholar] [CrossRef]
- Boudouresque, C.F.; Bernard, G.; Bonhomme, P.; Charbonnel, E.; Diviacco, G.; Meinesz, A.; Pergent, G.; Pergent-Martini, C.; Ruitton, S.; Tunesi, L. Protection and Conservation of Posidonia oceanica Meadows; RAMOGE and RAC/SPA Publications: Tunis, Tunisia, 2012; p. 202. [Google Scholar]
- Vassallo, P.; Paoli, C.; Rovere, A.; Montefalcone, M.; Morri, C.; Bianchi, N.B. The value of the seagrass Posidonia oceanica: A natural capital assessment. Mar. Pollut. Bull. 2013, 75, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Boudouresque, C.F.; Pergent, G.; Pergent-Martini, C.; Ruitton, S.; Thibaut, T.; Verlaque, V. The necromass of the Posidonia oceanica seagrass meadow: Fate, role, ecosystem services and vulnerability. Hydrobiologia 2016, 781, 25–42. [Google Scholar] [CrossRef] [Green Version]
- Mazzella, L.; Buia, M.C.; Gambi, M.C.; Lorenti, M.; Russo, G.F.; Scipione, M.B.; Zupo, V. Plant-animal trophic relationships in the Posidonia oceanica ecosystem of the Mediterranean Sea: A review. In Plant-Animal Interactions in Marine Benthos; John, D.M., Hawkins, S.J., Pric, J.H., Eds.; Clarendon Press: Oxford, UK, 1992; Volume 46, pp. 165–187. [Google Scholar]
- Montagne, C. Exploration Scientifique de l’Algérie Pendant les Années 1840, 1841, 1842; Durieu De Maisonneuve, M.C., Ed.; Imprimerie Royale: Paris, France, 1846; pp. 1–197. [Google Scholar]
- Johnson, T.W., Jr.; Sparrow, F.K. Marine Biology: Fungi in Oceans and Estuaries; J. Cramer, Lehere: Weinheim, Germany, 1961; pp. xxiv + 668. [Google Scholar]
- Kohlmeyer, J. Zwei neue Ascomycetes Gat tungen auf Posidonia-rhizomen. Nova Hedwig. 1963, 6, 5–13. [Google Scholar]
- Kohlmeyer, J. Intertidal and phycophilous fungi from Tenerife (Canary Islands). Trans. Br. Mycol. Soc. 1967, 50, 137–147. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Kohlmeyer, E. Marine fungi from tropical America and Africa. Mycologia 1971, 63, 831–861. [Google Scholar] [CrossRef]
- Cuomo, V.; Vanzanella, F.; Fresi, E.; Mazzella, L.; Scipione, M.B. Micoflora delle fenerogame dell ‘Isolad’ Ischia: Posidonia oceanica (L.) Delile Cymodocea nodosa (Ucria) Aschers; Bulletin Musea Institute Biologia, Universiti Genova: Genoa, Italy, 1982; Volume 50, pp. 162–166. [Google Scholar]
- Cuomo, V.; Vanzanella, F.; Fresi, E.; Cinelli, F.; Mazzella, L. Fungal flora of Posidonia oceanica and its ecological significance. Trans. Br. Mycol. Soc. 1985, 84, 35–40. [Google Scholar] [CrossRef]
- Garcias-Bonet, N.; Arrieta, J.M.; de Santana, C.N.; Duarte, C.M.; Marba, N. Endophytic bacterial community of a Mediterranean marine angiosperm (Posidonia oceanica). Front. Microbiol. 2012, 3, 342. [Google Scholar] [CrossRef] [Green Version]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 2013, 30, 686–694. [Google Scholar] [CrossRef]
- Torta, L.; Lo Piccolo, S.; Piazza, G.; Burruano, S.; Colombo, P.; Ottonello, D.; Perrone, R.; Di Maida, G.; Pirrotta, M.; Tomasello, A.; et al. Lulwoana sp., a dark septate endophyte in roots of Posidoniaoceanica (L.) Delile seagrass. Plant Biol. 2015, 17, 505–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, J.; Volkmann-Kohlmeyer, B.; Gräfenhan, T.; Spatafora, J.W.; Kohlmeyer, J. A re-evaluation of Lulworthiales: Relationships based on 18S and 28S rDNA. Mycol. Res. 2005, 109, 556–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumpponen, A.; Trappe, J.M. Dark septate endophytes: A review of facultative biotrophic root-colonizing fungi. New Phytol. 1998, 140, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Sieber, T.N.; Grünig, C.R. Fungal Root Endophytes. In Plant Roots—The Hidden Half, 4th ed.; Eshel, A., Beeckman, T., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2013; pp. 31–49. [Google Scholar]
- Vohník, M.; Borovec, O.; Župan, I.; Vondrášek, D.; Petrtýl, M.; Sudová, R. Anatomically and morphologically unique dark septate endophytic association in the roots of the Mediterranean endemic seagrass Posidonia oceanica. Mycorrhiza 2015, 25, 663–672. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Kolařík, M. Communities of cultivable root mycobionts of the seagrass Posidonia oceanica in the Northwest Mediterranean Sea are dominated by a hitherto undescribed pleosporalean dark septate endophyte. Microb. Ecol. 2016, 71, 442–451. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Župan, I.; Kolařík, M.; Sudová, R. Fungal root symbionts of the seagrass Posidonia oceanica in the central Adriatic Sea revealed by microscopy, culturing and pyrosequencing. Mar. Ecol. Prog. Ser. 2017, 583, 107–112. [Google Scholar] [CrossRef]
- Lefebvre, L.; Gobert, S. How Does Endophytic Fungi Transform the Posidonia oceanica (L.) Delile (1813) Meadow into the Aegagropiles? CIBM: Liège, Belgium, 2018. [Google Scholar]
- Tomasello, A.; Perrone, R.; Colombo, P.; Pirrotta, M.; Calvo, S. Variability of root hair anatomy and morphology in Posidonia oceanica (L.) Delile and substratum typology: First novel on spiral form. Aquat. Bot. 2018, 145, 45–48. [Google Scholar] [CrossRef]
- Vohník, M. Are lulworthioid fungi dark septate endophytes of the dominant Mediterranean seagrass Posidonia oceanica? Plant Biol. 2021, 24, 127–133. [Google Scholar] [CrossRef]
- Saikkonen, K.; Saari, S.; Helander, M. Defensive mutualism between plants and endophytic fungi? Fungal Divers. 2010, 41, 101–113. [Google Scholar] [CrossRef]
- Sarkar, S.; Dey, A.; Kumar, V.; El-Saber, B.G.; El-Esawi, M.A.; Tomczyk, M.; Ray, P. Fungal endophyte: An interactive endosymbiont with the capability of modulating host physiology. Front. Plant Sci. 2021, 12, 701800. [Google Scholar] [CrossRef]
- Calvo, S.; Tomasello, A.; Di Maida, G.; Pirrotta, M.; Buia, M.C.; Cinelli, F.; Cormaci, M.; Furnari, G.; Giaccone, G.; Luzzu, F.; et al. Seagrasses along the Sicilian coasts. Chem. Ecol. 2010, 26, 249–266. [Google Scholar] [CrossRef]
- Jones, E.B.J.; Sakayaroj, J.; Suetrong, S.; Somrithipol, S.; Pang, K. Classification of marine Ascomycota, anamorphic taxa and Basidiomycota. Fungal Divers. 2009, 35, 1–187. [Google Scholar]
- Garzoli, L.; Gnavi, G.; Tamma, F.; Tosi, S.; Varese, G.C.; Picco, A.M. Sink or swim: Updated knowledge on marine fungi associated with wood substrates in the Mediterranean Sea and hints about their potential to remediate hydrocarbons. Progr. Oceanogr. 2015, 137, 140–148. [Google Scholar] [CrossRef]
- Poli, A.; Prigione, V.; Bovio, E.; Perugini, I.; Varese, G.C. Insights on Lulworthiales inhabiting the Mediterranean Sea and description of three novel species of the genus Paralulworthia. J. Fungi. 2021, 7, 940. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, G.; Sirchia, B.; Ruvolo, V. Monitoring of Posidonia oceanica meadows in the Sicilian coasts under the Water Framework Directive (WFD). In Proceedings of the 8th International Symposium “Monitoring of Mediterranean coastal areas: Problems and measurement techniques”, Livorno, Italy, 16–18 June 2020; Firenze University Press: Livorno, Italy; pp. 510–518. [Google Scholar]
- Bellissimo, G.; Sirchia, B.; Ruvolo, V. Assessment of the ecological status of Sicilian coastal waters according to a macroalgae based index (CARLIT). In Proceedings of the 8th International Symposium “Monitoring of Mediterranean coastal areas: Problems and measurement techniques”, Livorno, Italy, 16–18 June 2020; Firenze University Press: Livorno, Italy; pp. 519–528. [Google Scholar] [CrossRef]
- Giovannetti, E.; Lasagna, R.; Montefalcone, M.; Bianchi, C.N.; Albertelli, G.; Morri, C. Inconsistent responses to substratum nature in Posidonia oceanica meadows: An integration through complexity levels? Chem. Ecol. 2008, 24, 83–91. [Google Scholar] [CrossRef]
- Di Maida, G.; Tomasello, A.; Sciandra, M.; Pirrotta, M.; Milazzo, M.; Calvo, S. Effect of different substrates on rhizome growth: Leaf biometry and shoot density of Posidonia oceanica. Mar. Environ. Res. 2013, 87–88, 96–102. [Google Scholar] [CrossRef]
- Balestri, E.; Cinelli, F.; Lardicci, C. Spatial variation in Posidonia oceanica structural, morphological and dynamic features in a northwestern Mediterranean coastal area: A multi-scale analysis. Mar. Ecol. Prog. Ser. 2003, 250, 51–60. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. Fungal diversity in the Neptune Forest: Comparison of the mycobiota of Posidonia oceanica, Flabellia petiolata, and Padina pavonica. Front. Microbiol. 2020, 11, 933. [Google Scholar] [CrossRef]
- Samerpitak, K.A.; Duarte, A.P.M.; Attili-Angelis, D.; Pagnocca, F.C.; Heinrichs, G.; Rijs, A.J.M.M.; Alfjorden, A.; Gerrits van den Ende, A.H.G.; Menken, S.B.J.; de Hoog, G.S. A new species of the oligotrophic genus Ochroconis (Sympoventuriaceae). Mycol. Progress. 2015, 14, 6. [Google Scholar] [CrossRef] [Green Version]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Dayarathne, M.C.; Devedatha, B.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Becker, K.; Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 2021, 74, 1–23. [Google Scholar] [CrossRef]
- Cheng, X.; Li, W.; Cai, L. Molecular phylogeny of Ascotricha, including two new marine algae-associated species. Mycologia 2015, 107, 490–504. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Visagie, C.M.; Meijer, M.; Frisvad, J.C.; Busby, P.E.; Pitt, J.I.; Seifert, K.A.; Louis-Seize, G.; Demirel, R.; Yilmaz, N.; et al. A taxonomic and phylogenetic revision of Penicillium section Aspergilloides. Stud. Mycol. 2014, 78, 373–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Lei, X.; Shao, S.; Zhou, X.; Li, Y.; Yang, B. Azaphilones and meroterpenoids from the soft coral-derived fungus Penicillium glabrum glmu003. Chem. Biodivers. 2021, 18, e2100663. [Google Scholar] [CrossRef]
- Zhang, Y.; Mu, J.; Feng, Y.; Kang, Y.; Zhang, J.; Gu, P.; Wang, Y.; Ma, L.; Zhu, Y. Broad-spectrum antimicrobial epiphytic and endophytic fungi from marine organisms: Isolation, bioassay and taxonomy. Mar. Drugs 2009, 7, 97–112. [Google Scholar] [CrossRef] [Green Version]
- Wiese, J.; Ohlendorf, B.; Blümel, M.; Schmaljohann, R.; Imhoff, J.F. Phylogenetic identification of fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Mar. Drugs 2011, 9, 561–585. [Google Scholar] [CrossRef]
- Kirichuka, N.N.; Pivkin, M.V. Filamentous fungi associated with the seagrass Zostera marina Linnaeus, 1753 of Rifovaya Bay (Peter the Great Bay, the Sea of Japan). Russ. J. Mar. Biol. 2015, 41, 351–355. [Google Scholar] [CrossRef]
- Knob, A.; Beitel, S.M.; Fortkamp, D.; Terrasan, C.R.; de Almeida, A.F. Production, purification, and characterization of a major Penicillium glabrum xylanase using Brewer’s spent grain as substrate. BioMed Res. Int. 2013, 2013, 728735. [Google Scholar] [CrossRef] [Green Version]
- Borovec, O.; Vohník, M. Ontogenetic transition from specialized root hairs to specific root-fungus symbiosis in the dominant Mediterranean seagrass Posidonia oceanica. Sci. Rep. 2018, 8, 10773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Piccolo, S.; Mondello, V.; Giambra, S.; Conigliaro, G.; Torta, L.; Burruano, S. Arthrinium phaeospermum, Phoma cladoniicola and Ulocladium consortiale, new olive pathogens in Italy. J. Phytopathol. 2014, 162, 258–263. [Google Scholar] [CrossRef]
- Ragazzi, A.; Mancini, F.; Dellavalle, I.; Capretti, P.; Moricca, S. Endophytic fungi in Quercus cerris: Isolation frequency in relation to phenological phase, tree health and the organ affected. Phytopathol. Mediterr. 2001, 40, 165–171. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; Academic Press: London, UK, 1980; p. 406. [Google Scholar]
- Barnett, H.L.; Hunter, B. Illustrated Genera of Imperfect Fungi; APS Press: St. Paul, MN, USA, 1998; p. 220. [Google Scholar]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage, 2nd ed.; Aspen Publishers Inc.: Gaithersburg, MD, USA, 1999; p. 234. [Google Scholar]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Alves, A.; Correia, A.; Luque, J.; Phillips, A. Botryosphaeria corticola, sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia 2004, 96, 598–613. [Google Scholar] [CrossRef]
- Giambra, S.; Piazza, G.; Alves, A.; Mondello, V.; Berbegal, M.; Burruano, S. Botryosphaeriaceae species associated with diseased loquat trees in Italy and description of Diplodia rosacearum sp. nov. Mycosphere 2016, 7, 978–989. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Giraud, G. Essai de classement des herbiers de Posidonia oceanica (L.) Delile. Bot. Mar. 1977, 20, 487–491. [Google Scholar] [CrossRef]
- Pergent-Martini, C.; Leoni, V.; Pasqualini, V.; Ardizzone, G.D.; Balestri, E.; Bedini, R.; Belluscio, A.; Belsher, T.; Borg, J.A.; Bordouresque, C.; et al. Descriptors of Posidonia oceanica meadows: Use and application. Ecol. Indic. 2005, 5, 213–230. [Google Scholar] [CrossRef]
- Pergent, G.; Pergent-Martini, C.; Boudouresque, C. Utilisation de l’herbier à Posidonia oceanica comme indicateur biologique de la qualité du milieu littoral en Méditerranée: État des connaissances. Mésogée 1995, 54, 3–27. [Google Scholar]
- Underwood, A.J. Experiments in Ecology: Their Logical Design and Interpretation Using Analysis of Variance; Cambridge University Press: Cambridge, UK, 1997; p. 524. [Google Scholar]
| Isolate Code | Isolate Identity | GenBank Accession No. | Blast Match Sequence | ||
|---|---|---|---|---|---|
| Reference Accession No. | Coverage (%) | Identity (%) | |||
| PO1 | Cordycipitaceae | KY463483 | MH231248 | 99 | 96 |
| PO2 | Fusarium sp. | KY463484 | MK589327 | 100 | 100 |
| PO3 | Lulwoana sp. | KY463485 | KF719966 | 100 | 100 |
| PO4 | Ochroconis sp. | KY463486 | MH063201 | 100 | 100 |
| PO5 | Paecilomyces sp. | KY463487 | KF871460 | 100 | 99 |
| PO6 | Penicillium glabrum | KY463488 | MH864674 | 100 | 100 |
| PO7 | Sordariomycetes | KY463489 | GQ153240 | 100 | 100 |
| PO8 | Thielavia microspora | KY463490 | JN709490 | 100 | 100 |
| PO9 | Xylariaceae sp. | KY463491 | MK334345 | 100 | 100 |
| PO10 | Lulwoana sp. | KY463492 | KF719966 | 100 | 100 |
| PO11 | Lulwoana sp. | KY463493 | KF719966 | 100 | 100 |
| PO12 | Lulwoana sp. | KY463494 | KF719966 | 100 | 100 |
| PO13 | Lulwoana sp. | KY463495 | KF719966 | 100 | 100 |
| Taxa | No. of Isolates | IF% |
|---|---|---|
| Penicillium glabrum | 331 | 65.7 |
| Lulwoana sp. | 116 | 23.0 |
| Xylariaceae | 40 | 7.9 |
| Ochroconis sp. | 10 | 2.0 |
| Cordycipitaceae | 3 | 0.6 |
| Fusarium sp. | 1 | 0.2 |
| Paecilomyces sp. | 1 | 0.2 |
| Sordariomycetes | 1 | 0.2 |
| Thielavia microspora | 1 | 0.2 |
| Taxa | No. of Isolates | IFo% | IFs% | ||||
|---|---|---|---|---|---|---|---|
| Roots | Rhizome | Leaf | Bonagia | Ognina | Sciacca | ||
| P. glabrum | 331 | 100 | 0 | 0 | 0 | 100 | 0 |
| Lulwoana sp. | 116 | 44.8 | 54.3 | 0.9 | 19.8 | 43.1 | 37.1 |
| Xylariaceae | 40 | 42.5 | 57.5 | 0 | 72.5 | 27.5 | 0 |
| Ochroconis sp. | 10 | 70 | 20 | 10 | 80 | 10 | 10 |
| Isolate Number | Isolate Identity | Host | Country | ITS GenBank Acc. No. |
|---|---|---|---|---|
| RP2 | Lulwoana sp. | Posidonia oceanica | Italy | KF719965 |
| RP5 | Lulwoana sp. | Posidonia oceanica | Italy | KF719968 |
| RP6 | Lulwoana sp. | Posidonia oceanica | Italy | KF719969 |
| RP1 | Lulwoana sp. | Posidonia oceanica | Italy | KF719964 |
| P12 | Lulworthiales | Posidonia oceanica | Spain | KC145431 |
| P03 | Lulwoana sp. | Posidonia oceanica | Italy | KY463485 |
| P13 | Lulworthiales | Posidonia oceanica | France | KC145432 |
| RP4 | Lulwoana sp. | Posidonia oceanica | Italy | KF719967 |
| P03 | Lulworthiales | Posidonia oceanica | Spain | KC145423 |
| RP3 | Lulwoana sp. | Posidonia oceanica | Italy | KF719966 |
| P010 | Lulwoana sp. | Posidonia oceanica | Italy | KY463492 |
| P011 | Lulwoana sp. | Posidonia oceanica | Italy | KY463493 |
| PO12 | Lulwoana sp. | Posidonia oceanica | Italy | KY463494 |
| PO13 | Lulwoana sp. | Posidonia oceanica | Italy | KY463495 |
| P32 | Lulworthiales | Posidonia oceanica | Croatia | KC412721 |
| MUT 5413 | Lulwoana sp. | Posidonia oceanica | Italy | MK578237 |
| P02 | Lulworthiales | Posidonia oceanica | Italy | KC145422 |
| MUT 1483 | Lulwoana sp. | Driftwood | Italy | KF915986 |
| NBRC 32137 | Lulwoana uniseptata | Submerged wood | Japan | LC146746 |
| ATCC62580 | Zalerion maritimum | Driftwood | U.S.A. | AF169305 |
| CBS 280.54 | Lulwoana uniseptata | Unknown | Unknown | MH857330 |
| FCUL210208SF10 | Lulworthia atlantica | Sea water | Portugal | KT347215 |
| CBS 139632 | Lulworthia atlantica | Fagus sylvatica | Portugal | KY202429 |
| FCUL090707CF10 | Lulworthia atlantica | Sea water | Portugal | KT347213 |
| FCUL061107CP4 | Lulworthia atlantica | Sea water | Portugal | KT347206 |
| FCUL210208SP4 | Lulworthia atlantica | Sea water | Portugal | KT347205 |
| CBS 541.66 | Leptosphaeria doliolum | Rudbeckia sp. | Netherlands | MH858879 |
| Leaf Length | Shoot Surface | Brown Tissue | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Source of Variation | Df | MS | F | Df | MS | F | Df | MS | F |
| Locality | 2 | 1863.9 | 9.5 *** | 2 | 24,980.1 | 4.8 * | 2 | 6.1 | 4.9 * |
| RES | 47 | 197.1 | 47 | 5155.8 | 33 | 1.3 | |||
| Levene’s test | ns | Ns | * | ||||||
| Transformation | None | Ln | |||||||
| Post hoc test | |||||||||
| 1 = 3 > 2 | 1 = 3 > 2 | 1 > 2 = 3 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torta, L.; Burruano, S.; Giambra, S.; Conigliaro, G.; Piazza, G.; Mirabile, G.; Pirrotta, M.; Calvo, R.; Bellissimo, G.; Calvo, S.; et al. Cultivable Fungal Endophytes in Roots, Rhizomes and Leaves of Posidonia oceanica (L.) Delile along the Coast of Sicily, Italy. Plants 2022, 11, 1139. https://doi.org/10.3390/plants11091139
Torta L, Burruano S, Giambra S, Conigliaro G, Piazza G, Mirabile G, Pirrotta M, Calvo R, Bellissimo G, Calvo S, et al. Cultivable Fungal Endophytes in Roots, Rhizomes and Leaves of Posidonia oceanica (L.) Delile along the Coast of Sicily, Italy. Plants. 2022; 11(9):1139. https://doi.org/10.3390/plants11091139
Chicago/Turabian StyleTorta, Livio, Santella Burruano, Selene Giambra, Gaetano Conigliaro, Gaia Piazza, Giulia Mirabile, Maria Pirrotta, Roberta Calvo, Giancarlo Bellissimo, Sebastiano Calvo, and et al. 2022. "Cultivable Fungal Endophytes in Roots, Rhizomes and Leaves of Posidonia oceanica (L.) Delile along the Coast of Sicily, Italy" Plants 11, no. 9: 1139. https://doi.org/10.3390/plants11091139
APA StyleTorta, L., Burruano, S., Giambra, S., Conigliaro, G., Piazza, G., Mirabile, G., Pirrotta, M., Calvo, R., Bellissimo, G., Calvo, S., & Tomasello, A. (2022). Cultivable Fungal Endophytes in Roots, Rhizomes and Leaves of Posidonia oceanica (L.) Delile along the Coast of Sicily, Italy. Plants, 11(9), 1139. https://doi.org/10.3390/plants11091139










