CRISPR/Cas9 in Planta Hairy Root Transformation: A Powerful Platform for Functional Analysis of Root Traits in Soybean
Abstract
:1. Introduction
2. Hairy Root Transformation
3. CRISPR/Cas9 System for Functional Analysis in Soybean
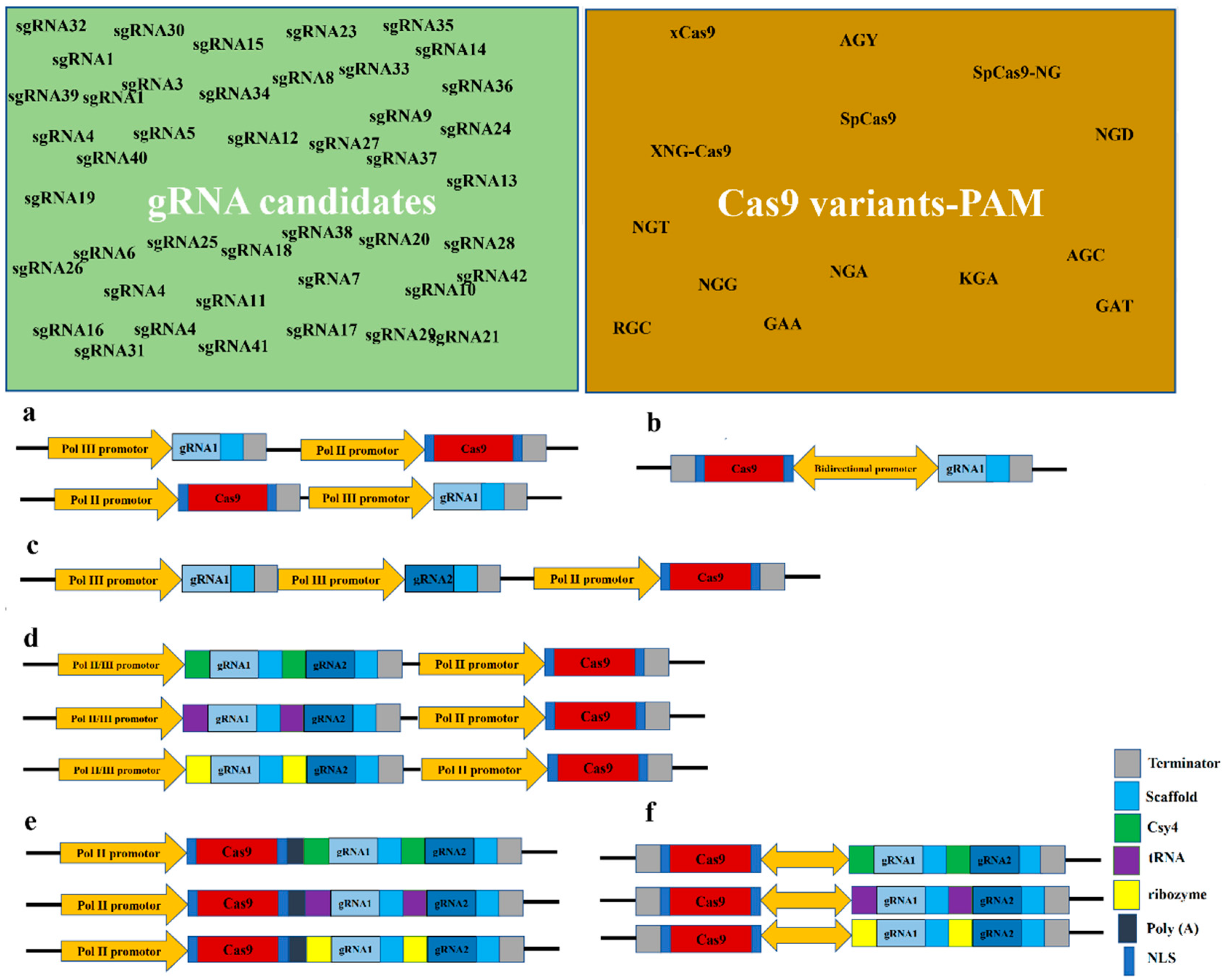
3.1. gRNA and Its Components
3.2. Critical Criteria in Designing gRNA
3.2.1. Proper Selection of the Target Site(s) within the Locus of Interest
3.2.2. Determination and Prediction of Off-Target Activity
3.2.3. Nucleotide Features of the Designed gRNA and its Associated Secondary Structure
3.3. Number of Designed gRNAs
3.4. Assembly of Multiple gRNAs and gRNA Processing Strategies
3.5. Cas9 Nucleases
3.6. Chimeric Deactivated Cas9 Proteins and Their Applications
3.7. Gene Regulatory Elements (GREs) of gRNAs and Cas9 Nuclease Cassettes
3.8. Configuration of gRNA and Cas9 Cassettes
4. Examples of Agrobacterium rhizogenes-Mediated Delivery of CRISPR/Cas9 Reagents for Gene Functional Analysis in Soybean
4.1. Modifications in In Vitro Inoculation
4.2. Modifications in in Planta Inoculation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABE | Adenine base editor |
| BE | Base editor |
| CBE | Cytosine base editor |
| CRISPR/Cas | Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein |
| DSB | Double-strand breaks |
| gRNA | Guide RNA |
| NGS | Next-generation sequencing |
| NLSs | Nuclear localization signals |
| PAM | Protospacer adjacent motif |
| PTG | Polycistronic tRNA-gRNA vector |
| RNAi | RNA interference |
| sgRNA | Single guide RNA |
| TALENs | Transcription activator-like effector nucleases |
| ZFNs | Zinc finger nucleases |
References
- Kuma, K.M.; Lopes-Caitar, V.S.; Romero, C.C.T.; Silva, S.M.H.; Kuwahara, M.K.; Carvalho, M.C.C.G.; Abdelnoor, R.V.; Dias, W.P.; Marcelino-Guimarães, F.C. A High Efficient Protocol for Soybean Root Transformation by Agrobacterium rhizogenes and Most Stable Reference Genes for RT-QPCR Analysis. Plant Cell Rep. 2015, 34, 1987–2000. [Google Scholar] [CrossRef]
- Sahu, P.K.; Sao, R.; Mondal, S.; Vishwakarma, G.; Gupta, S.K.; Kumar, V.; Singh, S.; Sharma, D.; Das, B.K. Next Generation Sequencing Based Forward Genetic Approaches for Identification and Mapping of Causal Mutations in Crop Plants: A Comprehensive Review. Plants 2020, 9, 1355. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-Throughput Functional Genomics Using CRISPR–Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Alonso, J.M.; Ecker, J.R. Moving Forward in Reverse: Genetic Technologies to Enable Genome-Wide Phenomic Screens in Arabidopsis. Nat. Rev. Genet. 2006, 7, 524–536. [Google Scholar] [CrossRef]
- Bezie, Y.; Tilahun, T.; Atnaf, M.; Taye, M. The Potential Applications of Site-Directed Mutagenesis for Crop Improvement: A Review. J. Crop Sci. Biotechnol. 2021, 24, 229–244. [Google Scholar] [CrossRef]
- Anjanappa, R.B.; Gruissem, W. Current Progress and Challenges in Crop Genetic Transformation. J. Plant Physiol. 2021, 261, 153411. [Google Scholar] [CrossRef]
- Joung, Y.H.; Choi, P.-S.; Kwon, S.-Y.; Harn, C.H. Plant Transformation Methods and Applications. In Current Technologies in Plant Molecular Breeding; Springer: Dordrecht, The Netherlands, 2015; pp. 297–343. [Google Scholar]
- Rivera, A.L.; Gómez-Lim, M.; Fernández, F.; Loske, A.M. Physical Methods for Genetic Plant Transformation. Phys. Life Rev. 2012, 9, 308–345. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat Noori, S.A.; Galuszka, P.; Mortazavian, S.M.M. Tissue Culture-Based Agrobacterium-Mediated and in Planta Transformation Methods. Czech J. Genet. Plant Breed. 2017, 53, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to Produce T-DNA Free CRISPRed Fruit Trees via Agrobacterium Tumefaciens Stable Gene Transfer. Sci. Rep. 2020, 10, 20155. [Google Scholar] [CrossRef]
- Opabode, J.T. Agrobacterium-Mediated Transformation of Plants: Emerging Factors That Influence Efficiency. Biotechnol. Mol. Biol. Rev. 2006, 1, 12–20. [Google Scholar]
- Atif, R.M.; Patat-Ochatt, E.M.; Svabova, L.; Ondrej, V.; Klenoticova, H.; Jacas, L.; Griga, M.; Ochatt, S.J. Gene Transfer in Legumes; Springer: Berlin/Heidelberg, Germany, 2013; pp. 37–100. [Google Scholar]
- Collado, R.; Bermúdez-Caraballoso, I.; García, L.R.; Veitía, N.; Torres, D.; Romero, C.; Angenon, G. Epicotyl Sections as Targets for Plant Regeneration and Transient Transformation of Common Bean Using Agrobacterium Tumefaciens. Vitr. Cell. Dev. Biol. Plant 2016, 52, 500–511. [Google Scholar] [CrossRef]
- Tyurin, A.A.; Suhorukova, A.V.; Kabardaeva, K.V.; Goldenkova-Pavlova, I.V. Transient Gene Expression Is an Effective Experimental Tool for the Research into the Fine Mechanisms of Plant Gene Function: Advantages, Limitations, and Solutions. Plants 2020, 9, 1187. [Google Scholar] [CrossRef] [PubMed]
- Paolis, A.; Frugis, G.; Giannino, D.; Iannelli, M.; Mele, G.; Rugini, E.; Silvestri, C.; Sparvoli, F.; Testone, G.; Mauro, M.; et al. Plant Cellular and Molecular Biotechnology: Following Mariotti’s Steps. Plants 2019, 8, 18. [Google Scholar] [CrossRef] [Green Version]
- Iantcheva, A.; Mysore, K.S.; Ratet, P. Transformation of Leguminous Plants to Study Symbiotic Interactions. Int. J. Dev. Biol. 2013, 57, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Nambiar-Veetil, M.; Bogusz, D.; Franche, C. Hairy Roots as a Tool for the Functional Analysis of Plant Genes. In Hairy Roots; Springer: Singapore, 2018; pp. 275–292. [Google Scholar]
- Alagarsamy, K.; Shamala, L.F.; Wei, S. Protocol: High-Efficiency in-Planta Agrobacterium-Mediated Transgenic Hairy Root Induction of Camellia Sinensis Var. Sinensis. Plant Methods 2018, 14, 17. [Google Scholar] [CrossRef]
- Niazian, M. Application of Genetics and Biotechnology for Improving Medicinal Plants. Planta 2019, 249, 953–973. [Google Scholar] [CrossRef]
- Smith, J. Requirements for Double-Strand Cleavage by Chimeric Restriction Enzymes with Zinc Finger DNA-Recognition Domains. Nucleic Acids Res. 2000, 28, 3361–3369. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A Widely Applicable Technology for Targeted Genome Editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef] [Green Version]
- Voytas, D.F. Plant Genome Engineering with Sequence-Specific Nucleases. Annu. Rev. Plant Biol. 2013, 64, 327–350. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.-S. A Guide to Genome Engineering with Programmable Nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef]
- Scheben, A.; Wolter, F.; Batley, J.; Puchta, H.; Edwards, D. Towards CRISPR/Cas Crops—Bringing Together Genomics and Genome Editing. New Phytol. 2017, 216, 682–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Q.S.M.; Tian, L. An Efficient and Specific CRISPR-Cas9 Genome Editing System Targeting Soybean Phytoene Desaturase Genes. BMC Biotechnol. 2022, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.U.; McCoy, E.; Raza, G.; Ali, Z.; Mansoor, S.; Amin, I. Improvement of Soybean; A Way Forward Transition from Genetic Engineering to New Plant Breeding Technologies. Mol. Biotechnol. 2022, 1–19. [Google Scholar] [CrossRef]
- Alok, A.; Kumar, J.; Upadhyay, S.K. Engineering in Hairy Roots Using CRISPR/Cas9-Mediated Editing. In Hairy Roots; Springer: Singapore, 2018; pp. 329–342. [Google Scholar]
- Cai, Y.; Chen, L.; Liu, X.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-Mediated Genome Editing in Soybean Hairy Roots. PLoS ONE 2015, 10, e0136064. [Google Scholar] [CrossRef]
- Boobalan, S.; Kamalanathan, D. Tailoring Enhanced Production of Aervine in Aerva Lanata (L.) Juss. Ex Schult by Agrobacterium rhizogenes- Mediated Hairy Root Cultures. Ind. Crops Prod. 2020, 155, 112814. [Google Scholar] [CrossRef]
- Demirci, T.; Akçay, U.Ç.; Göktürk Baydar, N. Physical and Biochemical Differences in Agrobacterium rhizogenes-Mediated Transgenic Hairy Root Lines of Echinacea Purpurea. Vitr. Cell. Dev. Biol. Plant 2020, 56, 875–881. [Google Scholar] [CrossRef]
- Daspute, A.A.; Yunxuan, X.; Gu, M.; Kobayashi, Y.; Wagh, S.; Panche, A.; Koyama, H. Agrobacterium rhizogenes—Mediated Hairy Roots Transformation as a Tool for Exploring Aluminum-Responsive Genes Function. Future Sci. OA 2019, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-Mediated Transformation of Soybean to Study Root Biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. Soybean Hairy Roots Produced in Vitro by Agrobacterium rhizogenes-Mediated Transformation. Crop J. 2018, 6, 162–171. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, X.; Cao, L.; Ji, J.; Liu, T.; Duan, K. Highly Efficient Agrobacterium rhizogenes-Mediated Hairy Root Transformation for Gene Functional and Gene Editing Analysis in Soybean. Plant Methods 2021, 17, 73. [Google Scholar] [CrossRef]
- Sun, T.; Ma, N.; Wang, C.; Fan, H.; Wang, M.; Zhang, J.; Cao, J.; Wang, D. A Golgi-Localized Sodium/Hydrogen Exchanger Positively Regulates Salt Tolerance by Maintaining Higher K+/Na+ Ratio in Soybean. Front. Plant Sci. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Piya, S.; Hawk, T.; Patel, B.; Baldwin, L.; Rice, J.H.; Stewart, C.N.; Hewezi, T. Kinase-dead Mutation: A Novel Strategy for Improving Soybean Resistance to Soybean Cyst Nematode Heterodera Glycines. Mol. Plant Pathol. 2022, 23, 417–430. [Google Scholar] [CrossRef]
- Yang, R.; Li, S.; Yang, X.; Zhu, X.; Fan, H.; Xuan, Y.; Chen, L.; Liu, X.; Wang, Y.; Duan, Y. Fluorescent Soybean Hairy Root Construction and Its Application in the Soybean—Nematode Interaction: An Investigation. Biology 2021, 10, 1353. [Google Scholar] [CrossRef]
- Indrasumunar, A.; Wilde, J.; Hayashi, S.; Li, D.; Gresshoff, P.M. Functional Analysis of Duplicated Symbiosis Receptor Kinase (SymRK) Genes during Nodulation and Mycorrhizal Infection in Soybean (Glycine Max). J. Plant Physiol. 2015, 176, 157–168. [Google Scholar] [CrossRef]
- Indrasumunar, A.; Kereszt, A.; Searle, I.; Miyagi, M.; Li, D.; Nguyen, C.D.T.; Men, A.; Carroll, B.J.; Gresshoff, P.M. Inactivation of Duplicated Nod Factor Receptor 5 (NFR5) Genes in Recessive Loss-of-Function Non-Nodulation Mutants of Allotetraploid Soybean (Glycine Max L. Merr.). Plant Cell Physiol. 2010, 51, 201–214. [Google Scholar] [CrossRef] [Green Version]
- Kiryushkin, A.S.; Ilina, E.L.; Guseva, E.D.; Pawlowski, K.; Demchenko, K.N. Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation. Plants 2021, 11, 51. [Google Scholar] [CrossRef]
- Lin, M.-H.; Gresshoff, P.M.; Indrasumunar, A.; Ferguson, B.J. PHairyRed: A Novel Binary Vector Containing the DsRed2 Reporter Gene for Visual Selection of Transgenic Hairy Roots. Mol. Plant 2011, 4, 537–545. [Google Scholar] [CrossRef]
- Bajaj, R.; Irvin, L.M.; Vaidya, B.N.; Dhekney, S.A.; Joshee, N. Optimizing Plant Regeneration and Genetic Transformation of Paulownia Elongata. Biocatal. Agric. Biotechnol. 2021, 33, 101970. [Google Scholar] [CrossRef]
- Stoykova, P.; Ohkawa, H.; Inui, H. Simple Monitoring of Endocrine-Disrupting Chemicals Using Transgenic Arabidopsis Plants Expressing Medaka Estrogen Receptor. Chemosphere 2022, 286, 131633. [Google Scholar] [CrossRef]
- Garcia-Parajo, M.F.; Koopman, M.; van Dijk, E.M.H.P.; Subramaniam, V.; van Hulst, N.F. The Nature of Fluorescence Emission in the Red Fluorescent Protein DsRed, Revealed by Single-Molecule Detection. Proc. Natl. Acad. Sci. 2001, 98, 14392–14397. [Google Scholar] [CrossRef] [Green Version]
- Berg, R.H.; Beachy, R.N. Fluorescent Protein Applications in Plants. Methods Cell Biol. 2008, 85, 153–177. [Google Scholar]
- He, R.; Zhang, P.; Yan, Y.; Yu, C.; Jiang, L.; Zhu, Y.; Wang, D. Expanding the Range of CRISPR/Cas9-Directed Genome Editing in Soybean. aBIOTECH 2021, 1–10. [Google Scholar] [CrossRef]
- Yuan, G.; Lu, H.; Tang, D.; Hassan, M.M.; Li, Y.; Chen, J.-G.; Tuskan, G.A.; Yang, X. Expanding the Application of a UV-Visible Reporter for Transient Gene Expression and Stable Transformation in Plants. Hortic. Res. 2021, 8, 234. [Google Scholar] [CrossRef]
- Subramanian, S.; Stacey, G.; Yu, O. Endogenous Isoflavones Are Essential for the Establishment of Symbiosis between Soybean and Bradyrhizobium Japonicum. Plant J. 2006, 48, 261–273. [Google Scholar] [CrossRef]
- Hao, Y.-J.; Wei, W.; Song, Q.-X.; Chen, H.-W.; Zhang, Y.-Q.; Wang, F.; Zou, H.-F.; Lei, G.; Tian, A.-G.; Zhang, W.-K.; et al. Soybean NAC Transcription Factors Promote Abiotic Stress Tolerance and Lateral Root Formation in Transgenic Plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Pan, W.-J.; Tao, J.-J.; Cheng, T.; Bian, X.-H.; Wei, W.; Zhang, W.-K.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Soybean MiR172a Improves Salt Tolerance and Can Function as a Long-Distance Signal. Mol. Plant 2016, 9, 1337–1340. [Google Scholar] [CrossRef] [Green Version]
- White, L.J.; Ge, X.; Brözel, V.S.; Subramanian, S. Root Isoflavonoids and Hairy Root Transformation Influence Key Bacterial Taxa in the Soybean Rhizosphere. Environ. Microbiol. 2017, 19, 1391–1406. [Google Scholar] [CrossRef]
- Du, W.; Ning, L.; Liu, Y.; Zhang, S.; Yang, Y.; Wang, Q.; Chao, S.; Yang, H.; Huang, F.; Cheng, H.; et al. Identification of Loci and Candidate Gene GmSPX-RING1 Responsible for Phosphorus Efficiency in Soybean via Genome-Wide Association Analysis. BMC Genom. 2020, 21, 725. [Google Scholar] [CrossRef]
- Yu, Q.; Liu, Y.-L.; Sun, G.-Z.; Liu, Y.-X.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; Lan, J.-H. Genome-Wide Analysis of the Soybean Calmodulin-Binding Protein 60 Family and Identification of GmCBP60A-1 Responses to Drought and Salt Stresses. Int. J. Mol. Sci. 2021, 22, 13501. [Google Scholar] [CrossRef]
- Amritha, P.P.; Shah, J.M. Can Genetic Engineering-Based Methods for Gene Function Identification Be Eclipsed by Genome Editing in Plants? A Comparison of Methodologies. Mol. Genet. Genom. 2021, 296, 485–500. [Google Scholar] [CrossRef]
- Alok, A.; Sharma, S.; Kumar, J.; Verma, S.; Sood, H. Engineering in Plant Genome Using Agrobacterium: Progress and Future. In Metabolic Engineering for Bioactive Compounds; Springer: Singapore, 2017; pp. 91–111. [Google Scholar]
- Small, I. RNAi for Revealing and Engineering Plant Gene Functions. Curr. Opin. Biotechnol. 2007, 18, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Zhou, Y.; Duanmu, D. Use of CRISPR/Cas9 for Symbiotic Nitrogen Fixation Research in Legumes. Prog. Mol. Biol. Trans. Sci. 2017, 149, 187–213. [Google Scholar]
- Fu, M.; Sun, J.; Li, X.; Guan, Y.; Xie, F. Asymmetric Redundancy of Soybean Nodule Inception ( NIN ) Genes in Root Nodule Symbiosis. Plant Physiol. 2022, 188, 477–489. [Google Scholar] [CrossRef]
- Bao, A.; Zhang, C.; Huang, Y.; Chen, H.; Zhou, X.; Cao, D. Genome Editing Technology and Application in Soybean Improvement. Oil Crop Sci. 2020, 5, 31–40. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Tyagi, S.; Kumar, R.; Das, A.; Won, S.Y.; Shukla, P. CRISPR-Cas9 System: A Genome-Editing Tool with Endless Possibilities. J. Biotechnol. 2020, 319, 36–53. [Google Scholar] [CrossRef]
- Rao, M.J.; Wang, L. CRISPR/Cas9 Technology for Improving Agronomic Traits and Future Prospective in Agriculture. Planta 2021, 254, 68. [Google Scholar] [CrossRef]
- Hassan, M.M.; Zhang, Y.; Yuan, G.; De, K.; Chen, J.-G.; Muchero, W.; Tuskan, G.A.; Qi, Y.; Yang, X. Construct Design for CRISPR/Cas-Based Genome Editing in Plants. Trends Plant Sci. 2021, 26, 1133–1152. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Hou, W. Generation of Knockout and Fragment Deletion Mutants in Soybean by CRISPR-Cas9; Humana: New York, NY, USA, 2021; pp. 123–135. [Google Scholar]
- Karmakar, S.; Behera, D.; Baig, M.J.; Molla, K.A. In Vitro Cas9 Cleavage Assay to Check Guide RNA Efficiency; Humana: New York, NY, USA, 2021; pp. 23–39. [Google Scholar]
- Molla, K.A.; Yang, Y. Predicting CRISPR/Cas9-Induced Mutations for Precise Genome Editing. Trends Biotechnol. 2020, 38, 136–141. [Google Scholar] [CrossRef]
- Gayen, D.; Karmakar, S. Designing, Performing, and Analyzing CRISPR-Cas9-Mediated Genome Editing Experiments in Leguminous Plants; Humana: New York, NY, USA, 2021; pp. 103–122. [Google Scholar]
- Sun, X.; Hu, Z.; Chen, R.; Jiang, Q.; Song, G.; Zhang, H.; Xi, Y. Targeted Mutagenesis in Soybean Using the CRISPR-Cas9 System. Sci. Rep. 2015, 5, 10342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, B.; Lucas, S.J.; Budak, H. CRISPR/Cas9 in Plants: At Play in the Genome and at Work for Crop Improvement. Brief. Funct. Genom. 2018, 17, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Chowdhury, A.K.; Islam, T. In Silico Analysis of GRNA Secondary Structure to Predict Its Efficacy for Plant Genome Editing. Front. Oncol. 2021, 15–22. [Google Scholar] [CrossRef]
- Miki, D.; Zinta, G.; Zhang, W.; Peng, F.; Feng, Z.; Zhu, J.-K. CRISPR/Cas9-Based Genome Editing Toolbox for Arabidopsis Thaliana. In Arabidopsis Protocols; Humana: New York, NY, USA, 2021; pp. 121–146. [Google Scholar]
- Zheng, N.; Li, T.; Dittman, J.D.; Su, J.; Li, R.; Gassmann, W.; Peng, D.; Whitham, S.A.; Liu, S.; Yang, B. CRISPR/Cas9-Based Gene Editing Using Egg Cell-Specific Promoters in Arabidopsis and Soybean. Front. Plant Sci. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic Assembly of DNA Molecules up to Several Hundred Kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Sarrion-Perdigones, A.; Falconi, E.E.; Zandalinas, S.I.; Juárez, P.; Fernández-del-Carmen, A.; Granell, A.; Orzaez, D. GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. PLoS ONE 2011, 6, e21622. [Google Scholar] [CrossRef] [Green Version]
- Aliaga-Franco, N.; Zhang, C.; Presa, S.; Srivastava, A.K.; Granell, A.; Alabadí, D.; Sadanandom, A.; Blázquez, M.A.; Minguet, E.G. Identification of Transgene-Free CRISPR-Edited Plants of Rice, Tomato, and Arabidopsis by Monitoring DsRED Fluorescence in Dry Seeds. Front. Plant Sci. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Pavese, V.; Moglia, A.; Corredoira, E.; Martínez, M.T.; Torello Marinoni, D.; Botta, R. First Report of CRISPR/Cas9 Gene Editing in Castanea Sativa Mill. Front. Plant Sci. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Wei, Z.; Rohila, J.S.; Zhao, K. Multiplex Genome-Editing Technologies for Revolutionizing Plant Biology and Crop Improvement. Front. Plant Sci. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Raitskin, O.; Patron, N.J. Multi-Gene Engineering in Plants with RNA-Guided Cas9 Nuclease. Curr. Opin. Biotechnol. 2016, 37, 69–75. [Google Scholar] [CrossRef]
- Luo, Y.; Na, R.; Nowak, J.S.; Qiu, Y.; Lu, Q.S.; Yang, C.; Marsolais, F.; Tian, L. Development of a Csy4-Processed Guide RNA Delivery System with Soybean-Infecting Virus ALSV for Genome Editing. BMC Plant Biol. 2021, 21, 419. [Google Scholar] [CrossRef] [PubMed]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Gao, H.; Wang, H.; Guo, Y.; He, M.; Peng, Y.; Wang, X. GSK3-Mediated Stress Signaling Inhibits Legume–Rhizobium Symbiosis by Phosphorylating GmNSP1 in Soybean. Mol. Plant 2021, 14, 488–502. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-N.; Kim, H.-J.; Chung, Y.-S.; Kim, H.-U. Construction of Multiple Guide RNAs in CRISPR/Cas9 Vector Using Stepwise or Simultaneous Golden Gate Cloning: Case Study for Targeting the FAD2 and FATB Multigene in Soybean. Plants 2021, 10, 2542. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Zheng, L.; Zhao, Y.; Jiang, J.; Zhang, E.J.; Liu, T.; Gu, H.; Qu, L. Engineered XCas9 and SpCas9-NG Variants Broaden PAM Recognition Sites to Generate Mutations in Arabidopsis Plants. Plant Biotechnol. J. 2019, 17, 1865–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grützner, R.; Martin, P.; Horn, C.; Mortensen, S.; Cram, E.J.; Lee-Parsons, C.W.T.; Stuttmann, J.; Marillonnet, S. High-Efficiency Genome Editing in Plants Mediated by a Cas9 Gene Containing Multiple Introns. Plant Commun. 2021, 2, 100135. [Google Scholar] [CrossRef]
- Molla, K.A.; Yang, Y. CRISPR/Cas-Mediated Base Editing: Technical Considerations and Practical Applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Bharat, S.S.; Li, S.; Li, J.; Yan, L.; Xia, L. Base Editing in Plants: Current Status and Challenges. Crop J. 2020, 8, 384–395. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Hou, W. Targeted Base Editing in Soybean Using a CRISPR-Cas9 Cytidine Deaminase Fusion; Humana: New York, NY, USA, 2021; pp. 137–148. [Google Scholar]
- Kang, B.-C.; Woo, J.W.; Kim, S.-T.; Bae, S.-J.; Choi, M.; Kim, J.-S.; Kim, S.-G. Guidelines for C to T Base Editing in Plants: Base-Editing Window, Guide RNA Length, and Efficient Promoter. Plant Biotechnol. Rep. 2019, 13, 533–541. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Zhang, Y.; Yuan, S.; Su, Q.; Sun, S.; Wu, C.; Yao, W.; Han, T.; Hou, W. Target Base Editing in Soybean Using a Modified CRISPR/Cas9 System. Plant Biotechnol. J. 2020, 18, 1996–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Zeng, X.; Zhao, M.; Cui, X.; Wang, Q.; Yang, H.; Cheng, H.; Yu, D. Efficient Targeted Mutagenesis in Soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 2016, 217, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Hsieh-Feng, V.; Yang, Y. Efficient Expression of Multiple Guide RNAs for CRISPR/Cas Genome Editing. aBIOTECH 2020, 1, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Liu, S.; Liu, X.; Liu, B.; Tang, X.; Ren, Q.; Zhou, J.; Zheng, X.; Qi, Y.; Zhang, Y. Intron-Based Single Transcript Unit CRISPR Systems for Plant Genome Editing. Rice 2020, 13, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrijo, J.; Illa-Berenguer, E.; LaFayette, P.; Torres, N.; Aragão, F.J.L.; Parrott, W.; Vianna, G.R. Two Efficient CRISPR/Cas9 Systems for Gene Editing in Soybean. Transgenic Res. 2021, 30, 239–249. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, J.; Lyu, S.; Wang, Q.; Yang, S.; Zhu, H. The Soybean Rfg1 Gene Restricts Nodulation by Sinorhizobium Fredii USDA193. Front. Plant Sci. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Gao, J.-P.; Xu, P.; Wang, M.; Zhang, X.; Yang, J.; Zhou, Y.; Murray, J.D.; Song, C.-P.; Wang, E. Nod Factor Receptor Complex Phosphorylates GmGEF2 to Stimulate ROP Signaling during Nodulation. Curr. Biol. 2021, 31, 3538–3550.e5. [Google Scholar] [CrossRef]
- Nguyen, C.X.; Dohnalkova, A.; Hancock, C.N.; Kirk, K.R.; Stacey, G.; Stacey, M.G. Critical Role for Uricase and Xanthine Dehydrogenase in Soybean Nitrogen Fixation and Nodule Development. Plant Genome 2021, e20171. [Google Scholar] [CrossRef]
- Yun, J.; Sun, Z.; Jiang, Q.; Wang, Y.; Wang, C.; Luo, Y.; Zhang, F.; Li, X. The MiR156b-GmSPL9d Module Modulates Nodulation by Targeting Multiple Core Nodulation Genes in Soybean. New Phytol. 2022, 233, 1881–1899. [Google Scholar] [CrossRef]
- Dong, J.; Hudson, M.E. WI12Rhg1 Interacts with DELLAs and Mediates Soybean Cyst Nematode Resistance through Hormone Pathways. Plant Biotechnol. J. 2022, 20, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Zou, J.; Wang, J.; Zhu, R.; Qi, Z.; Jiang, H.; Hu, Z.; Yang, M.; Zhao, Y.; Wu, X.; et al. A Soybean NAC Homolog Contributes to Resistance to Phytophthora Sojae Mediated by Dirigent Proteins. Crop J. 2021, 10, 332–341. [Google Scholar] [CrossRef]
- Jacobs, T.B.; LaFayette, P.R.; Schmitz, R.J.; Parrott, W.A. Targeted Genome Modifications in Soybean with CRISPR/Cas9. BMC Biotechnol. 2015, 15, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michno, J.-M.; Wang, X.; Liu, J.; Curtin, S.J.; Kono, T.J.; Stupar, R.M. CRISPR/Cas Mutagenesis of Soybean and Medicago Truncatula Using a New Web-Tool and a Modified Cas9 Enzyme. GM Crops Food 2015, 6, 243–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, F.; Yang, S.; Liu, J.; Zhu, H. Rj4, a Gene Controlling Nodulation Specificity in Soybeans, Encodes a Thaumatin-Like Protein But Not the One Previously Reported. Plant Physiol. 2016, 170, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.-T.; Zhao, M.-J.; Wang, C.-T.; Gao, Y.; Wang, Y.-X.; Liu, Y.-W.; Chen, M.; Chen, J.; Zhou, Y.-B.; Xu, Z.-S.; et al. Identification and Characterization of GmMYB118 Responses to Drought and Salt Stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Di, Y.-H.; Sun, X.-J.; Hu, Z.; Jiang, Q.-Y.; Song, G.-H.; Zhang, B.; Zhao, S.-S.; Zhang, H. Enhancing the CRISPR/Cas9 System Based on Multiple GmU6 Promoters in Soybean. Biochem. Biophys. Res. Commun. 2019, 519, 819–823. [Google Scholar] [CrossRef]
- Liu, J.; Gunapati, S.; Mihelich, N.T.; Stec, A.O.; Michno, J.-M.; Stupar, R.M. Genome Editing in Soybean with CRISPR/Cas9; Humana Press: New York, NY, USA, 2019; pp. 217–234. [Google Scholar]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light- and Temperature-entrainable Circadian Clock in Soybean Development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef]
- Butler, K.J.; Fliege, C.; Zapotocny, R.; Diers, B.; Hudson, M.; Bent, A.F. Soybean Cyst Nematode Resistance Quantitative Trait Locus CqSCN-006 Alters the Expression of a γ-SNAP Protein. Mol. Plant-Microbe Interact. 2021, 34, 1433–1445. [Google Scholar] [CrossRef]
- Li, M.; Chen, R.; Jiang, Q.; Sun, X.; Zhang, H.; Hu, Z. GmNAC06, a NAC Domain Transcription Factor Enhances Salt Stress Tolerance in Soybean. Plant Mol. Biol. 2021, 105, 333–345. [Google Scholar] [CrossRef]
- Xiao, Y.; Karikari, B.; Wang, L.; Chang, F.; Zhao, T. Structure Characterization and Potential Role of Soybean Phospholipases A Multigene Family in Response to Multiple Abiotic Stress Uncovered by CRISPR/Cas9 Technology. Environ. Exp. Bot. 2021, 188, 104521. [Google Scholar] [CrossRef]
- Xu, H.; Li, Y.; Zhang, K.; Li, M.; Fu, S.; Tian, Y.; Qin, T.; Li, X.; Zhong, Y.; Liao, H. MiR169c-NFYA-C-ENOD40 Modulates Nitrogen Inhibitory Effects in Soybean Nodulation. New Phytol. 2021, 229, 3377–3392. [Google Scholar] [CrossRef] [PubMed]
- Niazian, M.; Sadat-Noori, S.A.; Tohidfar, M.; Galuszka, P.; Mortazavian, S.M.M. Agrobacterium-Mediated Genetic Transformation of Ajowan (Trachyspermum Ammi (L.) Sprague): An Important Industrial Medicinal Plant. Ind. Crops Prod. 2019, 132, 29–40. [Google Scholar] [CrossRef]



| Cultivar/Line | Agrobacterium rhizogenes strain | Targeted Gene(s) | Inoculation Technique | Explant Type | Supplementary Treatment | Massive Hairy Root Formation | Efficiency | Reference |
|---|---|---|---|---|---|---|---|---|
| Williams 82 | NCPPB2659 (K599) | GmFEI2, GmSHR | In vitro | Cotyledonary node with hypocotyl | Acetosyringone | 2 weeks | 0.6–0.95% | [28] |
| Jack | NCPPB2659 (K599) | Glyma01g38150, Glyma11g07220 | In vitro | Cotyledonary node | Acetosyringone | - | 95% | [103] |
| Bert | ARqual | GS1, Glyma.18g041100 | In vitro | Cotyledon | Vacuum infiltration | - | - | [104] |
| Williams 82 | NCPPB2659 (K599) | Glyma06g14180, Glyma08g02290, Glyma12g37050 | Ex vitro | Hypocotyls | - | - | - | [69] |
| Jack | NCPPB2659 (K599) | GmPDS11, GmPDS18 | In vitro | Cotyledons | Acetosyringone | 2 weeks | 11.7–48.1% | [92] |
| Hill | NCPPB2659 (K599) | Glyma.01G165800, Glyma.01g165800-D | Ex vitro | Cotyledonary node | - | - | - | [105] |
| Williams 82 | NCPPB2659 (K599) | Rfg1 | Ex vitro | Cotyledonary node | - | 2–3 weeks | [97] | |
| Williams 82 | NCPPB2659 (K599) | GmMYB118 | Ex vitro | Cotyledonary node | - | - | 50% | [106] |
| Williams 82 | NCPPB2659 (K599) | Glyma03g36470, Glyma14g04180, Glyma06g136900 | Ex vitro | - | - | - | 2.8–20.6% | [107] |
| - | NCPPB2659 (K599) | - | In vitro | Cotyledons | Gentle shaking | 3 weeks | - | [108] |
| Williams 82 | NCPPB2659 (K599) | GmLCLa1, GmLCLa2, GmLCLb1, GmLCLb2 | In vitro | Cotyledons with hypocotyls | - | - | - | [109] |
| Williams 82 | NCPPB2659 (K599) | GmAGO7a, GmAGO7b | Ex vitro | Cotyledonary node | - | 2–3 weeks | 80–100% | [73] |
| LD10-30110 | ARqua | Glyma.15G191200 | In vitro | Cotyledon | Acetosyringone, cysteine, sodium thiosulfate | - | - | [110] |
| Jack | NCPPB2659 (K599) | GmIPK1, GmIPK2 | In vitro | Cotyledonary node | Acetosyringone | - | 73.20% | [96] |
| Williams 82, Magellan, Zhonghuang13, Maverick | NCPPB2659 (K599) | GmNSF, GmSNAP | In vitro | Cotyledon | Acetosyringone, 6-BA, GA3 | 20 d | 69% | [34] |
| Fayette | NCPPB2659 (K599) | DELLA11, DELLA18 | In vitro | Cotyledons | - | - | - | [101] |
| Williams 82 | NCPPB2659 (K599) | GmROP6a/b, GmROP9a/b | Ex vitro | Hypocotyls | - | 2–3 weeks | 21–43% | [98] |
| Williams 82 | NCPPB2659 (K599) | miR156a, miR156c, miR156f, miR166a, miR167a, miR172a, miR172b, miR172c, miR172d, miR2118a, miR396a, miR396c, miR396e, miR397a, miR398a, miR399d, miR408a, FEI, NARK | Ex vitro | Hypocotyls | - | 2–3 weeks | - | [46] |
| Williams 82 | NSP1a, NSP1b | Ex vitro | Hypocotyls | - | - | - | [82] | |
| Williams 82 | NCPPB2659 (K599) | GmNAC06 | Ex vitro | Cotyledonary node | - | 4 weeks | - | [111] |
| Williams 82 | NCPPB2659 (K599) | Glyma.15G249000, Glyma.13G259100 | In vitro | Seed | - | 25 days | 45.3% | [80] |
| Williams 82 | GmUOX, GmXDH | Ex vitro | - | - | - | 54% | [99] | |
| Mustang | NCPPB2659 (K599) | GmNHX5 | In vitro | Cotyledonary node | MES + acetosyringone | 15 d | - | [35] |
| Tianlong 1 | NCPPB2659 (K599) | GmpPLA-IIε, GmpPLA-IIζ | - | - | - | - | - | [112] |
| Williams 82 | NCPPB2659 (K599) | GmNFYA-C, miR169c | In vitro | Cotyledonary node | Ammonium glufosinate | - | - | [113] |
| Tianlong 1, Suinong 10 | NCPPB2659 (K599) | GmDRR1 | Ex vitro | Cotyledonary node | - | - | - | [102] |
| Williams 82 | NCPPB2659 (K599) | GmSPL9d, miR156 | Ex vitro | Cotyledonary node | - | - | - | [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niazian, M.; Belzile, F.; Torkamaneh, D. CRISPR/Cas9 in Planta Hairy Root Transformation: A Powerful Platform for Functional Analysis of Root Traits in Soybean. Plants 2022, 11, 1044. https://doi.org/10.3390/plants11081044
Niazian M, Belzile F, Torkamaneh D. CRISPR/Cas9 in Planta Hairy Root Transformation: A Powerful Platform for Functional Analysis of Root Traits in Soybean. Plants. 2022; 11(8):1044. https://doi.org/10.3390/plants11081044
Chicago/Turabian StyleNiazian, Mohsen, François Belzile, and Davoud Torkamaneh. 2022. "CRISPR/Cas9 in Planta Hairy Root Transformation: A Powerful Platform for Functional Analysis of Root Traits in Soybean" Plants 11, no. 8: 1044. https://doi.org/10.3390/plants11081044
APA StyleNiazian, M., Belzile, F., & Torkamaneh, D. (2022). CRISPR/Cas9 in Planta Hairy Root Transformation: A Powerful Platform for Functional Analysis of Root Traits in Soybean. Plants, 11(8), 1044. https://doi.org/10.3390/plants11081044







