Non-Heading Chinese Cabbage Database: An Open-Access Platform for the Genomics of Brassica campestris (syn. Brassica rapa) ssp. chinensis
Abstract
:1. Introduction
2. Construction and Content
2.1. Genome Sequences and Annotation Resources
2.2. Transcription Factor Family Identification
2.3. Simple Sequence Repeat Finder and Primer Design
2.4. Syntenic Regions
2.5. Literature Collection
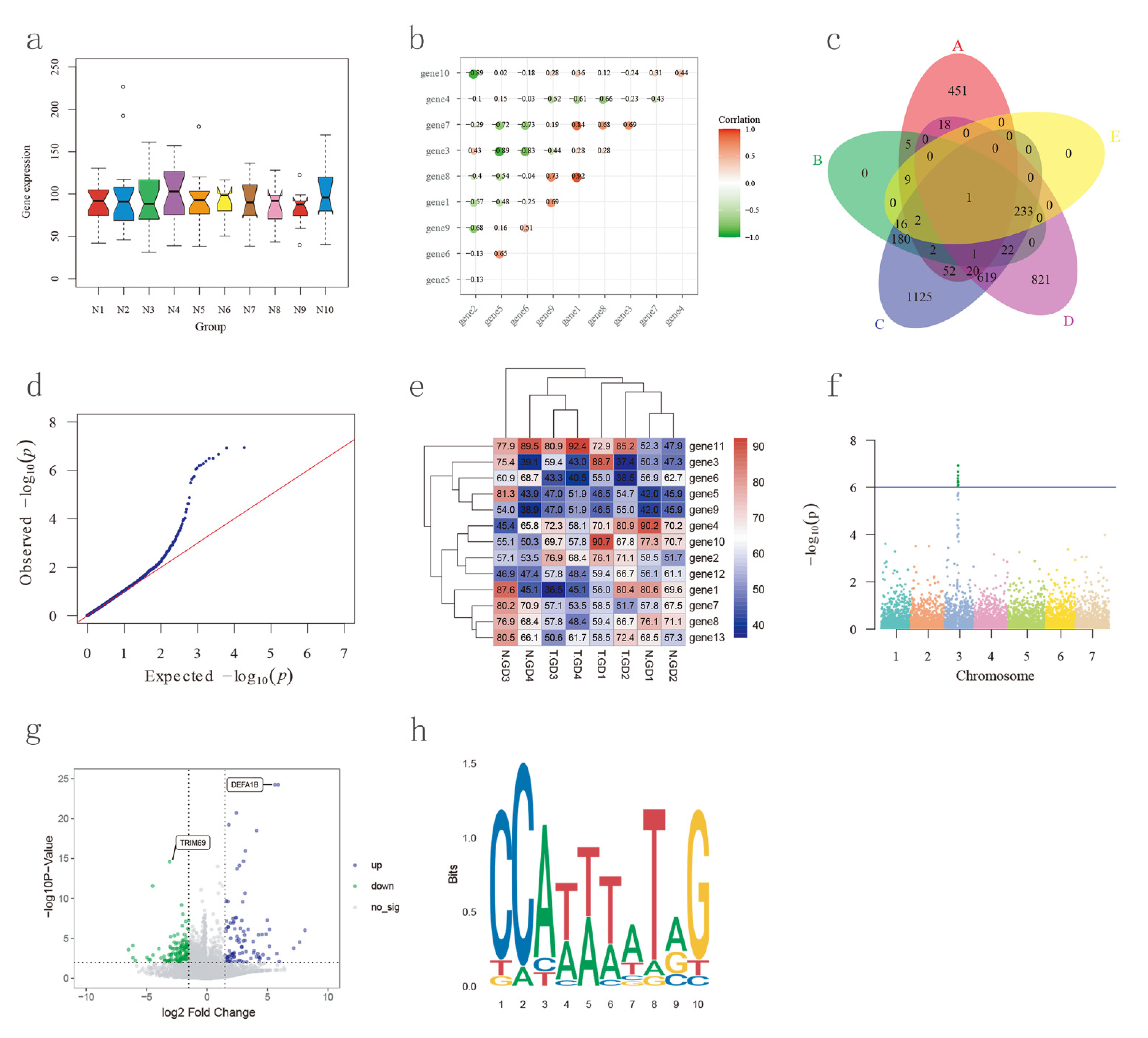
2.6. Biological Plot Platform Construction
2.7. Data Integration and Website Construction
3. Utility and Discussion
3.1. A Brief Introduction to the NHCCDB
3.2. Species Information
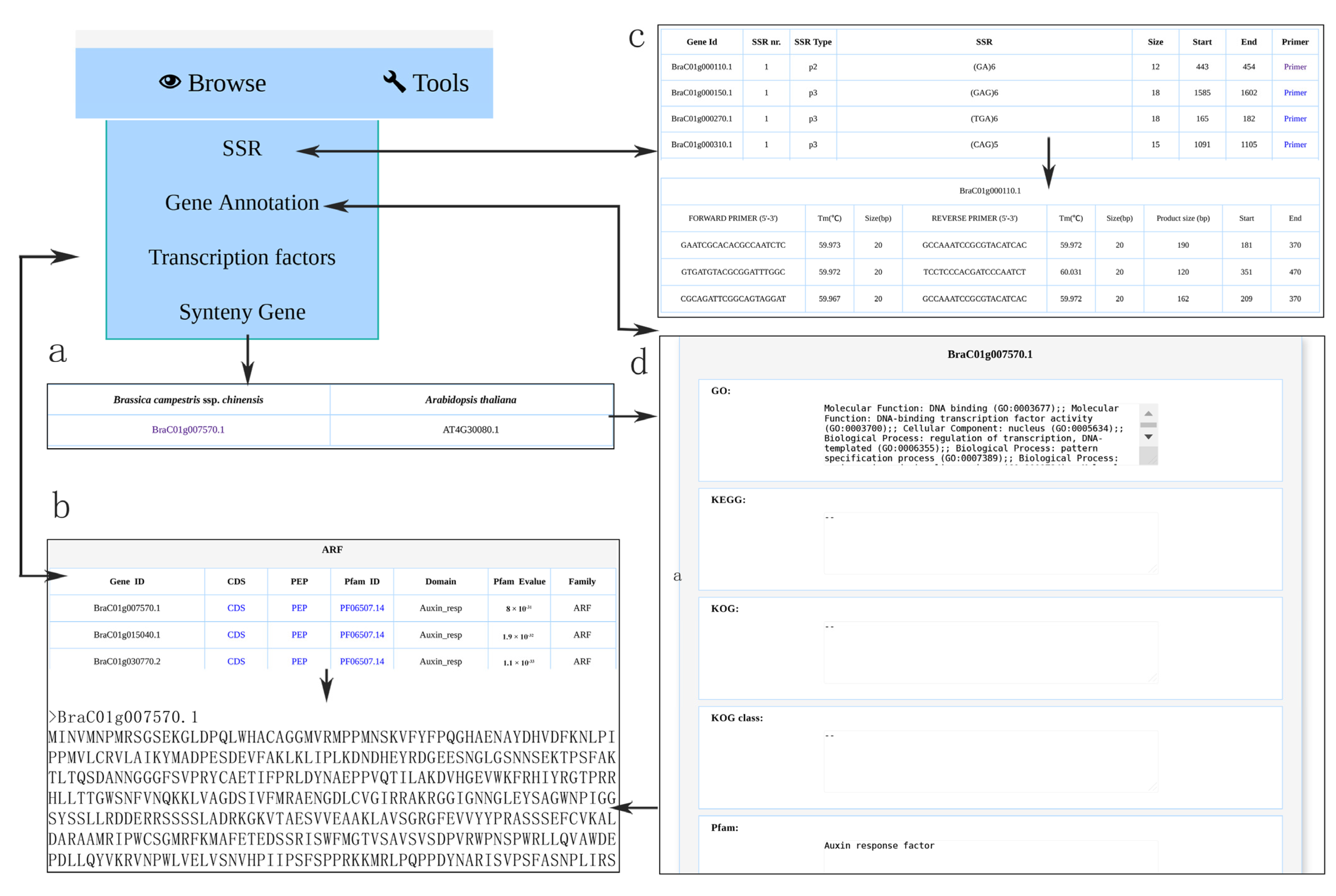
3.3. Browsing Gene Function Annotation and Simple Sequence Repeat
3.4. Browsing Syntenic Genes and Transcription Factor
3.5. Sequence Alignment Tools
3.6. Functional Genomics Tools
3.7. Primer Design and SSR Finder
3.8. JBrowse and Motif Analysis Tool
3.9. Biological Plot Platform
3.10. Search
3.11. Download
3.12. Submit
3.13. Home Module
4. Future Developments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagaharu, U. Genome analysis in Brassica carinata with special reference to the experimental formation of Brassica napus, a peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Qi, X.; An, H.; Ragsdale, A.P.; Hall, T.E.; Gutenkunst, R.N.; Chris Pires, J.; Barker, M.S. Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol. Ecol. 2017, 26, 3373–3388. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, X.; Chang, L.; Zhang, T.; Chen, H.; Zhang, L.; Lin, R.; Liang, J.; Wu, J.; Freeling, M.; Wang, X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.-C.; Falentin, C.; Genete, M.; Berrabah, W.; Chèvre, A.-M.; Delourme, R.; et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 2018, 4, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, G.; Ma, L.; Liu, T.; Zhang, C.; Xiao, D.; Zheng, H.; Chen, F.; Hou, X. A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic. Res. 2020, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Su, T.; Zhao, X.; Wang, W.; Zhang, D.; Yu, Y.; Bayer, P.E.; Edwards, D.; Yu, S.; Zhang, F. Assembly of the non-heading pak choi genome and comparison with the genomes of heading Chinese cabbage and the oilseed yellow sarson. Plant Biotechnol. J. 2021, 19, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Nie, F.; Chen, W.; Ma, X.; Gong, K.; Yang, Q.; Wang, J.; Li, N.; Sun, P.; Pei, Q.; et al. Coriander Genomics Database: A genomic, transcriptomic, and metabolic database for coriander. Hortic. Res. 2020, 7, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M.J.B. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorini, N.; Lipman, D.J.; Lu, Z. Towards PubMed 2.0. eLife 2017, 6, e28801. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinner, M.E.; Uzilov, A.V.; Stein, L.D.; Mungall, C.J.; Holmes, I.H. JBrowse: A next-generation genome browser. Genome Res. 2009, 19, 1630–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Li, Y.; Liu, T.; Zhang, C.; Xiao, D.; Hou, X. Non-Heading Chinese Cabbage Database: An Open-Access Platform for the Genomics of Brassica campestris (syn. Brassica rapa) ssp. chinensis. Plants 2022, 11, 1005. https://doi.org/10.3390/plants11081005
Li Z, Li Y, Liu T, Zhang C, Xiao D, Hou X. Non-Heading Chinese Cabbage Database: An Open-Access Platform for the Genomics of Brassica campestris (syn. Brassica rapa) ssp. chinensis. Plants. 2022; 11(8):1005. https://doi.org/10.3390/plants11081005
Chicago/Turabian StyleLi, Zhidong, Ying Li, Tongkun Liu, Changwei Zhang, Dong Xiao, and Xilin Hou. 2022. "Non-Heading Chinese Cabbage Database: An Open-Access Platform for the Genomics of Brassica campestris (syn. Brassica rapa) ssp. chinensis" Plants 11, no. 8: 1005. https://doi.org/10.3390/plants11081005
APA StyleLi, Z., Li, Y., Liu, T., Zhang, C., Xiao, D., & Hou, X. (2022). Non-Heading Chinese Cabbage Database: An Open-Access Platform for the Genomics of Brassica campestris (syn. Brassica rapa) ssp. chinensis. Plants, 11(8), 1005. https://doi.org/10.3390/plants11081005







