The Syngameon Enigma
Abstract
:Nearly all organisms, met with in nature as well as under cultivation, man included, are hybrids which were mistakenly considered to be specifically pure, so that their behaviour was unconsciously held to be that of specifically pure organisms, while it was that of hybrids; so it happened that segregation was mistaken for heredity.—Lotsy 1916
1. Introduction
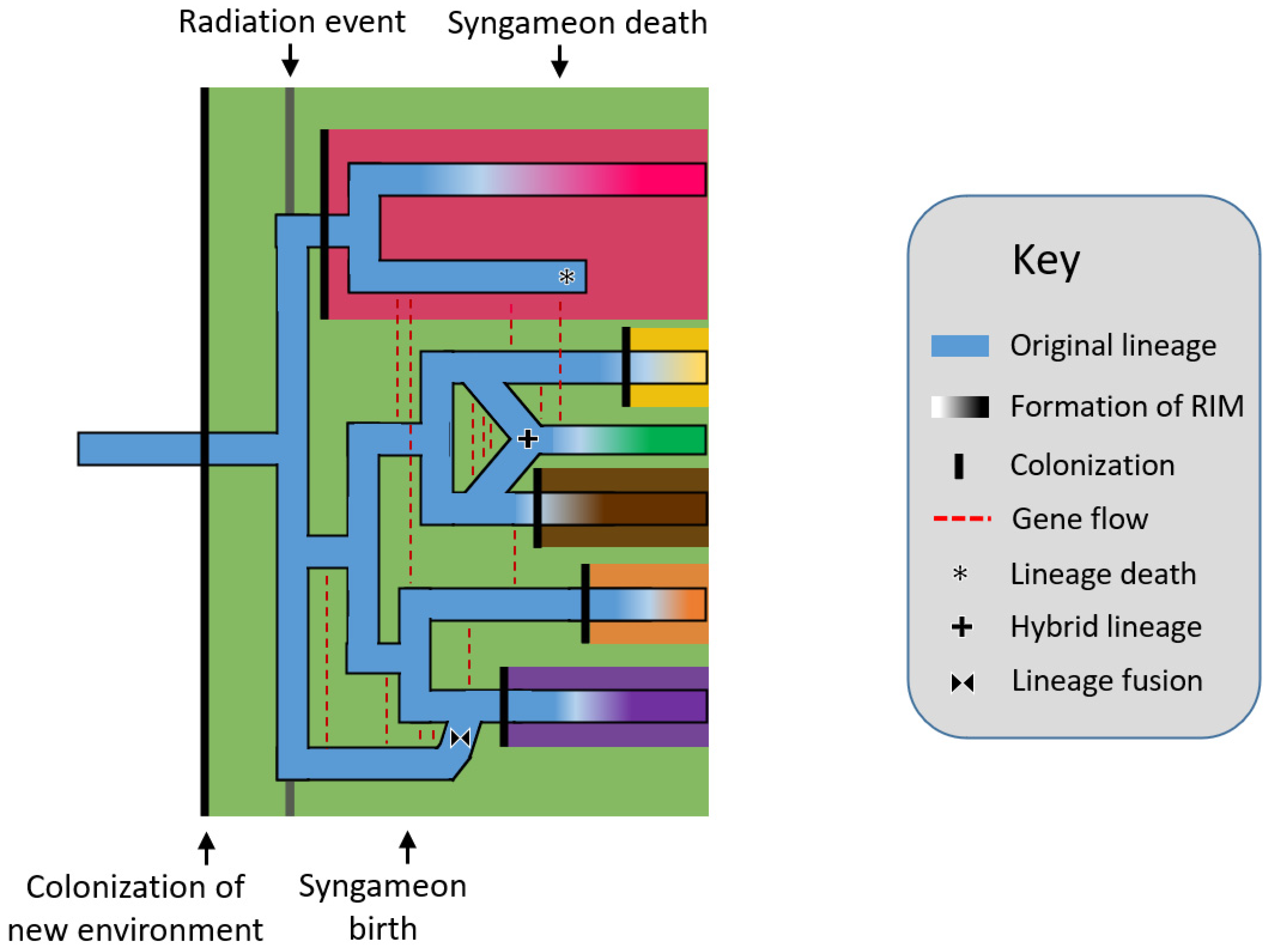
2. How Do Syngameons Form and Collapse?
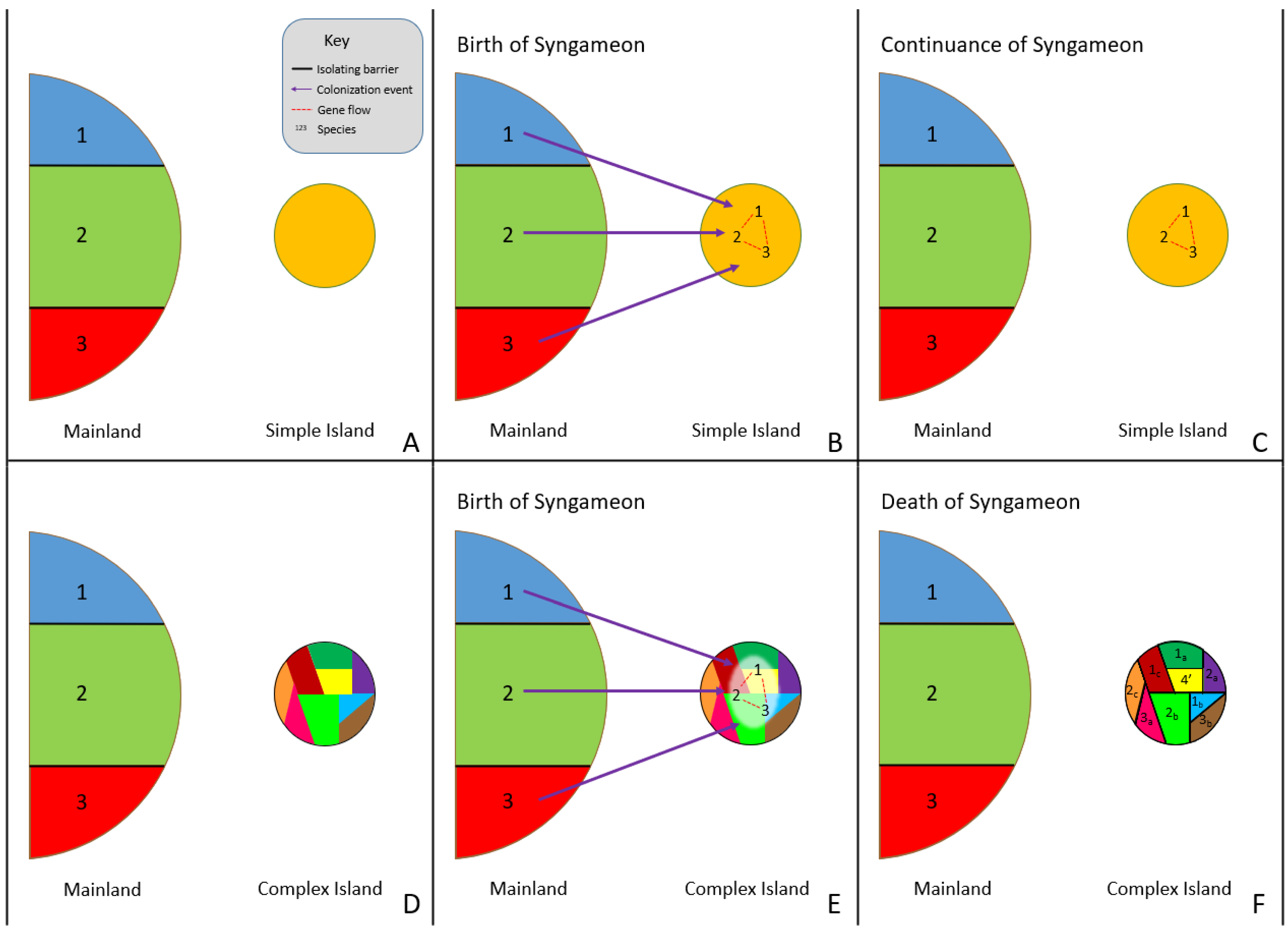
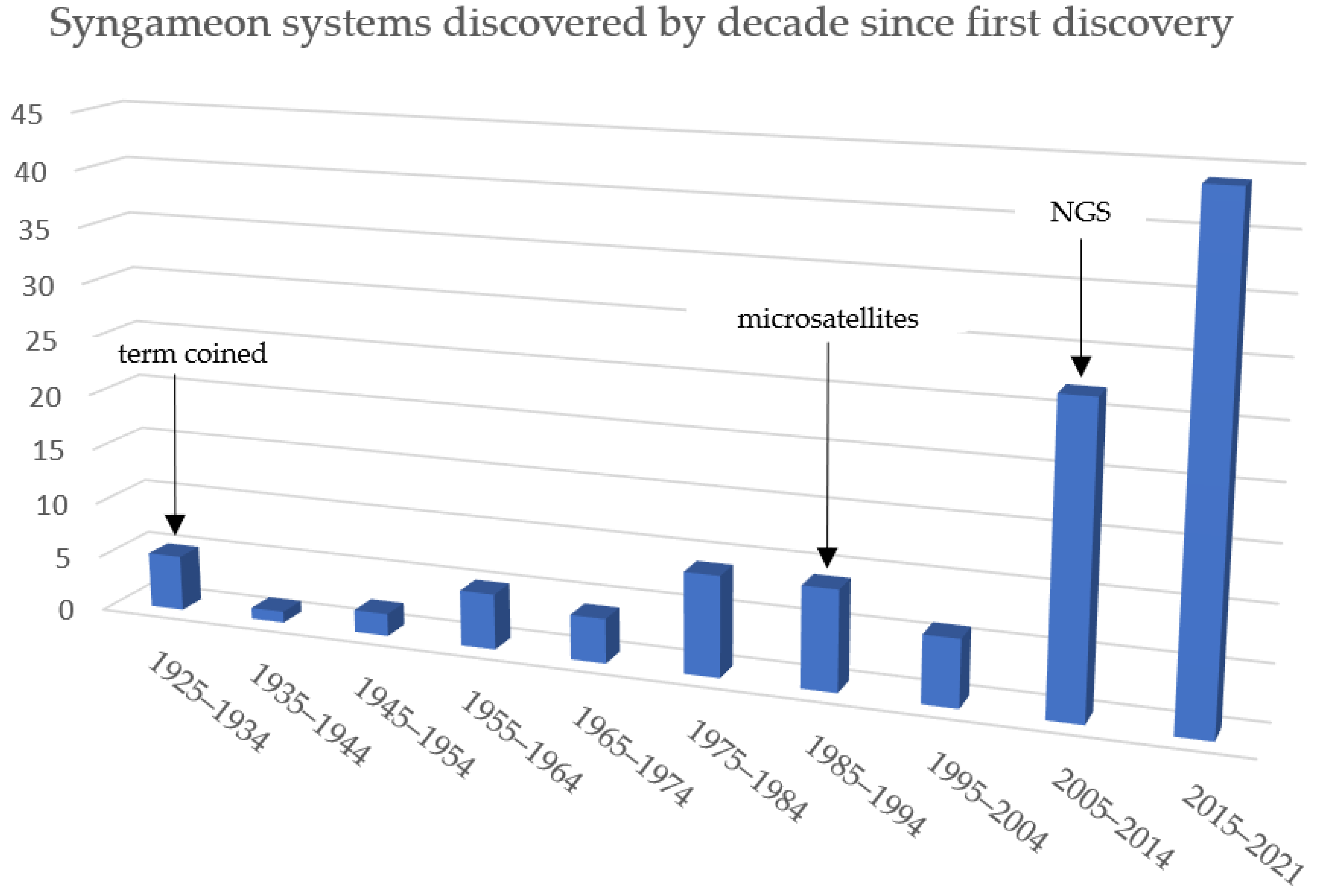
2.1. The Origin of Syngameons
2.2. The Birth and Death Hypotheses
2.2.1. The Rapid Radiation Hypothesis
2.2.2. Surfing Syngameon Hypothesis
2.2.3. Edge Range Hypothesis
2.2.4. Genomic Mutualist Hypothesis
2.3. Spatial Limitations
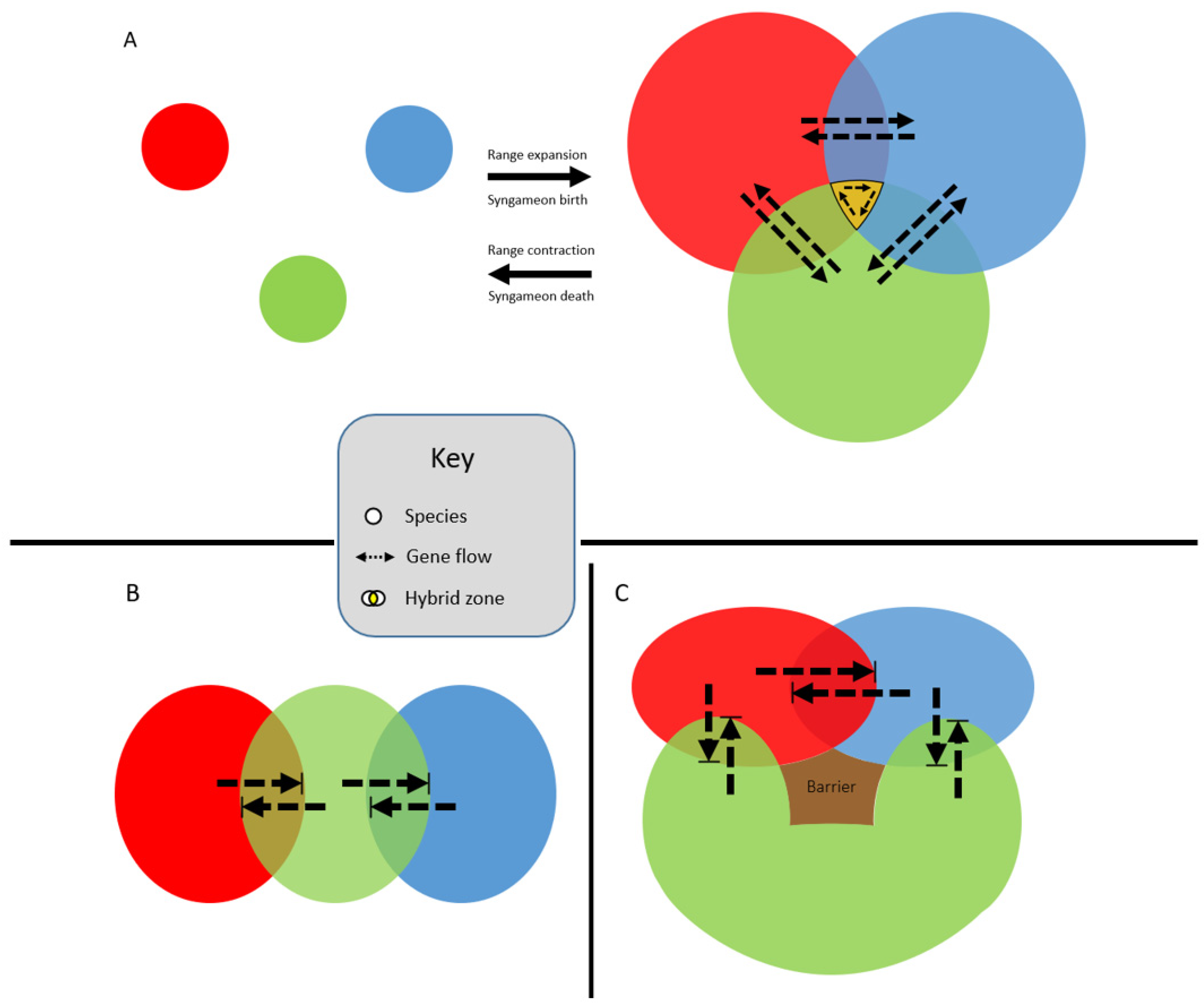
3. How Are Syngameons Maintained over Evolutionary Time?
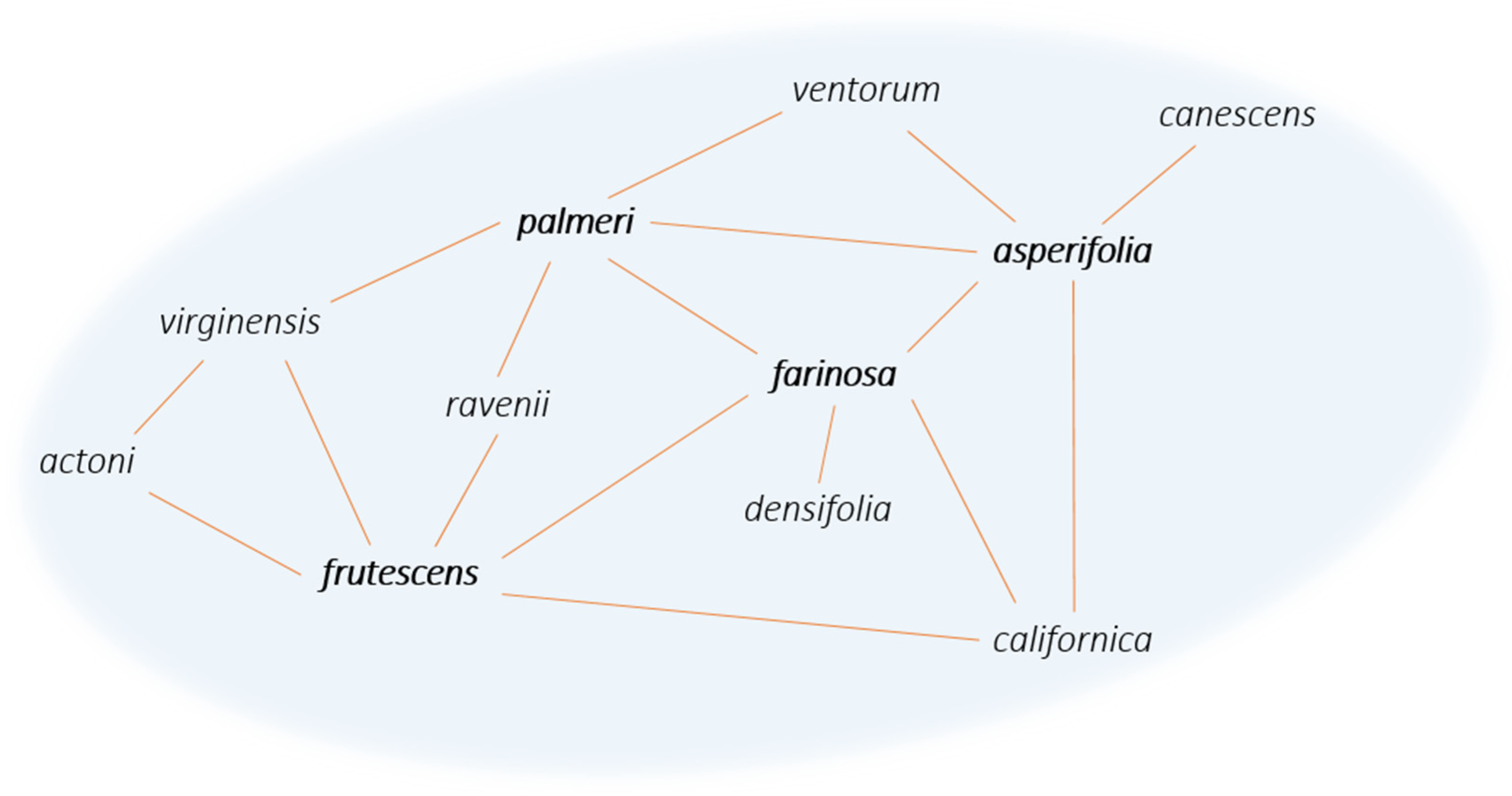
4. Why Are Syngameons So Rare?
5. The Conservation and Future of Syngameons
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Glossary
References
- Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005, 20, 229–237. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009, 60, 561–588. [Google Scholar] [CrossRef] [Green Version]
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [Green Version]
- Lotsy, J. Species or linneon. Genetica 1925, 7, 487–506. [Google Scholar] [CrossRef]
- Du Rietz, G.E. The Fundamental Units of Biological Taxonomy; Svensk Botaniska Foreningen: Uppsala, Sweden, 1930. [Google Scholar]
- Cuénot, L. L’Evolution Biologique: Les Faits, Les Incertitudes; Masson et Cie: Paris, France, 1951; p. 594. [Google Scholar]
- Grant, V. The plant species in theory and practice. In The Species Problem; Mayr, E., Ed.; American Association for the Advancement of Science: Washington, DC, USA, 1957; pp. 39–80. [Google Scholar]
- Beaudry, J.R. The species concept: Its evolution and present status. Rev. Can. Biol. 1960, 19, 219–240. [Google Scholar]
- Van Valen, L. Ecological species, multispecies, and oaks. Taxon 1976, 25, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Templeton, A.R. The meaning of species and speciation: A genetic perspective. In Speciation and Its Consequences; Sinauer Associates: Sunderland, MA, USA, 1989; pp. 3–27. [Google Scholar]
- Arduino, P.; Verra, F.; Cianchi, R.; Rossi, W.; Corrias, B.; Bullini, L. Genetic variation and natural hybridization between Orchis laxiflora and Orchis palustris (Orchidaceae). Plant Syst. Evol. 1996, 202, 87–109. [Google Scholar] [CrossRef]
- Holliday, T.W. Neanderthals and modern humans: An example of a mammalian syngameon? In Neanderthals Revisited: New Approaches and Perspectives; Springer: Berlin/Heidelberg, Germany, 2006; pp. 281–297. [Google Scholar]
- Barker, M.J. The empirical inadequacy of species cohesion by gene flow. Philos. Sci. 2007, 74, 654–665. [Google Scholar] [CrossRef]
- Caujapé-Castells, J.; García-Verdugo, C.; Marrero-Rodríguez, Á.; Fernández-Palacios, J.M.; Crawford, D.J.; Mort, M.E. Island ontogenies, syngameons, and the origins and evolution of genetic diversity in the Canarian endemic flora. Perspect. Plant Ecol. Evol. Syst. 2017, 27, 9–22. [Google Scholar] [CrossRef]
- Boecklen, W.J. Topology of syngameons. Ecol. Evol. 2017, 7, 10486–10491. [Google Scholar] [CrossRef] [Green Version]
- Cannon, C.H.; Petit, R.J. The oak syngameon: More than the sum of its parts. New Phytol. 2020, 226, 978–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turesson, G. The species and the variety as ecological units. Hereditas 1922, 3, 100–113. [Google Scholar] [CrossRef]
- Cannon, C.H.; Lerdau, M. Variable mating behaviors and the maintenance of tropical biodiversity. Front. Genet. 2015, 6, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bog, M. Hybridisation and Its Consequences: Population Genomics, Herbivory, and Phytochemistry in the Senecio nemorensis Syngameon. Ph.D. Thesis, Universität Regensburg, Regensburg, Germany, September 2016. [Google Scholar]
- Cannon, C.H.; Scher, C.L. Exploring the potential of gametic reconstruction of parental genotypes by F1 hybrids as a bridge for rapid introgression. Genome 2017, 60, 713–719. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, D.; Zhang, Q.; Song, C.; Zhong, C.; Zhang, X.; Wang, Y.; Yao, X.; Wang, Z.; Zeng, S. Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 2017, 215, 877–890. [Google Scholar] [CrossRef] [Green Version]
- Meier, J.I.; Marques, D.A.; Mwaiko, S.; Wagner, C.E.; Excoffier, L.; Seehausen, O. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 2017, 8, 14363. [Google Scholar] [CrossRef] [Green Version]
- Cronk, Q.; Suarez-Gonzalez, A. The role of interspecific hybridization in adaptive potential at range margins. Mol. Ecol. 2018, 27, 4653–4656. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Gonzalez, A.; Lexer, C.; Cronk, Q.C. Adaptive introgression: A plant perspective. Biol. Lett. 2018, 14, 20170688. [Google Scholar] [CrossRef]
- Cannon, C.H.; Lerdau, M.T. Demography and destiny: The syngameon in hyperdiverse systems. Proc. Natl. Acad. Sci. USA 2019, 116, 8105. [Google Scholar] [CrossRef] [Green Version]
- Levi, T.; Barfield, M.; Holt, R.D.; Terborgh, J. Reply to Cannon and Lerdau: Maintenance of tropical forest tree diversity. Proc. Natl. Acad. Sci. USA 2019, 116, 8106. [Google Scholar] [CrossRef] [Green Version]
- Van Oppen, M.J.; McDonald, B.J.; Willis, B.; Miller, D.J. The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: Reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol. 2001, 18, 1315–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladner, J.T. Hidden Diversity in Corals and Their Endosymbionts. Ph.D. Thesis, Stanford University, Stanford, CA, USA, March 2012. [Google Scholar]
- Ottenburghs, J.; van Hooft, P.; van Wieren, S.E.; Ydenberg, R.C.; Prins, H.H. Hybridization in geese: A review. Front. Zool. 2016, 13, 20. [Google Scholar] [CrossRef] [Green Version]
- Larsen, P.A.; Marchán-Rivadeneira, M.R.; Baker, R.J. Natural hybridization generates mammalian lineage with species characteristics. Proc. Natl. Acad. Sci. USA 2010, 107, 11447–11452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ackermann, R.R.; Arnold, M.L.; Baiz, M.D.; Cahill, J.A.; Cortés-Ortiz, L.; Evans, B.J.; Grant, B.R.; Grant, P.R.; Hallgrimsson, B.; Humphreys, R.A. Hybridization in human evolution: Insights from other organisms. Evol. Anthropol. Issues News Rev. 2019, 28, 189–209. [Google Scholar] [CrossRef] [Green Version]
- Wilson, P.J.; Grewal, S.K.; Mallory, F.F.; White, B.N. Genetic characterization of hybrid wolves across Ontario. J. Hered. 2009, 100 (Suppl. 1), S80–S89. [Google Scholar] [CrossRef] [Green Version]
- Rutledge, L.; Garroway, C.; Loveless, K.; Patterson, B. Genetic differentiation of eastern wolves in Algonquin Park despite bridging gene flow between coyotes and grey wolves. Heredity 2010, 105, 520–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, K.; Sota, T. Hybridization and speciation in the carabid beetles of the subgenus Ohomopterus (Coleoptera, Carabidae, genus Carabus). Res. Popul. Ecol. 1998, 40, 213–222. [Google Scholar] [CrossRef]
- Dowling, T.E.; Markle, D.F.; Tranah, G.J.; Carson, E.W.; Wagman, D.W.; May, B.P. Introgressive hybridization and the evolution of lake-adapted catostomid fishes. PLoS ONE 2016, 11, e0149884. [Google Scholar] [CrossRef] [Green Version]
- Gould, S.J.; Woodruff, D.S. History as a cause of area effects: An illustration from Cerion on Great Inagua, Bahamas. Biol. J. Linn. Soc. 1990, 40, 67–98. [Google Scholar] [CrossRef]
- Wheat, C.W.; Watt, W.B. A mitochondrial-DNA-based phylogeny for some evolutionary-genetic model species of Colias butterflies (Lepidoptera, Pieridae). Mol. Phylogenet. Evol. 2008, 47, 893–902. [Google Scholar] [CrossRef]
- Colbourne, J.; Wilson, C.; Hebert, P. The systematics of Australian Daphnia and Daphniopsis (Crustacea: Cladocera): A shared phylogenetic history transformed by habitat-specific rates of evolution. Biol. J. Linn. Soc. 2006, 89, 469–488. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.; Wilson, C.C. Provincialism in plankton: Endemism and allopatric speciation in Australian Daphnia. Evolution 1994, 48, 1333–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyron, R.A.; O’Connell, K.A.; Lemmon, E.M.; Lemmon, A.R.; Beamer, D.A. Phylogenomic data reveal reticulation and incongruence among mitochondrial candidate species in Dusky Salamanders (Desmognathus). Mol. Phylogenet. Evol. 2020, 146, 106751. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.D.; Orozco-ter Wengel, P.; Kreissl, M.; Schlötterer, C. Multiple hybridization events between Drosophila simulans and Drosophila mauritiana are supported by mtDNA introgression. Mol. Ecol. 2010, 19, 4695–4707. [Google Scholar] [CrossRef] [Green Version]
- Matute, D.R.; Ayroles, J.F. Hybridization occurs between Drosophila simulans and D. sechellia in the Seychelles archipelago. J. Evol. Biol. 2014, 27, 1057–1068. [Google Scholar] [CrossRef]
- Mallet, J.; Beltrán, M.; Neukirchen, W.; Linares, M. Natural hybridization in heliconiine butterflies: The species boundary as a continuum. BMC Evol. Biol. 2007, 7, 28. [Google Scholar] [CrossRef] [Green Version]
- Grant, B.R.; Grant, P.R. Evolutionary Dynamics of a Natural Population: The Large Cactus Finch of the Galápagos; University of Chicago Press: Chicago, IL, USA, 1989. [Google Scholar]
- Grant, P.R.; Grant, B.R. Hybridization of bird species. Science 1992, 256, 193–197. [Google Scholar] [CrossRef]
- Grant, P.R.; Grant, B.R. Triad hybridization via a conduit species. Proc. Natl. Acad. Sci. USA 2020, 117, 7888–7896. [Google Scholar] [CrossRef]
- Leduc-Robert, G.; Maddison, W.P. Phylogeny with introgression in Habronattus jumping spiders (Araneae: Salticidae). BMC Evol. Biol. 2018, 18, 24. [Google Scholar] [CrossRef] [Green Version]
- Hammer, M.F.; Woerner, A.E.; Mendez, F.L.; Watkins, J.C.; Wall, J.D. Genetic evidence for archaic admixture in Africa. Proc. Natl. Acad. Sci. USA 2011, 108, 15123–15128. [Google Scholar] [CrossRef] [Green Version]
- Schliewen, U.K.; Klee, B. Reticulate sympatric speciation in Cameroonian crater lake cichlids. Front. Zool. 2004, 1, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olave, M.; Avila, L.J.; Sites, J.W., Jr.; Morando, M. Hybridization could be a common phenomenon within the highly diverse lizard genus Liolaemus. J. Evol. Biol. 2018, 31, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Budd, A.F. Tracing the long-term evolution of a species complex: Examples from the Montastraea “annularis” complex. Palaeoworld 2010, 19, 348–356. [Google Scholar] [CrossRef]
- Fukami, H.; Budd, A.F.; Levitan, D.R.; Jara, J.; Kersanach, R.; Knowlton, N. Geographic differences in species boundaries among members of the Montastraea annularis complex based on molecular and morphological markers. Evolution 2004, 58, 324–337. [Google Scholar] [CrossRef]
- Granados Cifuentes, C.A. Interspecific and Intragenomic Variation of the Internal Transcribed Spacer2 (ITS2, rDNA) and Their Consequence in the Evolution of Eastern Pacific Octocorals. Master’s Thesis, Universidad de los Andes, Bogota, Colombia, 2008. [Google Scholar]
- Manrique Rodríguez, N.A. Phylogenetic Reconstruction of the Genus Pacifigorgia (Gorgoniidae: Octocorallia) Using the Internal Transcribed Spacer ITS2. Master’s Thesis, Universidad de los Andes, Bogota, Colombia, May 2008. [Google Scholar]
- Godfrey, L.; Marks, J. The nature and origins of primate species. Am. J. Phys. Anthropol. 1991, 34, 39–68. [Google Scholar] [CrossRef]
- Stefani, F.; Benzoni, F.; Pichon, M.; Cancelliere, C.; Galli, P. A multidisciplinary approach to the definition of species boundaries in branching species of the coral genus Psammocora (Cnidaria, Scleractinia). Zool. Scr. 2008, 37, 71–91. [Google Scholar] [CrossRef]
- Woodruff, D.S. Towards a genodynamics of hybrid zones: Studies of Australian frogs and West Indian land snails. In Evolution and Speciation: Essays in Honor of M.J.D. White; Atchley, W.R., Woodruff, D.S., Eds.; Cambridge University Press: Cambridge, UK, 1981; pp. 171–197. [Google Scholar]
- Schwarzer, J.; Misof, B.; Schliewen, U. Speciation within genomic networks: A case study based on Steatocranus cichlids of the lower Congo rapids. J. Evol. Biol. 2012, 25, 138–148. [Google Scholar] [CrossRef]
- Arrigoni, R.; Benzoni, F.; Terraneo, T.I.; Caragnano, A.; Berumen, M.L. Recent origin and semi-permeable species boundaries in the scleractinian coral genus Stylophora from the Red Sea. Sci. Rep. 2016, 6, 34612. [Google Scholar] [CrossRef] [Green Version]
- Frantz, L.A.; Schraiber, J.G.; Madsen, O.; Megens, H.-J.; Bosse, M.; Paudel, Y.; Semiadi, G.; Meijaard, E.; Li, N.; Crooijmans, R.P. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol. 2013, 14, R107. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Lammers, F.; Bidon, T.; Pfenninger, M.; Kolter, L.; Nilsson, M.A.; Janke, A. The evolutionary history of bears is characterized by gene flow across species. Sci. Rep. 2017, 7, 46487. [Google Scholar] [CrossRef] [Green Version]
- Cui, R.; Schumer, M.; Kruesi, K.; Walter, R.; Andolfatto, P.; Rosenthal, G.G. Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution 2013, 67, 2166–2179. [Google Scholar] [CrossRef] [PubMed]
- Cinget, B.; de Lafontaine, G.; Gérardi, S.; Bousquet, J. Integrating phylogeography and paleoecology to investigate the origin and dynamics of hybrid zones: Insights from two widespread North American firs. Mol. Ecol. 2015, 24, 2856–2870. [Google Scholar] [CrossRef] [PubMed]
- Beddows, I.; Rose, L.E. Factors determining hybridization rate in plants: A case study in Michigan. Perspect. Plant Ecol. Evol. Syst. 2018, 34, 51–60. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Huang, H. Genetic variation and natural hybridization among sympatric Actinidia species and the implications for introgression breeding of kiwifruit. Tree Genet. Genomes 2010, 6, 801–813. [Google Scholar] [CrossRef]
- dePamphilis, C.W.; Wyatt, R. Hybridization and introgression in buckeyes (Aesculus: Hippocastanaceae): A review of the evidence and a hypothesis to explain long-distance gene flow. Syst. Bot. 1989, 14, 593–611. [Google Scholar] [CrossRef]
- Nielsen, E.L. A taxonomic study of the genus Amelanchier in Minnesota. Am. Midl. Nat. 1939, 22, 160–206. [Google Scholar] [CrossRef]
- Grant, V. Plant Speciation; Columbia University Press: New York, NY, USA, 1971; p. 435. [Google Scholar]
- McElwee-Adame, A.; San Diego State University, San Diego, CA, USA. Personal communication, 2020.
- Gottlieb, L.D. Hybridization between Arctostaphylos viscida and A. canescens in Oregon. Brittonia 1968, 20, 83–93. [Google Scholar] [CrossRef]
- Schmid, R.; Mallory, T.E.; Tucker, J.M. Biosystematic evidence for hybridization between Arctostaphylos nissenana and A. viscida. Brittonia 1968, 20, 34–43. [Google Scholar] [CrossRef]
- Brownsey, P.J. A taxonomic revision of the New Zealand species of Asplenium. N. Z. J. Bot. 1977, 15, 39–86. [Google Scholar] [CrossRef] [Green Version]
- Barnes, B.V.; Dancik, B.P. Characteristics and origin of a new birch species, Betula murrayana, from southeastern Michigan. Can. J. Bot. 1985, 63, 223–226. [Google Scholar] [CrossRef]
- Walters, R.; Yawney, H. Silvics Manual: Volume 2: Hardwoods; United States Department of Agriculture, Forest Service: Washington, DC, USA, 2004.
- Alexander, P.J.; Windham, M.D.; Beck, J.B.; Al-Shehbaz, I.A.; Allphin, L.; Bailey, C.D. Weaving a tangled web: Divergent and reticulate speciation in Boechera fendleri sensu lato (Brassicaceae: Boechereae). Syst. Bot. 2015, 40, 572–596. [Google Scholar] [CrossRef]
- Hedrén, M. Species delimitation and the partitioning of genetic diversity—An example from the Carex flava complex (Cyperaceae). Biodivers. Conserv. 2004, 13, 293–316. [Google Scholar] [CrossRef]
- Chen, S.-C.; Cannon, C.H.; Kua, C.-S.; Liu, J.-J.; Galbraith, D.W. Genome size variation in the Fagaceae and its implications for trees. Tree Genet. Genomes 2014, 10, 977–988. [Google Scholar] [CrossRef]
- Grant, V. Plant Speciation, 2nd ed.; Columbia University Press: New York, NY, USA, 1981; p. 564. [Google Scholar]
- Bureš, P.; Šmarda, P.; Rotreklova, O.; Oberreiter, M.; Burešová, M.; Konečný, J.; Knoll, A.; Fajmon, K.; Šmerda, J. Pollen viability and natural hybridization of Central European species of Cirsium. Preslia 2010, 82, 391–422. [Google Scholar]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butelli, E.; Licciardello, C.; Ramadugu, C.; Durand-Hulak, M.; Celant, A.; Recupero, G.R.; Froelicher, Y.; Martin, C. Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Curr. Biol. 2019, 29, 158–164. [Google Scholar] [CrossRef] [Green Version]
- Papadopulos, A.; Price, Z.; Devaux, C.; Hipperson, H.; Smadja, C.; Hutton, I.; Baker, W.; Butlin, R.; Savolainen, V. A comparative analysis of the mechanisms underlying speciation on Lord Howe Island. J. Evol. Biol. 2013, 26, 733–745. [Google Scholar] [CrossRef]
- Wagner, N.D.; Clements, M.A.; Simpson, L.; Nargar, K. Conservation in the face of hybridisation: Genome-wide study to evaluate taxonomic delimitation and conservation status of a threatened orchid species. Conserv. Genet. 2021, 22, 151–168. [Google Scholar] [CrossRef]
- Marcks, B. Preliminary reports on the flora of Wisconsin. 66. Cyperaceae II—Sedge family II. the genus Cyperus: The umbrella sedges. Trans. Wis. Acad. Sci. Arts Lett. 1974, 62, 261–284. [Google Scholar]
- Voss, E.; Reznicek, A.A. Field Manual of Michigan Flora; University of Michigan Press: Ann Arbor, MI, USA, 2012. [Google Scholar]
- Linan, A.G.; Lowry, P.P.; Miller, A.J.; Schatz, G.E.; Sevathian, J.C.; Edwards, C.E. RAD-sequencing reveals patterns of diversification and hybridization, and the accumulation of reproductive isolation in a clade of partially sympatric, tropical island trees. Mol. Ecol. 2021, 30, 4520–4537. [Google Scholar] [CrossRef]
- Beeks, R.M. Variation and hybridization in southern California populations of Diplacus (Scrophulariaceae). Aliso J. Syst. Florist. Bot. 1962, 5, 83–122. [Google Scholar] [CrossRef] [Green Version]
- Wood, C.E. Evidence for the hybrid origin of Drosera anglica. Rhodora 1955, 57, 105–130. [Google Scholar]
- Crowder, A.; Pearson, M.; Grubb, P.; Langlois, P. Drosera L. J. Ecol. 1990, 78, 233–267. [Google Scholar] [CrossRef]
- Rünk, K.; Zobel, M.; Zobel, K. Biological Flora of the British Isles: Dryopteris carthusiana, D. dilatata and D. expansa. J. Ecol. 2012, 100, 1039–1063. [Google Scholar] [CrossRef]
- Carr, G.D. Hybridization of the Hawaiian Silversword complex. In Proceedings of the Second Conference in Natural Sciences, Honolulu, HI, USA, 1–3 June 1978. [Google Scholar]
- Singhal, S.; Roddy, A.B.; DiVittorio, C.; Sanchez-Amaya, A.; Henriquez, C.L.; Brodersen, C.R.; Fehlberg, S.; Zapata, F. Diversification, disparification and hybridization in the desert shrubs Encelia. New Phytol. 2021, 230, 1228–1241. [Google Scholar] [CrossRef]
- Schmitt, S.; Hérault, B.; Ducouret, É.; Baranger, A.; Tysklind, N.; Heuertz, M.; Marcon, É.; Cazal, S.O.; Derroire, G. Topography consistently drives intra-and inter-specific leaf trait variation within tree species complexes in a Neotropical forest. Oikos 2020, 129, 1521–1530. [Google Scholar] [CrossRef]
- Pineda Torres, Y.M. The Nature of Espeletia Species. Master’s Thesis, Universidad de Los Andes, Bogota, Colombia, August 2019. [Google Scholar]
- Rutherford, S.; Rossetto, M.; Bragg, J.G.; McPherson, H.; Benson, D.; Bonser, S.P.; Wilson, P.G. Speciation in the presence of gene flow: Population genomics of closely related and diverging Eucalyptus species. Heredity 2018, 121, 126–141. [Google Scholar] [CrossRef]
- Flores-Rentería, L.; Rymer, P.D.; Riegler, M. Unpacking boxes: Integration of molecular, morphological and ecological approaches reveals extensive patterns of reticulate evolution in box eucalypts. Mol. Phylogenet. Evol. 2017, 108, 70–87. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, X.; Herre, E.A.; McKey, D.; Machado, C.A.; Yu, W.-B.; Cannon, C.H.; Arnold, M.L.; Pereira, R.A.; Ming, R. Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig-wasp pollination mutualism. Nat. Commun. 2021, 12, 718. [Google Scholar] [CrossRef]
- Pryer, K.M.; Haufler, C.H. Isozymic and chromosomal evidence for the allotetraploid origin of Gymnocarpium dryopteris (Dryopteridaceae). Syst. Bot. 1993, 18, 150–172. [Google Scholar] [CrossRef]
- Heiser, C. Study in the evolution of the sunflower species Helianthus annuus and H. bolanderi. Univ. Calif. Publ. Bot. 1949, 23, 157–208. [Google Scholar]
- Heiser, C.B., Jr. Hybridization in the annual sunflowers: Helianthus annuus × H. debilis var. cucumerifolius. Evolution 1951, 5, 42–51. [Google Scholar] [CrossRef]
- Heiser, C.B., Jr. Hybridization in the annual sunflowers: Helianthus annuus × H. argophyllus. Am. Nat. 1951, 85, 65–72. [Google Scholar] [CrossRef]
- Heiser, C.B., Jr. Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution 1947, 1, 249–262. [Google Scholar] [CrossRef]
- Rieseberg, L.H. Hybridization in rare plants: Insights from case studies in Cercocarpus and Helianthus. In Genetics and Conservation of Rare Plants; Falk, D.A., Holsinger, K.E., Eds.; Oxford University Press: Oxford, UK, 1991; pp. 171–181. [Google Scholar]
- Lenz, L.W. Hybridization and speciation in the Pacific Coast irises. Aliso J. Syst. Florist. Bot. 1959, 4, 237–309. [Google Scholar] [CrossRef] [Green Version]
- Young, N.D. Pacific Coast Iris species delimitation using three species definitions: Biological, phylogenetic and genealogical. Biol. J. Linn. Soc. 1998, 63, 99–120. [Google Scholar] [CrossRef] [Green Version]
- Lint, H.L. A Revision of Juncus Subgenus Genuini (Juncaceae) in the Pacific States. Ph.D. Thesis, Oregon State University, Corvallis, OR, USA, April 1977. [Google Scholar]
- Flake, R.H.; Urbatsch, L.; Turner, B. Chemical documentation of allopatric introgression in Juniperus. Syst. Bot. 1978, 3, 129–144. [Google Scholar] [CrossRef]
- Palma-Otal, M.; Moore, W.; Adams, R.; Joswiak, G. Morphological, chemical, and biogeographical analyses of a hybrid zone involving Juniperus virginiana and J. horizontalis in Wisconsin. Can. J. Bot. 1983, 61, 2733–2746. [Google Scholar] [CrossRef]
- Urban, A.; Simelane, D.; Retief, E.; Heystek, F.; Williams, H.; Madire, L. The invasive ‘Lantana camara L.’ hybrid complex (Verbenaceae): A review of research into its identity and biological control in South Africa. Afr. Entomol. 2011, 19, 315–348. [Google Scholar] [CrossRef]
- Zhang, N.-N.; Yu, J.-J.; Wang, Y.-H.; Gong, X. Molecular evidence for asymmetric hybridization in three closely related sympatric species. AoB Plants 2018, 10, ply011. [Google Scholar] [CrossRef]
- Barkworth, M.E.; Adams, R.P. Flora of North America: Volume 2: Pteridophytes and Gymnosperms; Oxford University Press: Oxford, UK, 1993. [Google Scholar]
- Curto, M.; Puppo, P.; Kratschmer, S.; Meimberg, H. Genetic diversity and differentiation patterns in Micromeria from the Canary Islands are congruent with multiple colonization dynamics and the establishment of species syngameons. BMC Evol. Biol. 2017, 17, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stecconi, M.; Marchelli, P.; Puntieri, J.; Picca, P.; Gallo, L. Natural hybridization between a deciduous (Nothofagus antarctica, Nothofagaceae) and an evergreen (N. dombeyi) forest tree species: Evidence from morphological and isoenzymatic traits. Ann. Bot. 2004, 94, 775–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soliani, C.; Gallo, L.; Marchelli, P. Phylogeography of two hybridizing southern beeches (Nothofagus spp.) with different adaptive abilities. Tree Genet. Genomes 2012, 8, 659–673. [Google Scholar] [CrossRef]
- Grant, V.; Grant, K.A. Systematics of the Opuntia phaeacantha group in Texas. Bot. Gaz. 1979, 140, 199–207. [Google Scholar] [CrossRef]
- Debouck, D.G. Views on variability in Phaseolus beans. Ann. Rep. Bean Improv. Coop. 1992, 35, 9–10. [Google Scholar]
- Wyatt, R.; Antonovics, J. Butterflyweed re-revisited: Spatial and temporal patterns of leaf shape variation in Asclepias tuberosa. Evolution 1981, 35, 529–542. [Google Scholar]
- Hamilton, J.A.; de la Torre, A.R.; Aitken, S.N. Fine-scale environmental variation contributes to introgression in a three-species spruce hybrid complex. Tree Genet. Genomes 2015, 11, 817. [Google Scholar] [CrossRef]
- Buck, R.; Hyasat, S.; Hossfeld, A.; Flores-Rentería, L. Patterns of hybridization and cryptic introgression among one-and four-needled pinyon pines. Ann. Bot. 2020, 126, 401–411. [Google Scholar] [CrossRef]
- Buck, R.; Ortega-Del Vecchyo, D.; Whipple, A.V.; Gehring, C.; Michelson, R.; Flores-Rentería, D.; Klein, B.; Flores-Rentería, L. Sequential hybridization may have faciliated ecological transitions in the Southwestern pinyon pine syngameon. New Phytol. 2022, submitted.
- Wallace, L.E. Molecular evidence for allopolyploid speciation and recurrent origins in Platanthera huronensis (Orchidaceae). Int. J. Plant Sci. 2003, 164, 907–916. [Google Scholar] [CrossRef]
- Seybold, S. Schmeil-Fitschen: Flora von Deutschland und Angrenzender Länder; Quelle and Meyer: Heidelberg, Germany, 2009. [Google Scholar]
- Chhatre, V.E.; Evans, L.M.; DiFazio, S.P.; Keller, S.R. Adaptive introgression and maintenance of a trispecies hybrid complex in range-edge populations of Populus. Mol. Ecol. 2018, 27, 4820–4838. [Google Scholar] [CrossRef] [PubMed]
- Clapham, A.R.; Tutin, T.G.; Moore, D.M. Flora of the British Isles, 3rd ed.; Cambridge University Press: Cambridge, UK, 1990. [Google Scholar]
- Saidman, B.; Vilardi, J. Analysis of the genetic similarities among seven species of Prosopis (Leguminosae: Mimosoideae). Theor. Appl. Genet. 1987, 75, 109–116. [Google Scholar] [CrossRef]
- Torales, S.L.; Rivarola, M.; Pomponio, M.F.; Gonzalez, S.; Acuña, C.V.; Fernández, P.; Lauenstein, D.L.; Verga, A.R.; Hopp, H.E.; Paniego, N.B. De novo assembly and characterization of leaf transcriptome for the development of functional molecular markers of the extremophile multipurpose tree species Prosopis alba. BMC Genom. 2013, 14, 705. [Google Scholar] [CrossRef] [Green Version]
- Shaw, J.T. Systematics of the North American Plums (Prunus subgenus Prunus section Prunocerasus; Rosaceae). Ph.D. Thesis, The University of Tennessee, Knoxville, TN, USA, May 2005. [Google Scholar]
- Li, X.; Wei, G.; El-Kassaby, Y.A.; Fang, Y. Hybridization and introgression in sympatric and allopatric populations of four oak species. BMC Plant Biol. 2021, 21, 266. [Google Scholar] [CrossRef] [PubMed]
- Hardin, J.W. Hybridization and introgression in Quercus alba. J. Arnold Arbor. 1975, 56, 336–363. [Google Scholar] [CrossRef]
- Lewis, W.H. Rosa carolina (Rosaceae) subspecies and hybrids in eastern and midwestern United States, Canada, and Mexico. Novon J. Bot. Nomencl. 2008, 18, 192–198. [Google Scholar] [CrossRef]
- Brunsfeld, S.J.; Soltis, D.E.; Soltis, P.S. Evolutionary patterns and processes in Salix sect. Longifoliae: Evidence from chloroplast DNA. Syst. Bot. 1992, 17, 239–256. [Google Scholar]
- Weller, S.G.; Sakai, A.K.; Wagner, W.L. Artificial and natural hybridization in Schiedea and Alsinidendron (Caryophyllaceae: Alsinoideae): The importance of phylogeny, genetic divergence, breeding system, and population size. Syst. Bot. 2001, 26, 571–584. [Google Scholar]
- Laureto, P.J.; Barkman, T.J. Nuclear and chloroplast DNA suggest a complex single origin for the threatened allopolyploid Solidago houghtonii (Asteraceae) involving reticulate evolution and introgression. Syst. Bot. 2011, 36, 209–226. [Google Scholar] [CrossRef]
- Dreher, S.E. Interspecific Hybridization in Sphaeralcea (Malvaceae). Master’s Thesis, Claremont Graduate University, Claremont, CA, USA, 2014. [Google Scholar]
- Love, R.M. Interspecific hybridization in Stipa. II. Hybrids of S. cernua, S. lepida, and S. pulchra. Am. J. Bot. 1954, 41, 107–110. [Google Scholar] [CrossRef]
- Baiakhmetov, E.; Nowak, A.; Gudkova, P.D.; Nobis, M. Morphological and genome-wide evidence for natural hybridisation within the genus Stipa (Poaceae). Sci. Rep. 2020, 10, 13803. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, S.; Tysklind, N.; Hérault, B.; Heuertz, M. Topography drives microgeographic adaptations of closely related species in two tropical tree species complexes. Mol. Ecol. 2021, 30, 5080–5093. [Google Scholar] [CrossRef] [PubMed]
- Brouillet, L.; Semple, J.; Allen, G.; Chambers, K.; Sundberg, S. Symphyotrichum. Flora N. Am. 2006, 20, 465–539. [Google Scholar]
- Sauleda, R.P.; Hamilton, C.W. Tolumnia xpulchella (Hooker) Rafinesque (Orchidaceae) is Established as the Proper Name for a Jamaican Hybrid Swarm (Syngameon) and a New Species, One of The Parents of the Hybrid Swarm, Tolumnia hamiltonii Sauleda is Described. New World Orchid.—Nomencl. Notes 2019. Available online: https://www.researchgate.net/publication/341056945_Tolumnia_xpulchella_Hooker_Rafinesque_Orchidaceae_is_Established_as_the_Proper_Name_for_a_Jamaican_Hybrid_Swarm_Syngameon_and_a_New_Species_One_of_The_Parents_of_the_Hybrid_swarm_Tolumnia_hamiltonii_S (accessed on 24 March 2022).
- Ownbey, M. Natural hybridization and amphiploidy in the genus Tragopogon. Am. J. Bot. 1950, 37, 487–499. [Google Scholar] [CrossRef]
- Lipman, M.J.; Chester, M.; Soltis, P.S.; Soltis, D.E. Natural hybrids between Tragopogon mirus and T. miscellus (Asteraceae): A new perspective on karyotypic changes following hybridization at the polyploid level. Am. J. Bot. 2013, 100, 2016–2022. [Google Scholar] [CrossRef]
- Stoehrel, C.P. A Study of the Systematic Relationships between Members of the Trillium erectum Complex. Master’s Thesis, Western Carolina University, Cullowhee, NC, USA, November 2010. [Google Scholar]
- Case, F.J.; Case, R. Trilliums; Timber Press: Portland, OR, USA, 1997. [Google Scholar]
- Randolph, L. Variation among Tripsacum populations of Mexico and Guatemala. Brittonia 1970, 22, 305–337. [Google Scholar] [CrossRef]
- Widmer, A.; Lexer, C.; Cozzolino, S. Evolution of reproductive isolation in plants. Heredity 2009, 102, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Garner, A.G.; Goulet, B.E.; Farnitano, M.C.; Molina-Henao, Y.F.; Hopkins, R. Genomic signatures of reinforcement. Genes 2018, 9, 191. [Google Scholar] [CrossRef] [Green Version]
- Moran, B.M.; Payne, C.; Langdon, Q.; Powell, D.L.; Brandvain, Y.; Schumer, M. The genomic consequences of hybridization. eLife 2021, 10, e69016. [Google Scholar] [CrossRef]
- Seehausen, O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004, 19, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, K.; Seehausen, O. The propagation of admixture-derived adaptive radiation potential. Proc. R. Soc. B 2020, 287, 20200941. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Folk, R.; Zhang, N.; Gong, X. Homoploid hybridization of plants in the Hengduan mountains region. Ecol. Evol. 2019, 9, 8399–8410. [Google Scholar] [CrossRef] [PubMed]
- Beltrán, M.; Jiggins, C.D.; Bull, V.; Linares, M.; Mallet, J.; McMillan, W.O.; Bermingham, E. Phylogenetic discordance at the species boundary: Comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol. Biol. Evol. 2002, 19, 2176–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, L.E. Adaptive Novelty through Introgression in Heliconius Wing Patterns: Evidence for Shared Genetic “Tool Box” from Synthetic Hybrid Zones and a Theory of Diversification; University of Chicago Press: Chicago, IL, USA, 2003; pp. 281–318. [Google Scholar]
- Grant, B.R.; Grant, P.R. High survival of Darwin’s finch hybrids: Effects of beak morphology and diets. Ecology 1996, 77, 500–509. [Google Scholar] [CrossRef]
- Freeland, J.R.; Boag, P.T. The mitochondrial and nuclear genetic homogeneity of the phenotypically diverse Darwin’s ground finches. Evolution 1999, 53, 1553–1563. [Google Scholar] [CrossRef]
- Seehausen, O.; Alphen, J.J.V.; Witte, F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 1997, 277, 1808–1811. [Google Scholar] [CrossRef]
- Martin, S.H.; Davey, J.W.; Jiggins, C.D. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 2015, 32, 244–257. [Google Scholar] [CrossRef] [Green Version]
- Twyford, A.D.; Ennos, R. Next-generation hybridization and introgression. Heredity 2012, 108, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Hey, J.; Chung, Y.; Sethuraman, A.; Lachance, J.; Tishkoff, S.; Sousa, V.C.; Wang, Y. Phylogeny estimation by integration over isolation with migration models. Mol. Biol. Evol. 2018, 35, 2805–2818. [Google Scholar] [CrossRef]
- Ottenburghs, J. Ghost introgression: Spooky gene flow in the distant past. Bioessays 2020, 42, 2000012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caujapé-Castells, J.; Bramwell, D. Jesters, red queens, boomerangs and surfers: A molecular outlook on the diversity of the Canarian endemic flora. In The Biology of Island Floras; Bramwell, D., Caujapé-Castells, J., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 284–324. [Google Scholar]
- Pfennig, K.S.; Kelly, A.L.; Pierce, A.A. Hybridization as a facilitator of species range expansion. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottenburghs, J. The genic view of hybridization in the Anthropocene. Evol. Appl. 2021, 14, 2342–2360. [Google Scholar] [CrossRef] [PubMed]
- Todesco, M.; Pascual, M.A.; Owens, G.L.; Ostevik, K.L.; Moyers, B.T.; Hübner, S.; Heredia, S.M.; Hahn, M.A.; Caseys, C.; Bock, D.G. Hybridization and extinction. Evol. Appl. 2016, 9, 892–908. [Google Scholar] [CrossRef] [PubMed]
- Ingvarsson, P.K. Restoration of genetic variation lost—The genetic rescue hypothesis. Trends Ecol. Evol. 2001, 16, 62–63. [Google Scholar] [CrossRef]
- Carlson, S.M.; Cunningham, C.J.; Westley, P.A. Evolutionary rescue in a changing world. Trends Ecol. Evol. 2014, 29, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Barraclough, T.G.; Vogler, A.P. Detecting the geographical pattern of speciation from species-level phylogenies. Am. Nat. 2000, 155, 419–434. [Google Scholar] [CrossRef]
- Foote, A.D. Sympatric speciation in the genomic era. Trends Ecol. Evol. 2018, 33, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.M. Sympatric speciation. Am. Nat. 1966, 100, 637–650. [Google Scholar] [CrossRef]
- Schliewen, U.K.; Tautz, D.; Pääbo, S. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature 1994, 368, 629–632. [Google Scholar] [CrossRef]
- Nikolakis, Z.L.; Orton, R.; Crother, B.I. Fine scale population structure and extensive gene flow within an Eastern Nearctic snake complex (Pituophis melanoleucus). bioRxiv 2021, 51, 133–146. [Google Scholar]
- Larcombe, M.J.; Holland, B.; Steane, D.A.; Jones, R.C.; Nicolle, D.; Vaillancourt, R.E.; Potts, B.M. Patterns of reproductive isolation in Eucalyptus—A phylogenetic perspective. Mol. Biol. Evol. 2015, 32, 1833–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trucco, F.; Tatum, T.; Rayburn, A.L.; Tranel, P.J. Out of the swamp: Unidirectional hybridization with weedy species may explain the prevalence of Amaranthus tuberculatus as a weed. New Phytol. 2009, 184, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, D.; Garcia-Vazquez, E. Maintenance of asymmetric hybridization between Atlantic salmon (Salmo salar) and brown trout (Salmo trutta) via postzygotic barriers and paternal effects. Can. J. Fish. Aquat. Sci. 2011, 68, 593–602. [Google Scholar] [CrossRef]
- Del-Rio, G.; Rego, M.A.; Whitney, B.M.; Schunck, F.; Silveira, L.F.; Faircloth, B.C.; Brumfield, R.T. Displaced clines in an avian hybrid zone (Thamnophilidae: Rhegmatorhina) within an Amazonian interfluve. Evolution 2021, 76, 455–475. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H. On the completion of speciation. Philos. Trans. R. Soc. B 2020, 375, 20190530. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank, T.E.; Hahn, M.W. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol. Ecol. 2014, 23, 3133–3157. [Google Scholar] [CrossRef] [Green Version]
- Rieseberg, L.H.; Wendel, J.F. Introgression and its consequences in plants. Hybrid Zones Evol. Process 1993, 70, 109. [Google Scholar]
- Tallmon, D.A.; Luikart, G.; Waples, R.S. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 2004, 19, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Cannon, C. Is speciation an inexorable march to reproductive isolation? Mol. Ecol. 2021, 30, 4349–4352. [Google Scholar] [CrossRef]
- Rothfels, C.J.; Johnson, A.K.; Hovenkamp, P.H.; Swofford, D.L.; Roskam, H.C.; Fraser-Jenkins, C.R.; Windham, M.D.; Pryer, K.M. Natural hybridization between genera that diverged from each other approximately 60 million years ago. Am. Nat. 2015, 185, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H. The dynamics of hybrid zones. Heredity 1979, 43, 341–359. [Google Scholar] [CrossRef] [Green Version]
- Anderson, E. Introgressive Hybridization; Wiley: New York, NY, USA, 1949. [Google Scholar]
- Oreskes, N. The scientific consensus on climate change. Science 2004, 306, 1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmesan, C.; Yohe, G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 2003, 421, 37–42. [Google Scholar] [CrossRef]
- Williams, J.W.; Jackson, S.T.; Kutzbach, J.E. Projected distributions of novel and disappearing climates by 2100 AD. Proc. Natl. Acad. Sci. USA 2007, 104, 5738–5742. [Google Scholar] [CrossRef] [Green Version]
- Chunco, A.J. Hybridization in a warmer world. Ecol. Evol. 2014, 4, 2019–2031. [Google Scholar] [CrossRef]
- Stanford, P.K. For pluralism and against realism about species. Philos. Sci. 1995, 62, 70–91. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, Q.D.; Meier, R. Species Concepts and Phylogenetic Theory: A Debate; Columbia University Press: New York, NY, USA, 2000. [Google Scholar]
- Department of the Interior. Endangered Species Act of 1973; Department of the Interior: Washington, DC, USA, 1973; p. 44.
- Mayr, E. Systematics and the origin of species; Columbia University Press: New York, NY, USA, 1942. [Google Scholar]
- Rieseberg, L.H.; Beckstrom-Sternberg, S.M.; Liston, A.; Arias, D.M. Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus (Asteraceae). Syst. Bot. 1991, 16, 50–76. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Carter, R.; Zona, S. Molecular tests of the hypothesized hybrid origin of two diploid Helianthus species (Asteraceae). Evolution 1990, 44, 1498–1511. [Google Scholar] [CrossRef]
- Rieseberg, L.H. Homoploid reticulate evolution in Helianthus (Asteraceae): Evidence from ribosomal genes. Am. J. Bot. 1991, 78, 1218–1237. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Miller, J.M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2016, 30, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.Y.; Hoffmann, A.A.; van Oppen, M.J. Hybridization as a conservation management tool. Conserv. Lett. 2019, 12, e12652. [Google Scholar] [CrossRef]
- Quilodrán, C.S.; Montoya-Burgos, J.I.; Currat, M. Harmonizing hybridization dissonance in conservation. Commun. Biol. 2020, 3, 391. [Google Scholar] [CrossRef] [PubMed]

| Genera (Common Name) | Known Participants | Kingdom | Source |
|---|---|---|---|
| Acropora (Coral) | 8 | Animalia | [27,28] * |
| Anser + Branta (Geese) | 15 | Animalia | [29] |
| Artibeus (bats) | 3 | Animalia | [30] |
| Callithrix (marmosets) | 3 | Animalia | [31] |
| Canis | 3 | Animalia | [32,33] 2 |
| Carabus (Carabid beetles) | 6 | Animalia | [34] |
| Catostomus + Chasmistes + Deltistes (catostomid fish) | 4 | Animalia | [35] * |
| Cerion (snail) | Not specified | Animalia | [36] * |
| Colias (sulfur butterflies) | 3 | Animalia | [37] * |
| Daphnia (plankton) | 5 | Animalia | [38] (but see [39]) |
| Desmognathus (Dusky Salamanders) | 3 | Animalia | [40] |
| Drosophila | At least 3 | Animalia | [41,42] |
| Eueides (butterflies) | 5 | Animalia | [43] |
| Geospiza (Darwin’s finches) | Two sets of 3 | Animalia | [44,45,46] |
| Habronattus (jumping spiders) | At least 3 | Animalia | [47] |
| Heliconius (butterflies) | One set of 3; one set of 4; one set of 9 | Animalia | [43] |
| Homo | 3 | Animalia | [31,48] |
| Konia + Myaka + Pungu + Sarotherodon (Cichlid fish) | 8 | Animalia | [49] * |
| Liolaemus (lizard) | 4 | Animalia | [50] |
| Montastraea (coral) | 3 | Animalia | [51] * (but see [52]) |
| Pacifigorgia (octocorals) | Not specified | Animalia | [53,54] 2 |
| Papio + Theropithecus (baboons) | at least 3 | Animalia | [55] * |
| Psammocora (Indo-Pacific corals) | 3 | Animalia | [56] |
| Pseudophryne (frogs) | 3 | Animalia | [57] |
| Steatocranus (cichlid fish) | 18 | Animalia | [58] * |
| Stylophora (Red Sea coral) | Not specified | Animalia | [59] * |
| Sus (wild pigs) | 4 | Animalia | [60] |
| Ursa (bears) | 6 | Animalia | [61] |
| Xiphophorus (fishes) | 5 | Animalia | [62] |
| Abies (fir) | 3 | Plantae | [63] (in [64] *) |
| Actinidia (Kiwi) | 9 | Plantae | [21,65] 2 |
| Aesculus (buckeye) | 3 | Plantae | [66] (in [64] *) |
| Ajuga (bugleherb) or Amaranthus (amaranths) | 5 | Plantae | [64] * |
| Ambrosia (ragweed) | 3 | Plantae | [64] * |
| Amelanchier (serviceberry) | 5 | Plantae | [67] (in [64] *) |
| Aquilegia (Columbines) | Not specified | Plantae | [68] * |
| Arbutus (madrones) | 5 | Plantae | [69] |
| Arctostaphylos (manzanita) | At least 3 | Plantae | [70,71] |
| Asclepias (milkweed) | 4 | Plantae | [64] * |
| Asplenium (spleenworts) | 16 | Plantae | [72] (in [15] *) |
| Betula (birch) | One set of 4; one set of 6 | Plantae | Gunnarsson in [4] *; [73,74] (in [64] *) |
| Boechera (rockcress) | 58 | Plantae | [75]; D. Bailey (in [15] *) |
| Carex (true sedges) | Three sets of 3; two sets of 4 | Plantae | [76] * |
| Castanea (chestnut) | Not specified | Plantae | [77] * |
| Ceanothus (California lilac) | Not specified | Plantae | [78] * |
| Cirsium (plume thistle) | 17 | Plantae | [79] * |
| Citrus | 8 | Plantae | [80,81] 2 |
| Coprosma (stinkwood) | 6 | Plantae | [82] * |
| Cornus (dogwood) | 4 | Plantae | [64] * |
| Corybas (helmet orchid) | 3 | Plantae | [83] |
| Cyperus | 3 | Plantae | [84] (in [64] *) |
| Dichanthelium (rosette grass) | Two sets of 3; one set of 4 | Plantae | [64] *; [85] (in [64] *) |
| Diospyros (ebonies) | One set of 3, one set of 4 | Plantae | [86] * |
| Diplacus (monkey flower) | 5 | Plantae | [78,87] * |
| Drosera (sundew) | 4 | Plantae | [88] (in [64] *); [89] (in [64] *) |
| Dryopteris (woodfern) | 4 | Plantae | [90] (in [64] *) |
| Dubatia | 6 | Plantae | [91] |
| Elymus (wildrye) | 3 | Plantae | [64] * |
| Encelia (brittlebush) | 11 | Plantae | [92] * |
| Equisetum (horsetail) | 3 | Plantae | [64] * |
| Eschweilera | 3 | Plantae | [93] * |
| Espeletia (frailejones) | 3 | Plantae | [94] |
| Eucalyptus (Green ashes) | 4 | Plantae | [95] |
| Eucalyptus (Boxes) | ~10 | Plantae | [96] |
| Ficus (figs) | 13 | Plantae | [97] |
| Gentiana | 4 | Plantae | [64] * |
| Geum (avens) | Not specified | Plantae | [5] (in [78] *) |
| Gymnocarpium (oak fern) | 4 | Plantae | [98] (in [64] *) |
| Helianthus (sunflower) | One set of 4; one set of 6 | Plantae | [64,99,100,101,102,103] 1 |
| Hieracium (hawkweed) | One set of 4; one set of 5 | Plantae | [64] * |
| Huperzia (firmosses) | 3 | Plantae | [64] * |
| Hypericum (St. John’s wort) | 3 | Plantae | [64] * |
| Iris (California irises) | 12 | Plantae | [78,104] 2; [105] * (in [15] *) |
| Juncus (rushes) | 7 | Plantae | [106] * |
| Juniperus (junipers) | 3 | Plantae | [107,108] (in [64] *) |
| Lantana | 6 | Plantae | [109] * |
| Lespedeza (bush clovers) | 5 | Plantae | [64] * |
| Ligularia (leopard plants) | 3 | Plantae | [110] |
| Lycopodiella (bog clubmosses) | 4 | Plantae | [111] (in [64] *) |
| Lycopus | 4 | Plantae | [64] * |
| Lysimachia | 3 | Plantae | [64] * |
| Melandrium/Silene (campion) | Not specified | Plantae | [5] (in [78] *) |
| Micromeria | 20 | Plantae | [112] * |
| Nothofagus (southern beeches) | At least 3 | Plantae | [5] (in [78] *); [113,114] |
| Opuntia (prickly pear cactus) | At least 16 | Plantae | [115] * |
| Phaseolus (bean) | 3 | Plantae | [116] * |
| Phlox | 3 | Plantae | [117] (in [64]*) |
| Picea (spruces) | 3 | Plantae | [118] |
| Pinus (Southwestern pinyon pines) | 4 | Plantae | [119,120] * |
| Platanthera (butterfly orchids) | Two sets of 3 | Plantae | [121] (in [64] *) |
| Populus (cottonwood) | Three sets of 3 | Plantae | [122] (in [64] *); [123] (but see [23] *) |
| Potamogeton (pondweed) | 19 | Plantae | [78] * (in: [15] *); [124] |
| Prosopis (mesquite) | 7 | Plantae | [125,126] * |
| Prunus (plums) | 18 | Plantae | [127] * |
| Pycnanthemum (mountain mints) | 3 | Plantae | [64] * |
| Quercus (Chinese oaks) | 4 | Plantae | [128] |
| Quercus (Eastern white oaks) | 14 | Plantae | [78,129] * |
| Quercus (Southwestern white oaks) | 16 | Plantae | [78] *; R. Spellenberg (in: [15] *) |
| Rosa (rose) | 3 | Plantae | [130] (in [64] *) |
| Rubus (brambles) | 3 | Plantae | [64] * |
| Salix (willow) | Two sets of 3; one set of 6 | Plantae | [85] (in [64] *); [131] |
| Saxifraga (saxifrages) | 3 | Plantae | Lloyd in [4] * |
| Schiedea | 4 | Plantae | [132] |
| Scirpus (club-rush) | 3 | Plantae | [64] * |
| Senecio | 5 | Plantae | [19] * |
| Solidago (goldenrods) | One set of 4; one set of 5 | Plantae | [64] * (but see [133]) |
| Sphaeralcea (globemallows) | Not specified | Plantae | [134] |
| Stipa | Two sets of 3 | Plantae | [135,136] |
| Symphonia | 3 | Plantae | [137] * |
| Symphyotrichum | 8 | Plantae | [138] (in [64] *) |
| Thalictrum (meadow-rue) | 3 | Plantae | [64] * |
| Tolumnia (Dancing-lady orchid) | 4 | Plantae | [139] * |
| Tragopogon (salsifies) | 5 | Plantae | [140,141] |
| Trillium | One set of 3; one set of 4 | Plantae | [64] *; [142] * (but see [143]) |
| Tripsacum (gamagrass) | 7 | Plantae | [144] * |
| Verbascum (mullein) | 4 | Plantae | [64] * |
| Verbena (vervain) | 4 | Plantae | [64] * |
| Viola | One set of 4; one set of 5; one set of 7 | Plantae | [64] * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buck, R.; Flores-Rentería, L. The Syngameon Enigma. Plants 2022, 11, 895. https://doi.org/10.3390/plants11070895
Buck R, Flores-Rentería L. The Syngameon Enigma. Plants. 2022; 11(7):895. https://doi.org/10.3390/plants11070895
Chicago/Turabian StyleBuck, Ryan, and Lluvia Flores-Rentería. 2022. "The Syngameon Enigma" Plants 11, no. 7: 895. https://doi.org/10.3390/plants11070895
APA StyleBuck, R., & Flores-Rentería, L. (2022). The Syngameon Enigma. Plants, 11(7), 895. https://doi.org/10.3390/plants11070895






