Identification and Validation of a Chromosome 4D Quantitative Trait Locus Hotspot Conferring Heat Tolerance in Common Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Results
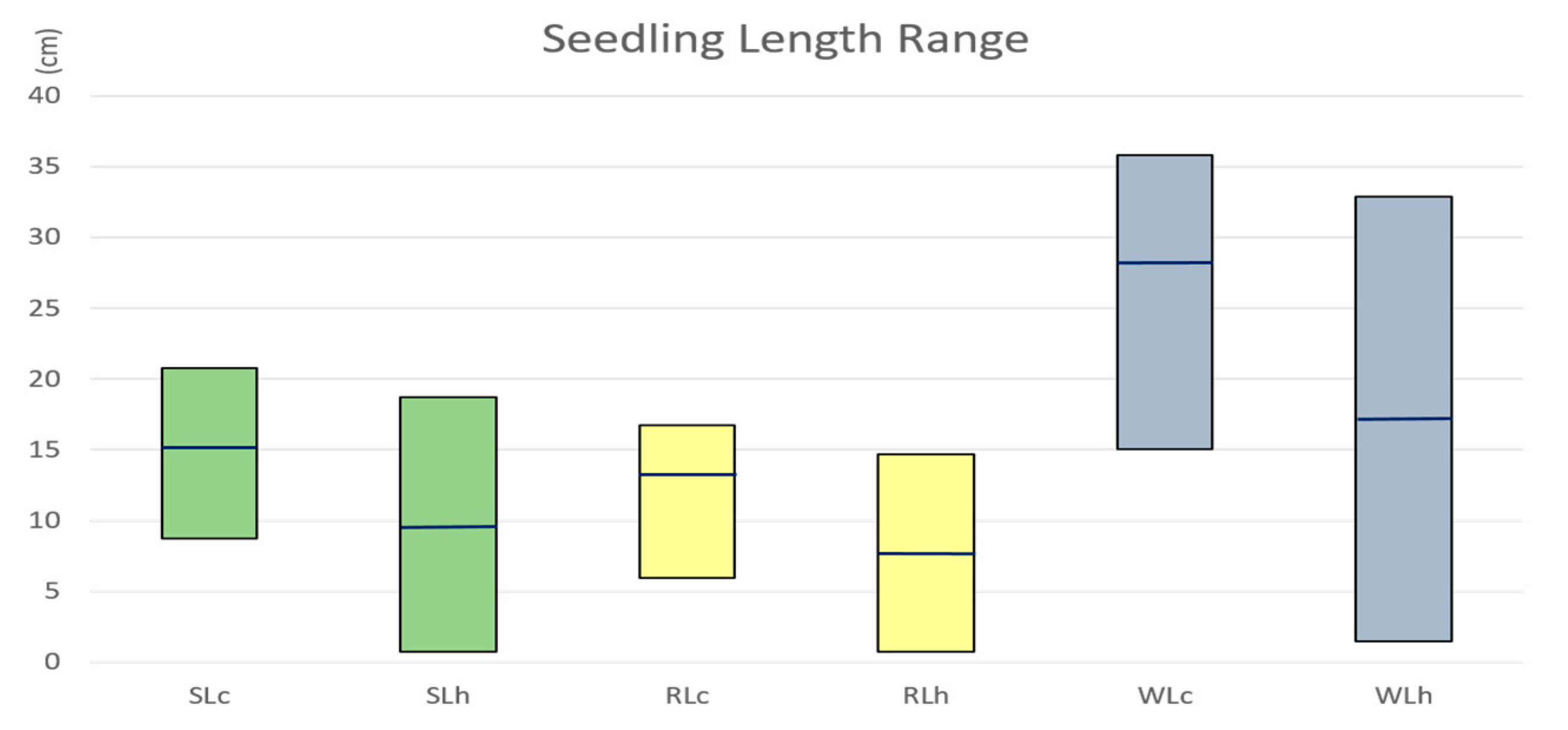
2.1. Seedling Length Reduced under Heat Stress
2.2. High Heritability of Seedling Length Traits under Heat Stress
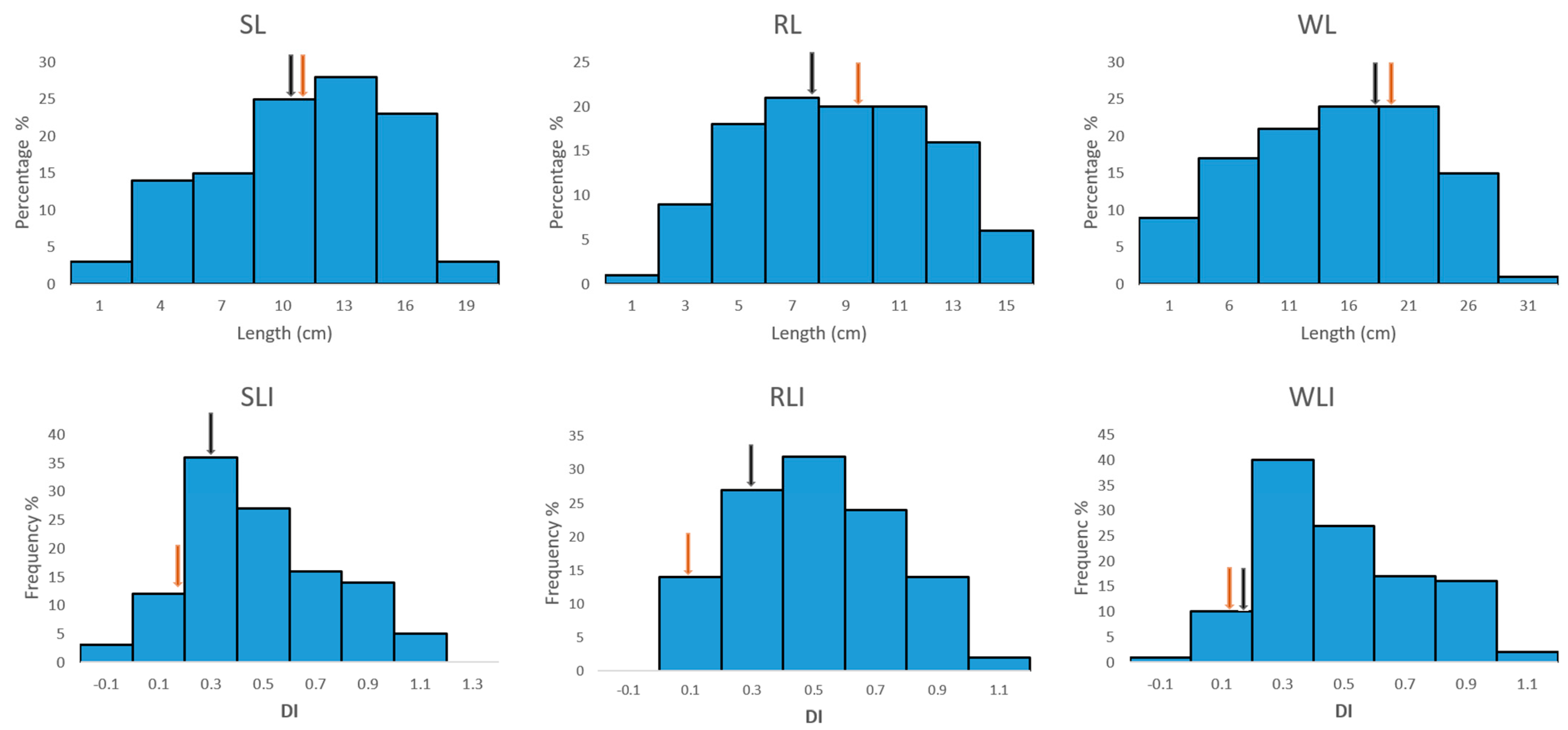
2.3. Phenotypic Variation in the Traits Were All Normally Distributed
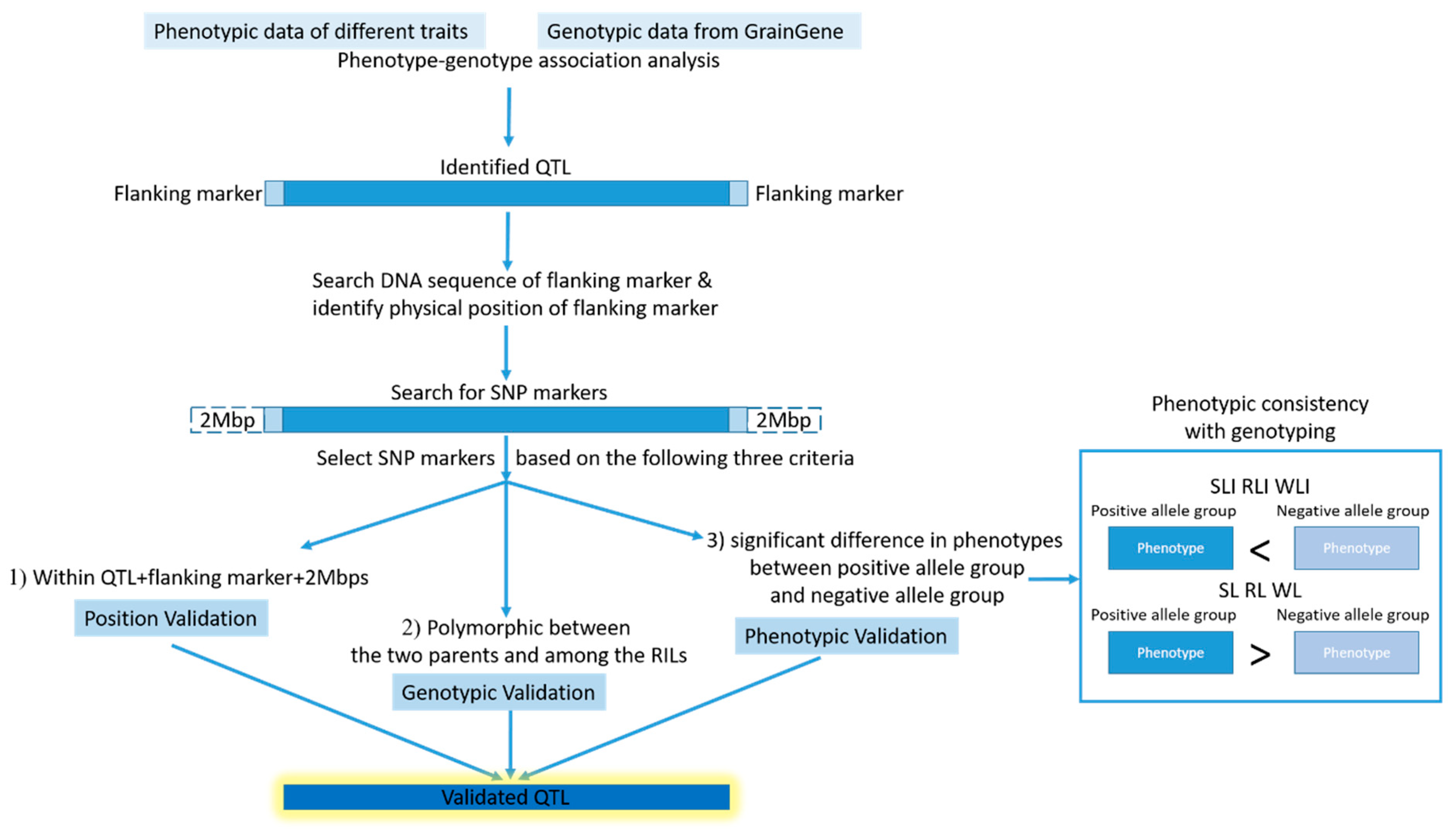
2.4. QTL Analysis Revealed Thirteen QTL and Six of Them Located on a Hotspot of Chromosome 4D Associated with HT
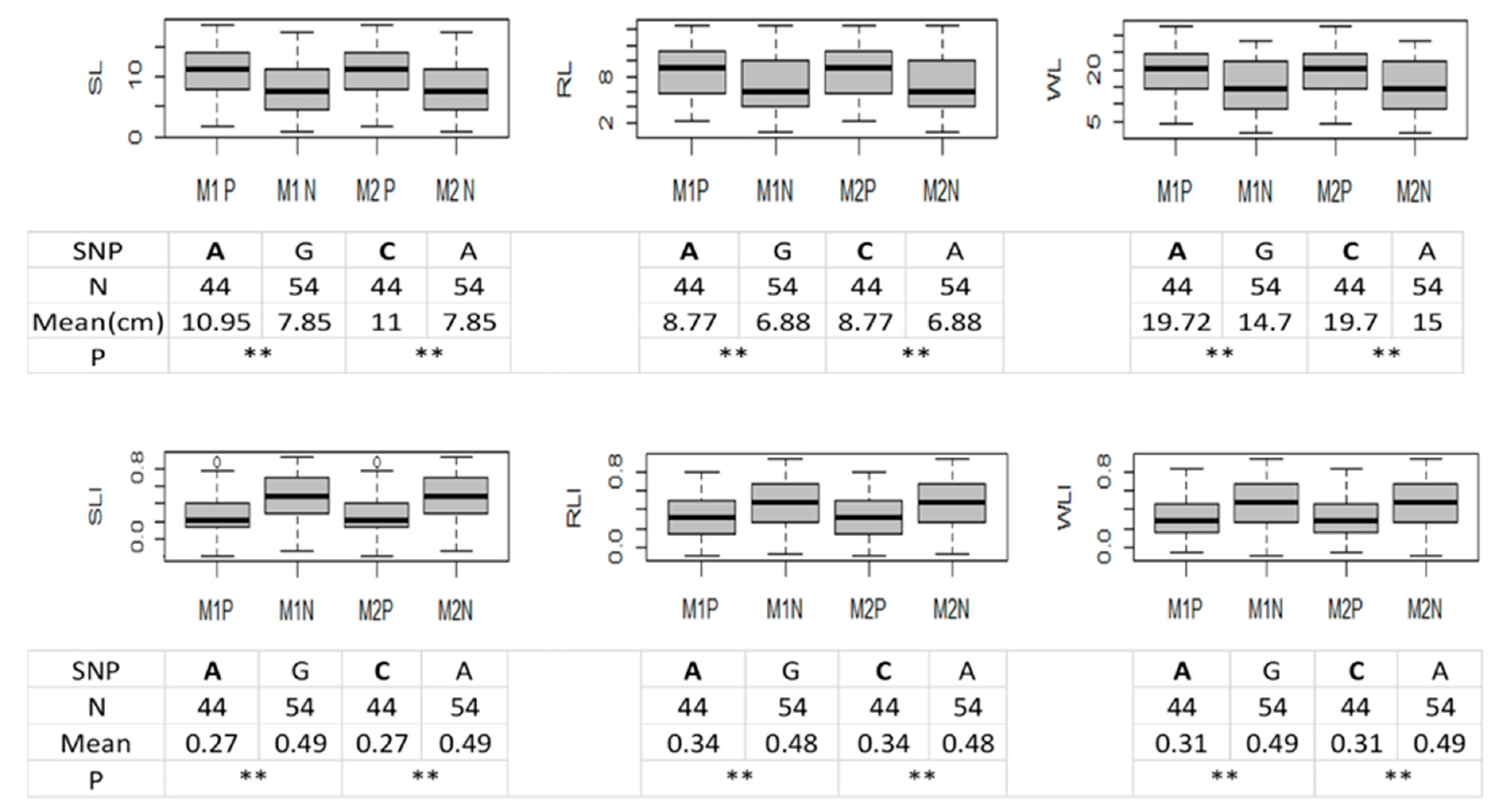
2.5. Validation for the QTL Hotspot
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Plant Growth and Treatment
4.3. Phenotypic Evaluation and Heritability Estimation
4.4. QTL Analysis and Mapping
4.5. Validation of Identified QTL
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Priya, M.; Dhanker, O.P.; Siddique, K.H.M.; HanumanthaRao, B.; Nair, R.M.; Pandey, S.; Singh, S.; Varshney, R.K.; Prasad, P.V.V.; Nayyar, H. Drought and heat stress-related proteins: An update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 2019, 132, 1607–1638. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Rehman, A.; Wahid, A.; Siddique, K.H.M. Photosynthesis under Heat Stress. In Handbook of Photosynthesis, 3rd ed.; Routledge: London, UK, 2016; pp. 697–701. [Google Scholar]
- Wardlaw, I.F.; Dawson, I.A.; Munibi, P.; Fewster, R. The tolerance of wheat to high temperatures during reproductive growth. I. Survey procedures and general response patterns. Aust. J. Agric. Res. 1989, 40, 965–980. [Google Scholar] [CrossRef]
- Wiegand, C.L.; Cuellar, J.A. Duration of grain filling and kernel weight of wheat as affected by temperature. Crop Sci. 1981, 21, 95–101. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 31, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maulana, F.; Ayalew, H.; Anderson, J.D.; Kumssa, T.T.; Huang, W.; Ma, X.F. Genome-wide association mapping of seedling heat tolerance in winter wheat. Front. Plant Sci. 2018, 9, 1272. [Google Scholar] [CrossRef]
- Spiertz, J.H.J.; Hamer, R.J.; Xu, H.; Primo-Martin, C.; Don, C.; van der Putten, P.E.L. Heat stress in wheat (Triticum aestivum L.): Effects on grain growth and quality traits. Eur. J. Agron. 2006, 25, 89–95. [Google Scholar] [CrossRef]
- El-Rawy, M.A.; Hassan, M.I. A diallel analysis of drought tolerance indices at seedling stage in bread wheat (Triticum aestivum L.). Plant Breed. Biotechnol. 2014, 2, 276–288. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, T.R.; Mahmoudi, T.R.; Yu, J.M.; Liu, S.Y.; Pierson, L.S.; Pierson, E.A. Drought-stress tolerance in wheat seedlings conferred by phenazine-producing rhizobacteria. Front. Microbiol. 2019, 10, 1590. [Google Scholar] [CrossRef] [Green Version]
- Liao, M.T.; Palta, J.A.; Fillery, I.R.P. Root characteristics of vigorous wheat improve early nitrogen uptake. Aust. J. Agric. Res. 2006, 57, 1097–1107. [Google Scholar] [CrossRef]
- Comas, L.H.; Becker, S.R.; Cruz, V.M.V.; Byrne, P.F.; Dierig, D.A. Root traits contributing to plant productivity under drought. Front. Plant Sci. 2013, 4, 442. [Google Scholar] [CrossRef] [Green Version]
- Kilasi, N.L.; Singh, J.; Vallejos, C.E.; Ye, C.; Jagadish, S.V.K.; Kusolwa, P.; Rathinasabapathi, B. Heat stress tolerance in Rice (Oryza sativa L.): Identification of quantitative trait loci and candidate genes for seedling growth under heat stress. Front. Plant Sci. 2018, 9, 1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Chang, X.; Wang, C.; Jing, R. Mapping QTLs for seedling traits and heat tolerance indices in common wheat. Acta Bot. Boreali-Occident. Sin. 2012, 32, 1525–1533. [Google Scholar]
- Ni, Z.; Li, H.; Zhao, Y.; Peng, H.; Hu, Z.; Xin, M.; Sun, Q. Genetic improvement of heat tolerance in wheat: Recent progress in understanding the underlying molecular mechanisms. Crop J. 2018, 6, 32–41. [Google Scholar] [CrossRef]
- Azam, F.; Chang, X.; Jing, R. Mapping QTL for chlorophyll fluorescence kinetics parameters at seedling stage as indicators of heat tolerance in wheat. Euphytica 2015, 202, 245–258. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, J.A.; Wingen, L.U.; Griffiths, M.; Pound, M.P.; Gaju, O.; Foulkes, M.J.; Le Gouis, J.; Griffiths, S.; Bennett, M.J.; King, J.; et al. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot. 2015, 66, 2283–2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarker, A.; Erskine, W.; Singh, M. Variation in shoot and root characteristics and their association with drought tolerance in lentil landraces. Genet. Resour. Crop Evol. 2005, 52, 89–97. [Google Scholar] [CrossRef]
- Lu, L.; Liu, H.; Wu, Y.; Yan, G. Wheat genotypes tolerant to heat at seedling stage tend to be also tolerant at adult stage: The possibility of early selection for heat tolerance breeding. Crop J. 2022; in press. [Google Scholar]
- Farooq, M.; Bramley, H.; Palta, J.A.; Siddique, K.H.M. Heat stress in wheat during reproductive and grain-filling phases. Crit. Rev. Plant Sci. 2011, 30, 491–507. [Google Scholar] [CrossRef]
- Yang, J.; Sears, R.G.; Gill, B.S.; Paulsen, G.M. Growth and senescence characteristics associated with tolerance of wheat-alien amphiploids to high temperature under controlled conditions. Euphytica 2002, 126, 185–193. [Google Scholar] [CrossRef]
- Mason, R.E.; Mondal, S.; Beecher, F.W.; Pacheco, A.; Jampala, B.; Ibrahim, A.M.H.; Hays, D.B. QTL associated with heat susceptibility index in wheat (Triticum aestivum L.) under short-term reproductive stage heat stress. Euphytica 2010, 174, 423–436. [Google Scholar] [CrossRef]
- Mason, R.E.; Mondal, S.; Beecher, F.W.; Hays, D.B. Genetic loci linking improved heat tolerance in wheat (Triticum aestivum L.) to lower leaf and spike temperatures under controlled conditions. Euphytica 2011, 180, 181–194. [Google Scholar] [CrossRef]
- Paliwal, R.; Roder, M.S.; Kumar, U.; Srivastava, J.P.; Joshi, A.K. QTL mapping of terminal heat tolerance in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2012, 125, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Vijayalakshmi, K.; Fritz, A.K.; Paulsen, G.M.; Bai, G.H.; Pandravada, S.; Gill, B.S. Modeling and mapping QTL for senescence-related traits in winter wheat under high temperature. Mol. Breed. 2010, 26, 163–175. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Singh, N.P. Heat stress in crop plants: Its nature, impacts and integrated breeding strategies to improve heat tolerance. Plant Breed. 2014, 133, 679–701. [Google Scholar] [CrossRef] [Green Version]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.H.; Leroy, P.; Ganal, M.W. A microsatellite map of wheat. Genetics 1998, 149, 2007–2023. [Google Scholar] [CrossRef]
- Deynze, A.E.; Nelson, J.C.; Sorrells, M.E.; McCouch, S.R.; Dubcovsky, J.; Dvorák, J.; Gill, K.S.; Gill, B.S.; Lagudah, E.S.; Appels, R. Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oat. Genome 1995, 38, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Song, Q.J.; Shi, J.R.; Singh, S.; Fickus, E.W.; Costa, J.M.; Lewis, J.; Gill, B.S.; Ward, R.; Cregan, P.B. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 2005, 110, 550–560. [Google Scholar] [CrossRef]
- Khalid, M.; Gul, A.; Amir, R.; Mohsin, A.; Afzal, F.; Umar, M.Q.; Rasheed, A. QTL mapping for seedling morphology under drought stress in wheat cross synthetic (W7984)/Opata. Plant Genet. Resour. 2018, 4, 359–366. [Google Scholar] [CrossRef]
- Onyemaobi, I.; Ayalew, H.; Liu, H.; Siddique, K.H.M.; Yan, G. Identification and validation of a major chromosome region for high grain number per spike under meiotic stage water stress in wheat (Triticum aestivum L.). PLoS ONE 2018, 13, e0194075. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Chen, G.Y. Conditional QTL mapping for waterlogging tolerance in two RILs populations of wheat. Springerplus 2013, 2, 245. [Google Scholar] [CrossRef] [Green Version]
- Ayalew, H.; Liu, H.; Yan, G. Identification and validation of root length QTLs for water stress resistance in hexaploid wheat (Titicum aestivum L.). Euphytica 2017, 213, 126. [Google Scholar] [CrossRef]
- Alsamadany, H. Diversity and Genetic Studies of Heat Tolerance in Wheat. Ph.D. Thesis, The University of Western Australia, Perth, Australia, 2015. [Google Scholar]
- Reynolds, M.; Manes, Y.; Izanloo, A.; Langridge, P. Phenotyping approaches for physiological breeding and gene discovery in wheat. Ann. Appl. Biol. 2009, 155, 139–320. [Google Scholar] [CrossRef]
- IWGSC; Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.L.; Jing, R.L.; Chang, X.P.; Li, W. Identification of quantitative trait loci and environmental interactions for accumulation and remobilization of water-soluble carbohydrates in wheat (Tiiticum aestivum L.) stems. Genetics 2007, 176, 571–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xin, F.; Zhu, T.; Wei, S.W.; Han, Y.C.; Zhao, Y.; Zhang, D.Z.; Ma, L.J.; Ding, Q. QTL mapping of kernel traits and validation of a major QTL for kernel length-width ratio using SNP and bulked segregant analysis in wheat. Sci. Rep. 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabral, A.L.; Jordan, M.C.; Larson, G.; Somers, D.J.; Humphreys, D.G.; McCartney, C.A. Relationship between QTL for grain shape, grain weight, test weight, milling yield, and plant height in the spring wheat cross RL4452/‘ACDomain’. PLoS ONE 2018, 13, e0190681. [Google Scholar] [CrossRef] [PubMed]
- Valluru, R.; Reynolds, M.P.; Davies, W.J.; Sukumaran, S. Phenotypic and genome-wide association analysis of spike ethylene in diverse wheat genotypes under heat stress. New Phytol. 2017, 214, 271–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, D.; Reynolds, M.; Mullan, D.; Izanloo, A.; Kuchel, H.; Langridge, P.; Schnurbusch, T. Detection of two major grain yield QTL in bread wheat (Triticum aestivum L.) under heat, drought and high yield potential environments. Theor. Appl. Genet. 2012, 125, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.S.; Le Gouis, J.; Leflon, M.; Rong, W.Y.; Laperche, A.; Brancourt-Hulmel, M. Using probe genotypes to dissect QTL × environment interactions for grain yield components in winter wheat. Theor. Appl. Genet. 2010, 121, 1501–1517. [Google Scholar] [CrossRef]
- Wu, P.Y.; Yang, M.H.; Kao, C.H. A statistical framework for QTL hotspot detection. G3 2021, 11, jkab056. [Google Scholar] [CrossRef]
- Bharadwaj, C.; Tripathi, S.; Soren, K.R.; Thudi, M.; Singh, R.K.; Sheoran, S.; Roorkiwal, M.; Patil, B.S.; Chitikineni, A.; Palakurthi, R.; et al. Introgression of “QTL-hotspot” region enhances drought tolerance and grain yield in three elite chickpea cultivars. Plant Genome 2021, 14, e20076. [Google Scholar] [CrossRef]
- Sinha, P.; Singh, V.K.; Saxena, R.K.; Khan, A.W.; Abbai, R.; Chitikineni, A.; Desai, A.; Molla, J.; Upadhyaya, H.D.; Kumar, A.; et al. Superior haplotypes for haplotype-based breeding for drought tolerance in pigeonpea (Cajanus cajan L.). Plant Biotechnol. J. 2020, 18, 2482–2490. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Voorrips, R.E.; van de Weg, E.; Peace, C.; Iezzoni, A. Genetic structure of a QTL hotspot on chromosome 2 in sweet cherry indicates positive selection for favorable haplotypes. Mol. Breed. 2017, 37, 85. [Google Scholar] [CrossRef]
- Li, S.P.; Chang, X.P.; Wang, C.S.; Jing, R.L. Mapping QTL for heat tolerance at grain filling stage in common wheat. Scientia Agricultura Sinica 2013, 46, 2119–2129. [Google Scholar]
- Mondal, S.; Mason, R.E.; Huggins, T.; Hays, D.B. QTL on wheat (Triticum aestivum L.) chromosomes 1B, 3D and 5A are associated with constitutive production of leaf cuticular wax and may contribute to lower leaf temperatures under heat stress. Euphytica 2015, 201, 123–130. [Google Scholar] [CrossRef]
- Acuña-Galindo, M.A.; Mason, R.E.; Subramanian, N.K.; Hays, D.B. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 2015, 55, 477–492. [Google Scholar] [CrossRef]
- Liu, H.; Mullan, D.; Zhang, C.; Zhao, S.; Li, X.; Zhang, A.; Lu, Z.; Wang, Y.; Yan, G. Major genomic regions responsible for wheat yield and its components as revealed by meta-QTL and genotype-phenotype association analyses. Planta 2020, 252, 65. [Google Scholar] [CrossRef]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Baenziger, P.S. Genome-wide association study reveals novel genomic regions for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat. Int. J. Mol. Sci. 2018, 19, 3011. [Google Scholar] [CrossRef] [Green Version]
- Czyczyło-Mysza, I.; Tyrka, M.; Marcińska, I.; Skrzypek, E.; Karbarz, M.; Dziurka, M.; Hura, T.; Dziurka, K.; Quarrie, S.A. Quantitative trait loci for leaf chlorophyll fluorescence parameters, chlorophyll and carotenoid contents in relation to biomass and yield in bread wheat and their chromosome deletion bin assignments. Mol. Breed. 2013, 32, 189–210. [Google Scholar] [CrossRef] [Green Version]
- Guan, P.; Lu, L.; Jia, L.; Kabir, M.R.; Zhang, J.; Lan, T.; Zhao, Y.; Xin, M.; Hu, Z.; Yao, Y.; et al. Global QTL Analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 529. [Google Scholar] [CrossRef] [Green Version]
- Juliana, P.; Poland, J.; Huerta-Espino, J.; Shrestha, S.; Crossa, J.; Crespo-Herrera, L.; Toledo, F.H.; Govindan, V.; Mondal, S.; Kumar, U.; et al. Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nat. Genet. 2019, 51, 1530–1539. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Mathews, K.L.; Rattey, A.; Chapman, S.C.; Drenth, J.; Ghaderi, M.; Reynolds, M.; Shorter, R. Molecular detection of genomic regions associated with grain yield and yield-related components in an elite bread wheat cross evaluated under irrigated and rainfed conditions. Theor. Appl. Genet. 2010, 120, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Tura, H.; Edwards, J.; Gahlaut, V.; Garcia, M.; Sznajder, B.; Baumann, U.; Shahinnia, F.; Reynolds, M.; Langridge, P.; Balyan, H.S.; et al. QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments. Theor. Appl. Genet. 2020, 133, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Amo, A.; Wei, D.; Chai, Y.; Zheng, J.; Qiao, P.; Cui, C.; Lu, S.; Chen, L.; Hu, Y.G. Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor. Appl. Genet. 2021, 134, 3083–3109. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mao, X.; Wang, J.; Chang, X.; Reynolds, M.; Jing, R. Genetic dissection of drought and heat-responsive agronomic traits in wheat. Plant Cell Environ. 2019, 42, 2540–2553. [Google Scholar] [CrossRef] [Green Version]
- Bhoite, R.; Onyemaobi, I.; Si, P.; Siddique, K.H.M.; Yan, G. Identification and validation of QTL and their associated genes for pre-emergent metribuzin tolerance in hexaploid wheat (Triticum aestivum L.). BMC Genet. 2018, 19, 102. [Google Scholar] [CrossRef] [Green Version]
- Jemaa, E.; Ammar, S.; Bouzid, S. Effect of temperature on root and shoot development in wheat seedlings during early growth stage. Asian J. Plant Sci. 2010, 9, 375–379. [Google Scholar]
- Medany, M.A.; Hegazy, A.K.; Kabiel, H.A.; Maez, M.M. Prediction of seed germination growth of four crop plants as affected by root zone temperature. World J. Agric. Res. 2007, 3, 714–720. [Google Scholar]
- Lei, L.; Zhu, X.K.; Wang, S.W.; Zhu, M.R.; Carver, B.F.; Yan, L.L. TaMFT-A1 is associated with seed germination sensitive to temperature in winter wheat. PLoS ONE 2013, 8, e73330. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer; Department of Statistics, North Carolina State University: Raleigh, NC, USA, 2012. [Google Scholar]
- Doerge, R.W.; Churchill, G.A. Permutation tests for multiple loci affecting a quantitative character. Genetics 1996, 142, 285–294. [Google Scholar] [CrossRef]
- Lee, S.; Mian, M.A.R.; McHale, L.K.; Wang, H.; Wijeratne, A.J.; Sneller, C.H.; Dorrance, A.E. Novel quantitative trait loci for partial resistance to Phytophthora sojae in soybean PI 398841. Theor. Appl. Genet. 2013, 126, 1121–1132. [Google Scholar] [CrossRef] [Green Version]
- Brodie, A.; Azaria, J.R.; Ofran, Y. How far from the SNP may the causative genes be? Nucleic Acids Res. 2016, 44, 6046–6054. [Google Scholar] [CrossRef] [PubMed]
| Traits | MSg | MSe | h2 | |||
|---|---|---|---|---|---|---|
| SLc | 25.45 ** | 3.16 | 5.80 | 8.06 | 13.86 | 0.42 |
| RLc | 12.72 ** | 4.24 | 3.24 | 2.99 | 6.24 | 0.52 |
| WLc | 62.77 ** | 4.13 | 15.86 | 15.19 | 31.05 | 0.51 |
| SLh | 57.03 ** | 8.67 | 16.82 | 6.58 | 23.39 | 0.72 |
| RLh | 37.06 ** | 9.58 | 11.07 | 3.87 | 14.93 | 0.74 |
| WLh | 176.43 ** | 11.56 | 53.72 | 15.26 | 68.98 | 0.78 |
| Trait | QTL Name | Position (cm) | Flanking Markers | LOD Score | Additive | Donor | R2 |
|---|---|---|---|---|---|---|---|
| Effect | |||||||
| SLI | QSli.4D | 18.2 | Xmwg634/Xbarc225 | 12.45 | 0.16 | Opata | 0.32 |
| RLI | Qrli.3D | 22.5 | Xbarc8/XksuA6 | 4.68 | 0.09 | Opata | 0.11 |
| Qrli.4D | 18.2 | Xmwg634/Xbarc225 | 11.61 | 0.15 | Opata | 0.33 | |
| Qrli.6D | 60.7 | Xcdo534/Xgwm325 | 3.19 | 0.07 | Opata | 0.07 | |
| WLI | Qwli.4D | 17.2 | Xmwg634/Xbarc225 | 11.72 | 0.15 | Opata | 0.31 |
| SLh | QSlh.4D | 16 | Xmwg634/Xbarc225 | 12.22 | −2.56 | Opata | 0.33 |
| QSlh.5A | 64 | Xbarc151/Xbod183 | 4.1 | 1.38 | Synthetic | 0.09 | |
| QSlh.6B | 61.3 | XksuG30/Xgwm219 | 3.31 | −1.19 | Opata | 0.07 | |
| RLh | QRlh.2D | 21.1 | Xbod611/Xcdo1379 | 4.74 | −1.24 | Opata | 0.11 |
| QRlh.4D | 17.2 | Xmwg634/Xbarc225 | 10.04 | −1.81 | Opata | 0.26 | |
| QRlh.5A | 64 | Xbarc151/Xbod183 | 3.3 | 1 | Synthetic | 0.08 | |
| WLh | QWlh.4D | 17 | Xmwg634/Xbarc225 | 12.32 | −4.35 | Opata | 0.32 |
| QWlh.5A | 64 | Xbarc151/Xbod183 | 4.39 | 2.47 | Synthetic | 0.1 | |
| SLc | QSlc.2B | 87.31 | Xcdo678/Xmwg660 | 3.59 | 0.91 | Synthetic | 0.1 |
| RLc | QRlc.2A | 100.31 | Xbarc353/Xgwm356 | 3.59 | 0.57 | Synthetic | 0.08 |
| QRlc.2B | 51.31 | Xgwm191/Xbcd1779 | 3.74 | 0.54 | Synthetic | 0.08 | |
| QRlc.5A | 87.41 | Xbarc319/Xabg366 | 6.58 | 0.81 | Synthetic | 0.18 | |
| WLc | QWlc.2A | 100.3 | Xbarc353/Xgwm356 | 5.17 | 1.56 | Synthetic | 0.13 |
| QWlc.2B | 47.11 | Xbcd445/Xcnl6 | 3.52 | 1.24 | Synthetic | 0.09 | |
| QWlc.5A | 77.71 | Xrz395/Xbarc230 | 5.33 | 1.87 | Synthetic | 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Liu, H.; Wu, Y.; Yan, G. Identification and Validation of a Chromosome 4D Quantitative Trait Locus Hotspot Conferring Heat Tolerance in Common Wheat (Triticum aestivum L.). Plants 2022, 11, 729. https://doi.org/10.3390/plants11060729
Lu L, Liu H, Wu Y, Yan G. Identification and Validation of a Chromosome 4D Quantitative Trait Locus Hotspot Conferring Heat Tolerance in Common Wheat (Triticum aestivum L.). Plants. 2022; 11(6):729. https://doi.org/10.3390/plants11060729
Chicago/Turabian StyleLu, Lu, Hui Liu, Yu Wu, and Guijun Yan. 2022. "Identification and Validation of a Chromosome 4D Quantitative Trait Locus Hotspot Conferring Heat Tolerance in Common Wheat (Triticum aestivum L.)" Plants 11, no. 6: 729. https://doi.org/10.3390/plants11060729
APA StyleLu, L., Liu, H., Wu, Y., & Yan, G. (2022). Identification and Validation of a Chromosome 4D Quantitative Trait Locus Hotspot Conferring Heat Tolerance in Common Wheat (Triticum aestivum L.). Plants, 11(6), 729. https://doi.org/10.3390/plants11060729






