Can Artificial Intelligence Help in the Study of Vegetative Growth Patterns from Herbarium Collections? An Evaluation of the Tropical Flora of the French Guiana Forest
Abstract
:1. Introduction
- Do herbaria contain relevant information for the study of tropical tree growth? If so, to what extent?
- Can deep learning-based automated approaches detect growing specimens?
- If so, which approaches are most relevant and which growth patterns are best detected?
2. Results
2.1. Herbarium Data
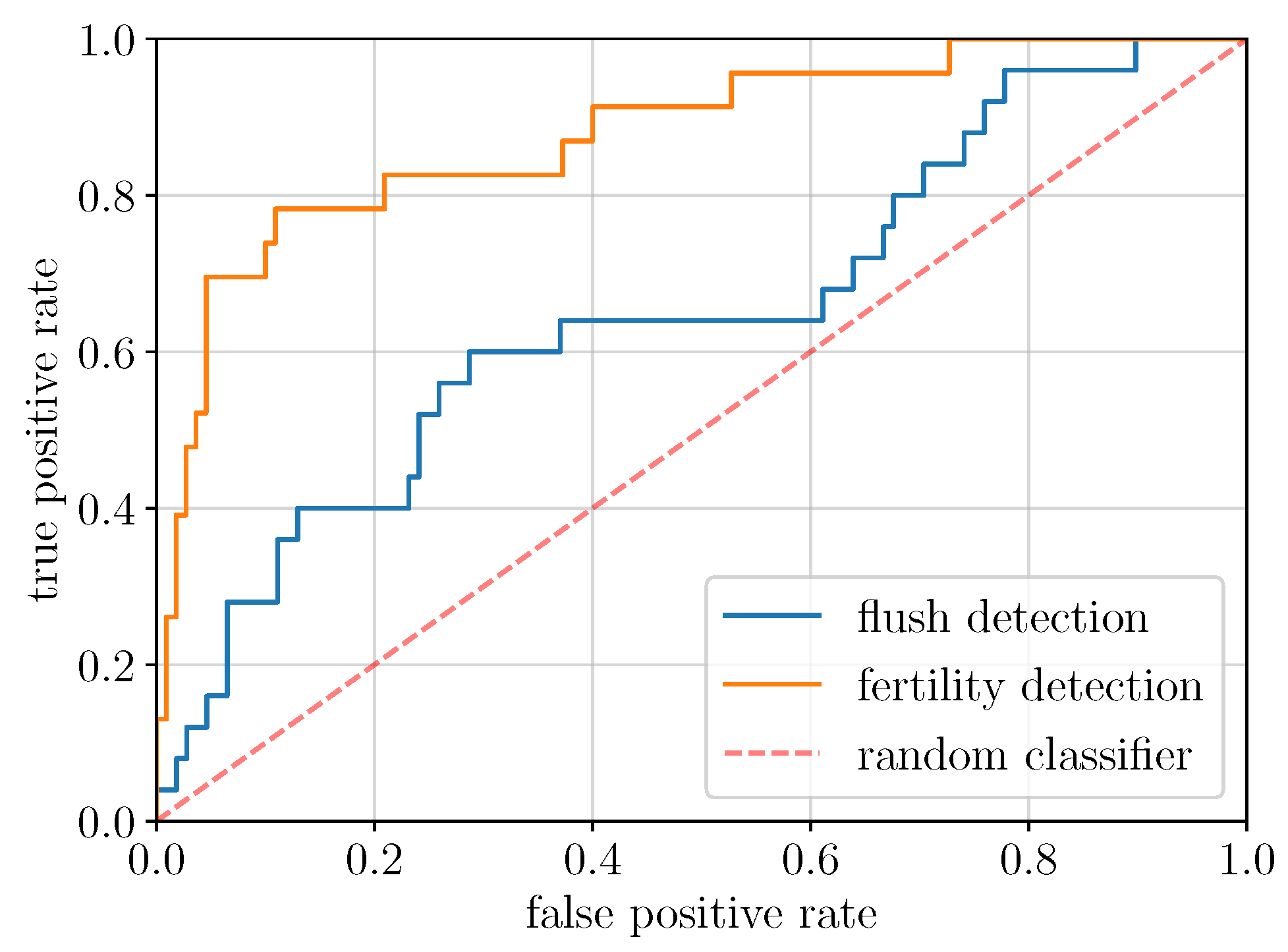
2.2. Global Model Detection Performance (EXP1, ResNet50)
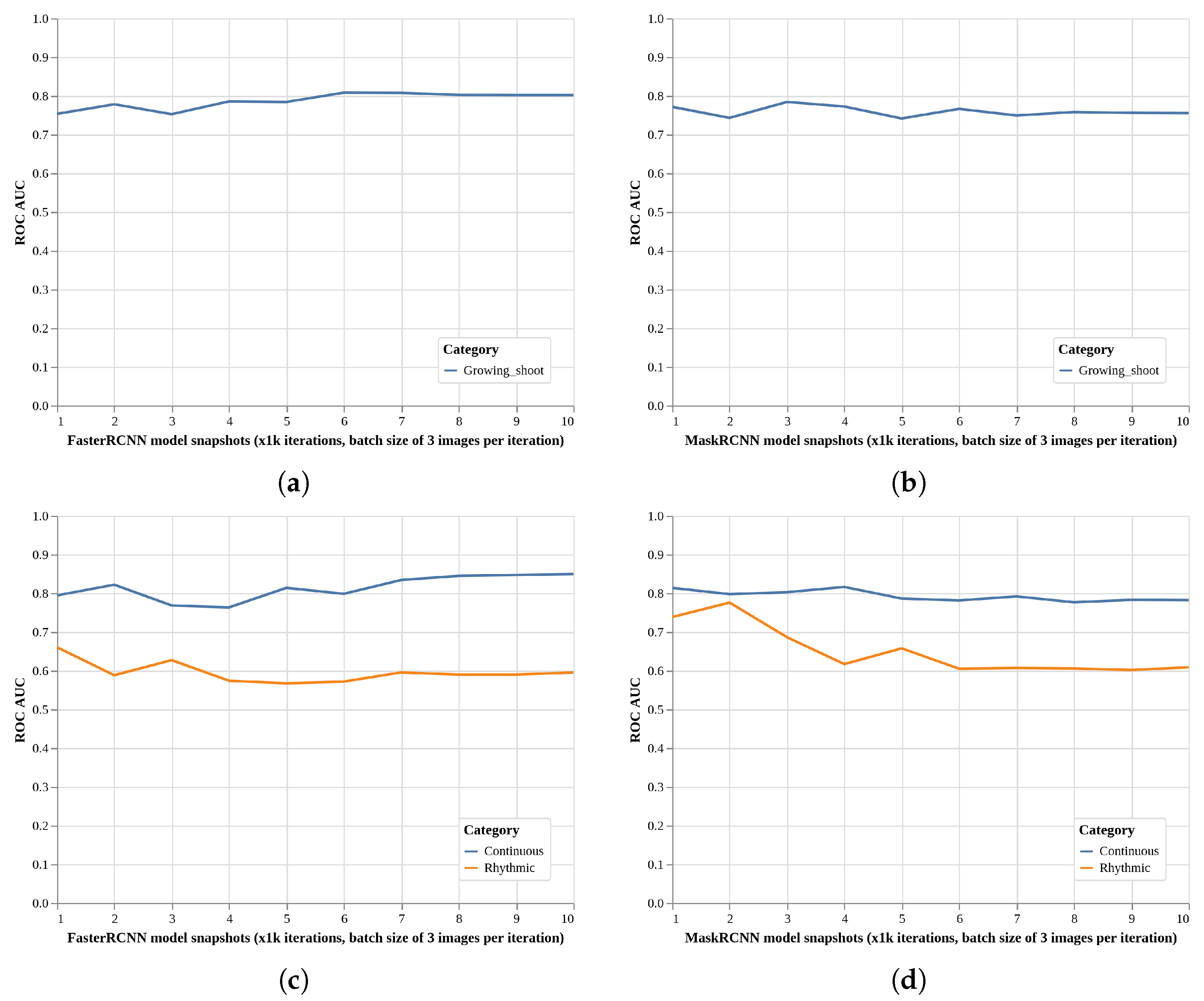
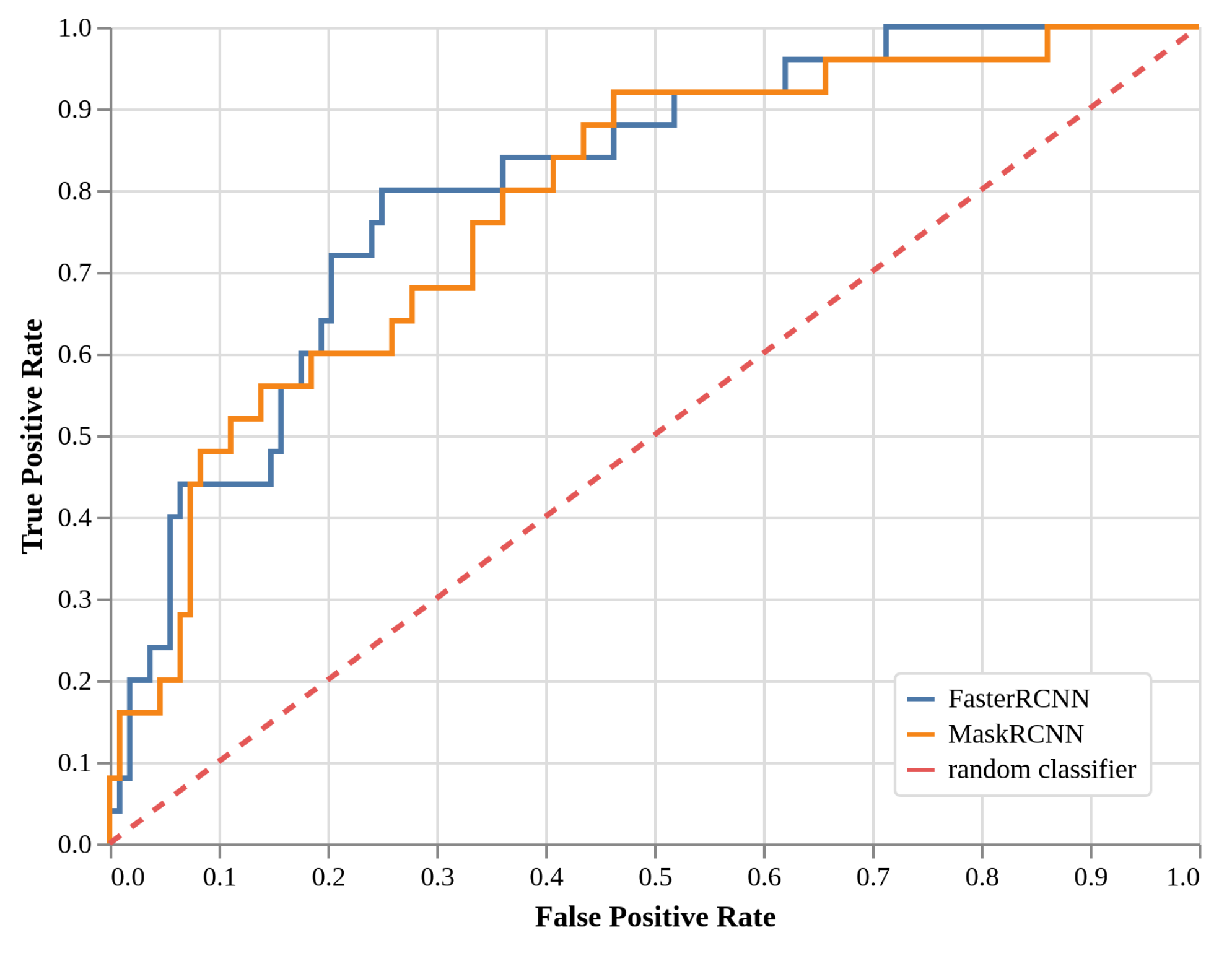
2.3. Local Models Detection Performance (EXP2, Faster R-CNN, and Mask R-CNN)
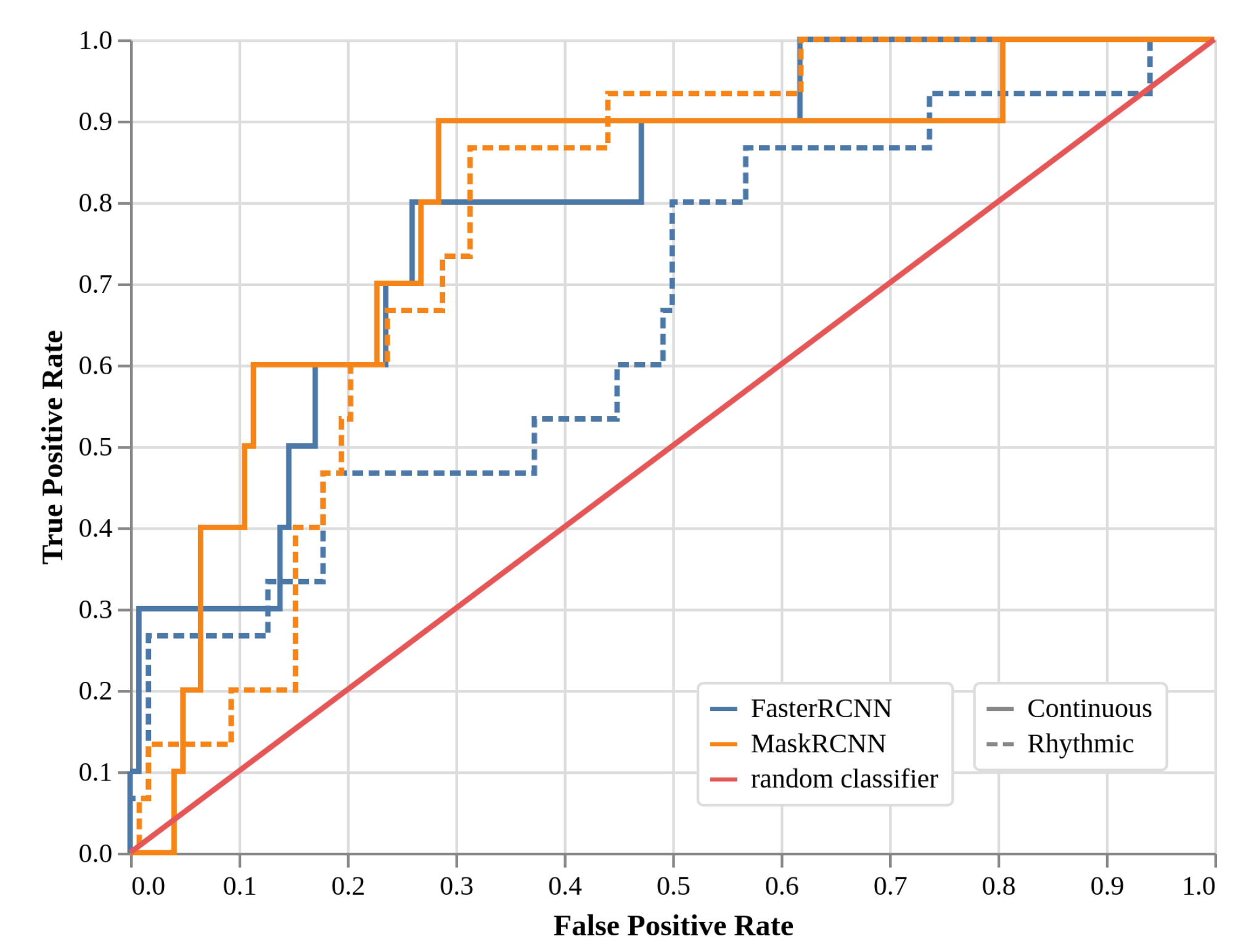
2.4. Local Models Two-Class Detection Performance (EXP3, Faster R-CNN, and Mask R-CNN)
3. Discussion
- Collect more specimens with growing shoots in addition to those with reproductive structures;
- Organize the specimens on the herbarium sheet in order to better visualize the ends of the axes, and to avoid leaf overlaps;
- Annotate the presence or absence of new growing shoots.
4. Materials and Methods
4.1. Assembling Herbarium Records and Manual Annotations
- “FullImage” level. All herbarium sheets used in this study were associated with a tag (Yes or No) indicating if the sheet had at least one recent growing shoot visible or not. No information on the location, number, or size of recent growing shoots was recorded.
- “Mask” level. In each herbarium sheet containing one or several recent growing shoots, each shoot was manually annotated and cross-validated by two co-authors using COCO Annotator (https://github.com/jsbroks/coco-annotator accessed on 10 May 2021) a web-based image annotation tool designed to efficiently label images and create training data for object detection tasks (see Figure 6). This allows for the precise capture of their full shape. It should be noted that it was not uncommon for growing shoots to be partially overlapped by other plant parts (leaves of adult sizes, branches, petioles) or sometimes by pieces of tape. In this case, only the visible part in the foreground was annotated, thus generating a mask potentially based on several disjointed polygons for a single growing shoot.
- “BoundingBox” level. Once the masks were created, the bounding boxes were directly deduced from the masks based on the min and max coordinates in x and y, respectively, for each mask. This gives us rectangles that capture both the growing shoots and the surrounding visual context such as parts of stems, leaves, textual description, and paper.
4.2. Preliminary Analysis of the Annotations
4.3. Description of Experiments
- Detection: What is the performance of automatic growing shoot detection in herbaria? Do we obtain performances comparable to the automatic detection of reproductive structures? With what proportions of missed growing shoots and detection errors?
- Detection and classification: Is it possible to both detect and classify different types of growing patterns (“Continuous” or “Rhythmic”) automatically?
4.4. Evaluated Deep Learning Architectures
- Global model (ResNet50). The first deep learning model that was trained is the Convolutional Neural Network (CNN) ResNet50 [59]. It was pretrained on the ImageNet dataset [60] and fine-tuned on our training dataset. ResNet50 is widely used in image classification tasks and research works for its good compromise between performance, memory use, and training time. Moreover, ResNet50 is often preferred to other recent architectures for a wide range of application studies because its architecture is rather simple, and it is relatively easy to find training hyperparameters that produce good and stable results. A CNN produces as outputs a list of classification scores (probabilities) related to the considered categories, but without any information about the sub-parts of the image that contributed to the prediction. Pre-trained models can be easily found for most Deep Learning frameworks, particularly for PyTorch (https://pytorch.org/, accessed on 10 May 2021), which was used for our experiments. Details on this model adaptation, data augmentation strategy, and the used hyperparameters are provided in Appendix A.
- Local model (Faster R-CNN). The second model that we evaluated is based on the Faster R-CNN architecture [61], which was chosen for its demonstrated efficiency in various object detection tasks and challenges such as MS COCO [62]. A trained Faster R-CNN model produces as outputs a list of bounding boxes associated with probabilities related to the considered categories for detection. We used the Detectron2 implementation [63] itself using the PyTorch framework, based on ResNet50 as the backbone CNN and the Feature Pyramid Network [64] as the Region Proposal Network for object detection. The total number of training iterations was made based on the empirical observation of the model’s training performance. A detailed description of the hyperparameters that were used to train the model is provided in Appendix B.
- Local model (Mask R-CNN). The third model that we evaluated is based on the Mask R-CNN architecture [65], which was chosen for its ability to perform an instance segmentation task by extending the Faster R-CNN approach to a pixel-level mask prediction task. A trained Mask R-CNN model produces as outputs a list of polygon sets, each associated with probabilities related to the considered categories. As for the Faster R-CNN, we used the Detectron2 implementation [63], using by default the same backbone ResNet50 and the Feature Pyramid Networks as for Faster R-CNN. A detailed description of the hyperparameters that were used to train the model is provided in Appendix B.
4.5. Assessing Raw Performances of Deep Learning Models
4.5.1. Training and Test Datasets
4.5.2. Tasks
- EXP1: Detection, Global model (ResNet50)
- EXP2: Detection, Local models (Faster R-CNN, Mask R-CNN)
- EXP3: Detection and Classification, Local models (Faster R-CNN, Mask R-CNN)
4.5.3. Metrics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TPR | True Positive Rate |
| FPR | False Positive Rate |
| ROC | Receiver Operating Characteristic |
| ROC AUC | Area Under the Receiver Operating Characteristic Curve |
| CNN | Convolutional Neural Network |
| ResNet | Residual Convolutional Neural Network |
| Faster R-CNN | Faster Region-Based Convolutional Neural Network |
| Mask R-CNN | Mask Region-Based Convolutional Neural Network |
Appendix A. Global Model (ResNet50)
Appendix B. Local Models (Faster R-CNN and Mask R-CNN)
- The images were resized with a minimum size of 1000 pixels;
- The anchor size values were set to [32; 64; 128; 256; 512] with aspect ratios of [0.5; 1; 2];
- An NMS threshold set to 0.7;
- An initial learning rate of 0.002, with a rate decay of 6:2:2 (the initial learning rate value was divided by 10 at six-tenths and eight-tenths of the training);
- Random horizontal flips and random rotations of ±45 degrees as data augmentation;
- Batch size of three images per iteration;
- A maximum of 10,000 iterations (approximately 74 epochs), with snaphots every 1 k iterations to select the best model (see below).
References
- Canadell, J.G.; Raupach, M.R. Managing forests for climate change mitigation. Science 2008, 320, 1456–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleland, E.E.; Chuine, I.; Menzel, A.; Mooney, H.A.; Schwartz, M.D. Shifting plant phenology in response to global change. Trends Ecol. Evol. 2007, 22, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Barthélémy, D.; Caraglio, Y. Plant architecture: A dynamic, multilevel and comprehensive approach to plant form, structure and ontogeny. Ann. Bot. 2007, 99, 375–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viémont, J.D.; Crabbé, J. Dormancy in Plants: From Whole Plant Behaviour to Cellular Control; CABI Publishing: Wallingford, UK, 2000. [Google Scholar]
- Spicer, M.E.; Mellor, H.; Carson, W.P. Seeing beyond the trees: A comparison of tropical and temperate plant growth forms and their vertical distribution. Ecology 2020, 101, e02974. [Google Scholar] [CrossRef] [PubMed]
- Newstrom, L.E.; Frankie, G.W.; Baker, H.G. A new classification for plant phenology based on flowering patterns in lowland tropical rain forest trees at La Selva, Costa Rica. Biotropica 1994, 26, 141–159. [Google Scholar] [CrossRef]
- Nemani, R.R.; Keeling, C.D.; Hashimoto, H.; Jolly, W.M.; Piper, S.C.; Tucker, C.J.; Myneni, R.B.; Running, S.W. Climate-driven increases in global terrestrial net primary production from 1982 to 1999. Science 2003, 300, 1560–1563. [Google Scholar] [CrossRef] [Green Version]
- Huete, A.R.; Didan, K.; Shimabukuro, Y.E.; Ratana, P.; Saleska, S.R.; Hutyra, L.R.; Yang, W.; Nemani, R.R.; Myneni, R. Amazon rainforests green-up with sunlight in dry season. Geophys. Res. Lett. 2006, 33. [Google Scholar] [CrossRef] [Green Version]
- Wagner, F.; Rossi, V.; Stahl, C.; Bonal, D.; Herault, B. Water availability is the main climate driver of neotropical tree growth. PLoS ONE 2012, 7, e34074. [Google Scholar] [CrossRef]
- Wagner, F.H.; Hérault, B.; Bonal, D.; Stahl, C.; Anderson, L.O.; Baker, T.R.; Becker, G.S.; Beeckman, H.; Boanerges Souza, D.; Botosso, P.C.; et al. Climate seasonality limits leaf carbon assimilation and wood productivity in tropical forests. Biogeosciences 2016, 13, 2537–2562. [Google Scholar] [CrossRef] [Green Version]
- Sabatier, D. Saisonnalité et déterminisme du pic de fructification en forêt guyanaise. Rev. d’Ecol. 1985, 40, 289–320. [Google Scholar]
- Loubry, D. Phenology of deciduous trees in a French-Guianan forest (5 degrees latitude north)-case of a determinism with endogenous and exogenous components. Can. J. Bot. 1994, 72, 1843–1857. [Google Scholar] [CrossRef]
- Pennec, A.; Gond, V.; Sabatier, D. Tropical forest phenology in French Guiana from MODIS time series. Remote. Sens. Lett. 2011, 2, 337–345. [Google Scholar] [CrossRef]
- Wagner, F.; Rossi, V.; Stahl, C.; Bonal, D.; Hérault, B. Asynchronism in leaf and wood production in tropical forests: A study combining satellite and ground-based measurements. Biogeosciences 2013, 10, 7307–7321. [Google Scholar]
- Nicolini, E.; Beauchêne, J.; de la Vallée, B.L.; Ruelle, J.; Mangenet, T.; Heuret, P. Dating branch growth units in a tropical tree using morphological and anatomical markers: The case of Parkia velutina Benoist (Mimosoïdeae). Ann. For. Sci. 2012, 69, 543–555. [Google Scholar] [CrossRef] [Green Version]
- Soltis, P.S.; Nelson, G.; Zare, A.; Meineke, E.K. Plants meet machines: Prospects in machine learning for plant biology. Appl. Plant Sci. 2020, 8, e11371. [Google Scholar] [CrossRef]
- Drew, J.A.; Moreau, C.S.; Stiassny, M.L. Digitization of museum collections holds the potential to enhance researcher diversity. Nat. Ecol. Evol. 2017, 1, 1789–1790. [Google Scholar] [CrossRef]
- Willis, C.G.; Ellwood, E.R.; Primack, R.B.; Davis, C.C.; Pearson, K.D.; Gallinat, A.S.; Yost, J.M.; Nelson, G.; Mazer, S.J.; Rossington, N.L.; et al. Old plants, new tricks: Phenological research using herbarium specimens. Trends Ecol. Evol. 2017, 32, 531–546. [Google Scholar] [CrossRef] [Green Version]
- Carine, M.A.; Cesar, E.A.; Ellis, L.; Hunnex, J.; Paul, A.M.; Prakash, R.; Rumsey, F.J.; Wajer, J.; Wilbraham, J.; Yesilyurt, J.C. Examining the spectra of herbarium uses and users. Bot. Lett. 2018, 165, 328–336. [Google Scholar] [CrossRef]
- Younis, S.; Weiland, C.; Hoehndorf, R.; Dressler, S.; Hickler, T.; Seeger, B.; Schmidt, M. Taxon and trait recognition from digitized herbarium specimens using deep convolutional neural networks. Bot. Lett. 2018, 165, 377–383. [Google Scholar] [CrossRef]
- López-Pujol, J.; López-Vinyallonga, S.; Susanna, A.; Ertuğrul, K.; Uysal, T.; Tugay, O.; Guetat, A.; Garcia-Jacas, N. Speciation and genetic diversity in Centaurea subsect. Phalolepis in Anatolia. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Gregor, T. The distribution of Galeopsis ladanum in Germany based on an analysis of herbarium material is smaller than that indicated in plant atlases. Preslia 2009, 84, 377–386. [Google Scholar]
- Geri, F.; Lastrucci, L.; Viciani, D.; Foggi, B.; Ferretti, G.; Maccherini, S.; Bonini, I.; Amici, V.; Chiarucci, A. Mapping patterns of ferns species richness through the use of herbarium data. Biodivers. Conserv. 2013, 22, 1679–1690. [Google Scholar] [CrossRef]
- Nualart, N.; Ibáñez Cortina, N.; Soriano, I.; López-Pujol, J. Assessing the relevance of herbarium collections as tools for conservation biology. Bot. Rev. 2020, 83, 303–325. [Google Scholar] [CrossRef]
- Borchert, R. Phenology and flowering periodicity of Neotropical dry forest species: Evidence from herbarium collections. J. Trop. Ecol. 1996, 12, 65–80. [Google Scholar] [CrossRef]
- MacGillivray, F.; Hudson, I.L.; Lowe, A.J. Herbarium collections and photographic images: Alternative data sources for phenological research. In Phenological Research; Springer: Dordrecht, The Netherlands, 2010; pp. 425–461. [Google Scholar]
- Zalamea, P.C.; Munoz, F.; Stevenson, P.R.; Paine, C.T.; Sarmiento, C.; Sabatier, D.; Heuret, P. Continental-scale patterns of Cecropia reproductive phenology: Evidence from herbarium specimens. Proc. R. Soc. B Biol. Sci. 2011, 278, 2437–2445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C.C.; Willis, C.G.; Connolly, B.; Kelly, C.; Ellison, A.M. Herbarium records are reliable sources of phenological change driven by climate and provide novel insights into species’ phenological cueing mechanisms. Am. J. Bot. 2015, 102, 1599–1609. [Google Scholar] [CrossRef]
- Feeley, K.J. Distributional migrations, expansions, and contractions of tropical plant species as revealed in dated herbarium records. Glob. Chang. Biol. 2012, 18, 1335–1341. [Google Scholar] [CrossRef]
- Lavoie, C. Biological collections in an ever changing world: Herbaria as tools for biogeographical and environmental studies. Perspect. Plant Ecol. Evol. Syst. 2013, 15, 68–76. [Google Scholar] [CrossRef]
- Primack, D.; Imbres, C.; Primack, R.B.; Miller-Rushing, A.J.; Del Tredici, P. Herbarium specimens demonstrate earlier flowering times in response to warming in Boston. Am. J. Bot. 2004, 91, 1260–1264. [Google Scholar] [CrossRef]
- Miller-Rushing, A.J.; Primack, R.B.; Primack, D.; Mukunda, S. Photographs and herbarium specimens as tools to document phenological changes in response to global warming. Am. J. Bot. 2006, 93, 1667–1674. [Google Scholar] [CrossRef]
- Gallagher, R.; Hughes, L.; Leishman, M. Phenological trends among Australian alpine species: Using herbarium records to identify climate-change indicators. Aust. J. Bot. 2009, 57, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Park, I.W. Digital herbarium archives as a spatially extensive, taxonomically discriminate phenological record; a comparison to MODIS satellite imagery. Int. J. Biometeorol. 2012, 56, 1179–1182. [Google Scholar] [CrossRef] [PubMed]
- Diskin, E.; Proctor, H.; Jebb, M.; Sparks, T.; Donnelly, A. The phenology of Rubus fruticosus in Ireland: Herbarium specimens provide evidence for the response of phenophases to temperature, with implications for climate warming. Int. J. Biometeorol. 2012, 56, 1103–1111. [Google Scholar] [CrossRef] [PubMed]
- Calinger, K.M.; Queenborough, S.; Curtis, P.S. Herbarium specimens reveal the footprint of climate change on flowering trends across north-central North America. Ecol. Lett. 2013, 16, 1037–1044. [Google Scholar] [CrossRef] [Green Version]
- Park, I.W.; Schwartz, M.D. Long-term herbarium records reveal temperature-dependent changes in flowering phenology in the southeastern USA. Int. J. Biometeorol. 2015, 59, 347–355. [Google Scholar] [CrossRef]
- Willis, C.G.; Law, E.; Williams, A.C.; Franzone, B.F.; Bernardos, R.; Bruno, L.; Hopkins, C.; Schorn, C.; Weber, E.; Park, D.S.; et al. CrowdCurio: An online crowdsourcing platform to facilitate climate change studies using herbarium specimens. New Phytol. 2017, 215, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Brenskelle, L.; Stucky, B.J.; Deck, J.; Walls, R.; Guralnick, R.P. Integrating herbarium specimen observations into global phenology data systems. Appl. Plant Sci. 2019, 7, e01231. [Google Scholar] [CrossRef] [Green Version]
- Meineke, E.K.; Davies, T.J. Museum specimens provide novel insights into changing plant–herbivore interactions. Philos. Trans. R. Soc. B 2019, 374, 20170393. [Google Scholar] [CrossRef] [Green Version]
- Beaulieu, C.; Lavoie, C.; Proulx, R. Bookkeeping of insect herbivory trends in herbarium specimens of purple loosestrife (Lythrum salicaria). Philos. Trans. R. Soc. B 2019, 374, 20170398. [Google Scholar] [CrossRef] [Green Version]
- Bonal, D.; Ponton, S.; Le Thiec, D.; Richard, B.; Ningre, N.; Hérault, B.; Ogée, J.; Gonzalez, S.; Pignal, M.; Sabatier, D.; et al. Leaf functional response to increasing atmospheric CO2 concentrations over the last century in two northern Amazonian tree species: A historical δ13C and δ18O approach using herbarium samples. Plant Cell Environ. 2011, 34, 1332–1344. [Google Scholar] [CrossRef]
- Daru, B.H.; Bowman, E.A.; Pfister, D.H.; Arnold, A.E. A novel proof of concept for capturing the diversity of endophytic fungi preserved in herbarium specimens. Philos. Trans. R. Soc. B 2019, 374, 20170395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, P.S. Digitization of herbaria enables novel research. Am. J. Bot. 2017, 104, 1281–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corney, D.P.; Clark, J.Y.; Tang, H.L.; Wilkin, P. Automatic extraction of leaf characters from herbarium specimens. Taxon 2012, 61, 231–244. [Google Scholar] [CrossRef]
- Younis, S.; Schmidt, M.; Weiland, C.; Dressler, S.; Seeger, B.; Hickler, T. Detection and annotation of plant organs from digitised herbarium scans using deep learning. Biodivers. Data J. 2020, 8, e57090. [Google Scholar] [CrossRef] [PubMed]
- Unger, J.; Merhof, D.; Renner, S. Computer vision applied to herbarium specimens of German trees: Testing the future utility of the millions of herbarium specimen images for automated identification. BMC Evol. Biol. 2016, 16, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carranza-Rojas, J.; Goeau, H.; Bonnet, P.; Mata-Montero, E.; Joly, A. Going deeper in the automated identification of Herbarium specimens. BMC Evol. Biol. 2017, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lorieul, T.; Pearson, K.D.; Ellwood, E.R.; Goëau, H.; Molino, J.f.; Sweeney, P.W.; Yost, J.M.; Sachs, J.; Mata-Montero, E.; Nelson, G.; et al. Toward a large-scale and deep phenological stage annotation of herbarium specimens: Case studies from temperate, tropical, and equatorial floras. Appl. Plant Sci. 2019, 7, e01233. [Google Scholar] [CrossRef] [Green Version]
- Goëau, H.; Mora-Fallas, A.; Champ, J.; Love, N.L.R.; Mazer, S.J.; Mata-Montero, E.; Joly, A.; Bonnet, P. A new fine-grained method for automated visual analysis of herbarium specimens: A case study for phenological data extraction. Appl. Plant Sci. 2020, 8, e11368. [Google Scholar] [CrossRef]
- Davis, C.C.; Champ, J.; Park, D.S.; Breckheimer, I.; Lyra, G.M.; Xie, J.; Joly, A.; Tarapore, D.; Ellison, A.M.; Bonnet, P. A new method for counting reproductive structures in digitized herbarium specimens using Mask R-CNN. Front. Plant Sci. 2020, 11, 1129. [Google Scholar] [CrossRef]
- Love, N.L.; Bonnet, P.; Goëau, H.; Joly, A.; Mazer, S.J. Machine Learning Undercounts Reproductive Organs on Herbarium Specimens but Accurately Derives Their Quantitative Phenological Status: A Case Study of Streptanthus tortuosus. Plants 2021, 10, 2471. [Google Scholar] [CrossRef]
- Mora-Fallas, A.; Goeau, H.; Mazer, S.J.; Love, N.; Mata-Montero, E.; Bonnet, P.; Joly, A. Accelerating the Automated Detection, Counting and Measurements of Reproductive Organs in Herbarium Collections in the Era of Deep Learning. Biodivers. Inf. Sci. Stand. 2019, 3, e37341. [Google Scholar] [CrossRef]
- Gonzalez, S.; Bilot-Guérin, V.; Delprete, P.G.; Geniez, C.; Molino, J.-F.; Smock, J.-L. L’herbier IRD de Guyane. Available online: https://herbier-guyane.ird.fr/ (accessed on 10 May 2021).
- Guitet, S.; Euriot, S.; Brunaux, O.; Baraloto, C.; Denis, T.; Dewynter, M.; Freycon, V.; Gonzales, S.; Jaouen, G.; Hansen, C.R.; et al. Catalogue des Habitats Forestiers de Guyane; National Forests Office (ONF): Guyane, France, 2015.
- Halle, F.; Martin, R. Study of the growth rhythm in Hevea brasiliensis (Euphorbiaceae Cronoideae). Andansonia 1968, 8, 475–503. [Google Scholar]
- Schoonderwoerd, K.M.; Friedman, W.E. Naked resting bud morphologies and their taxonomic and geographic distributions in temperate, woody floras. New Phytol. 2021, 232, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Liang, D.; Zhang, S.; Huang, X.; Li, B.; Hu, S. Traffic-sign detection and classification in the wild. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2110–2118. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.Y.; Maire, M.; Belongie, S.; Bourdev, L.; Girshick, R.; Hays, J.; Perona, P.; Ramanan, D.; Zitnick, C.L.; Dollár, P. Microsoft COCO: Common Objects in Context; Springer: Cham, Switzerland, 2014. [Google Scholar]
- Wu, Y.; Kirillov, A.; Massa, F.; Lo, W.Y.; Girshick, R. Detectron2. 2019. Available online: https://github.com/facebookresearch/detectron2 (accessed on 10 May 2021).
- Lin, T.; Dollár, P.; Girshick, R.B.; He, K.; Hariharan, B.; Belongie, S.J. Feature Pyramid Networks for Object Detection. arXiv 2016, arXiv:1612.03144. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R.B. Mask R-CNN. arXiv 2017, arXiv:1703.06870. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 38, 142–158. [Google Scholar] [CrossRef]
- Uijlings, J.; van de Sande, K.; Gevers, T.; Smeulders, A. Selective Search for Object Recognition. Int. J. Comput. Vis. 2013, 104, 154–171. [Google Scholar] [CrossRef] [Green Version]
- Girshick, R. Fast R-CNN. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar]


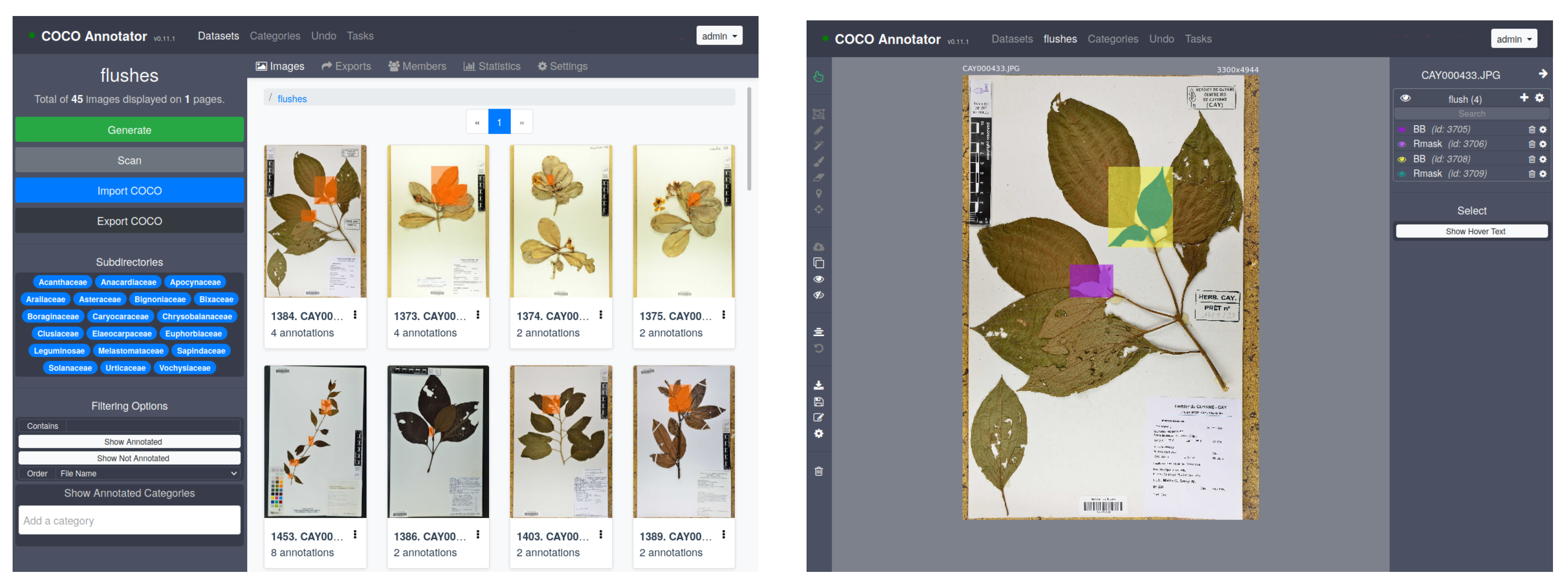
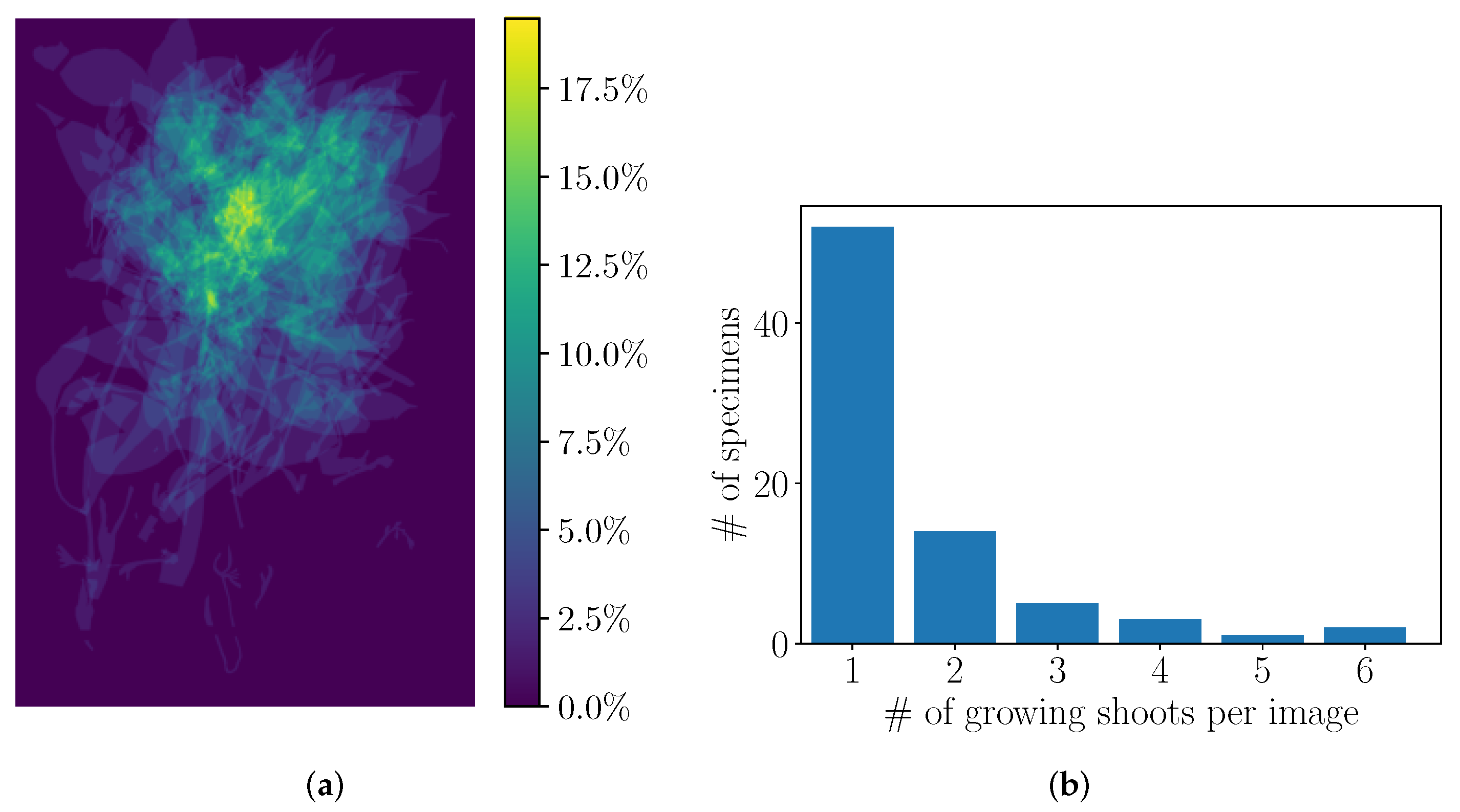
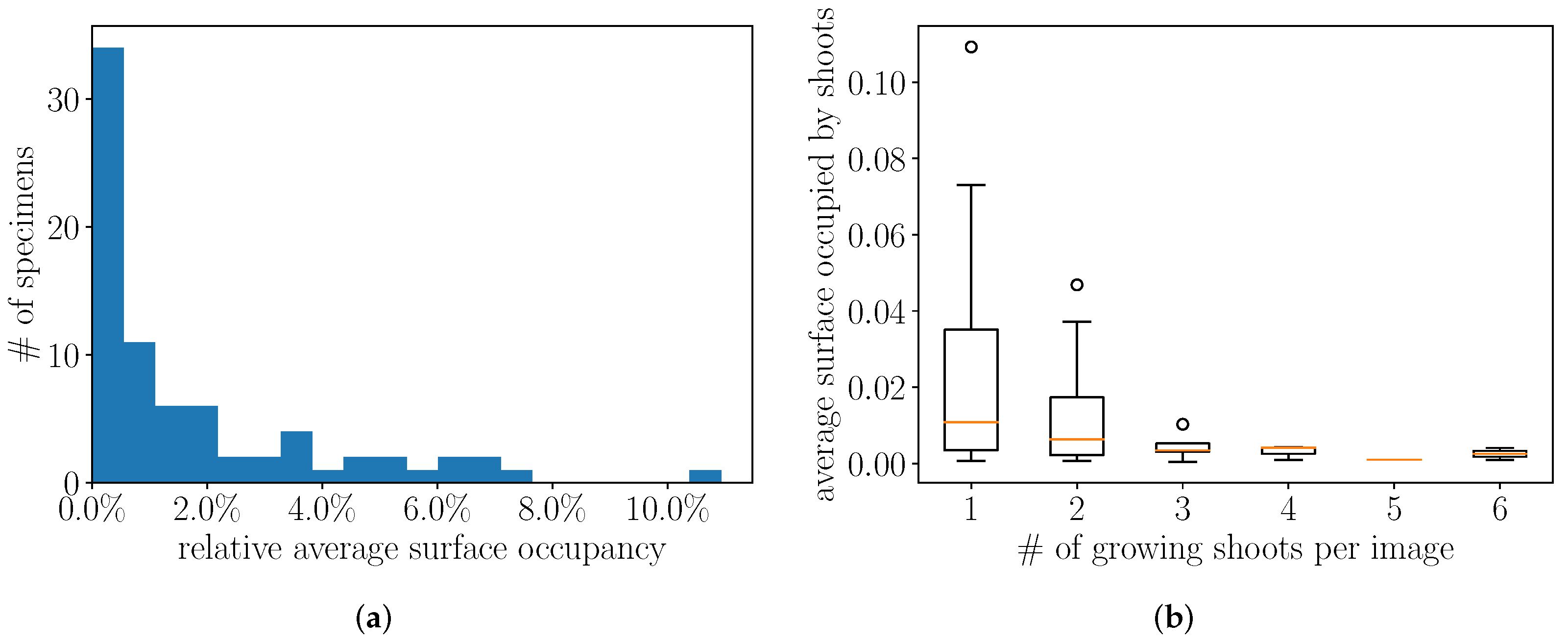
| Images | ||||||||
|---|---|---|---|---|---|---|---|---|
| Set | Fam. | Gen. | Sp. | Obs. | Total | w/Shoots | w/Cont. | w/Rhyt. |
| Total | 25 | 59 | 57 | 352 | 408 | 77 | 32 | 45 |
| Train | 23 | 49 | 42 | 235 | 275 | 52 | 22 | 30 |
| Test | 19 | 34 | 35 | 117 | 133 | 25 | 10 | 15 |
| FPR | ||||
|---|---|---|---|---|
| Model | 0.10 | 0.20 | 0.30 | |
| TPR | ResNet50 (global) | 0.24 | 0.40 | 0.60 |
| Faster R-CNN (local) | 0.44 | 0.80 | ||
| Mask R-CNN (local) | 0.48 | 0.60 | 0.68 | |
| FPR | |||||
|---|---|---|---|---|---|
| Growing Shoot Type | Model | 0.10 | 0.20 | 0.30 | |
| TPR | “Continuous” | Faster R-CNN | 0.30 | 0.60 | 0.80 |
| Mask R-CNN | 0.40 | 0.60 | 0.90 | ||
| “Rhythmic” | Faster R-CNN | 0.27 | 0.47 | 0.47 | |
| Mask R-CNN | 0.20 | 0.53 | 0.73 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goëau, H.; Lorieul, T.; Heuret, P.; Joly, A.; Bonnet, P. Can Artificial Intelligence Help in the Study of Vegetative Growth Patterns from Herbarium Collections? An Evaluation of the Tropical Flora of the French Guiana Forest. Plants 2022, 11, 530. https://doi.org/10.3390/plants11040530
Goëau H, Lorieul T, Heuret P, Joly A, Bonnet P. Can Artificial Intelligence Help in the Study of Vegetative Growth Patterns from Herbarium Collections? An Evaluation of the Tropical Flora of the French Guiana Forest. Plants. 2022; 11(4):530. https://doi.org/10.3390/plants11040530
Chicago/Turabian StyleGoëau, Hervé, Titouan Lorieul, Patrick Heuret, Alexis Joly, and Pierre Bonnet. 2022. "Can Artificial Intelligence Help in the Study of Vegetative Growth Patterns from Herbarium Collections? An Evaluation of the Tropical Flora of the French Guiana Forest" Plants 11, no. 4: 530. https://doi.org/10.3390/plants11040530
APA StyleGoëau, H., Lorieul, T., Heuret, P., Joly, A., & Bonnet, P. (2022). Can Artificial Intelligence Help in the Study of Vegetative Growth Patterns from Herbarium Collections? An Evaluation of the Tropical Flora of the French Guiana Forest. Plants, 11(4), 530. https://doi.org/10.3390/plants11040530







