Accumulation of Salicylic Acid and Related Metabolites in Selaginella moellendorffii
Abstract
:1. Introduction
2. Results and Discussion
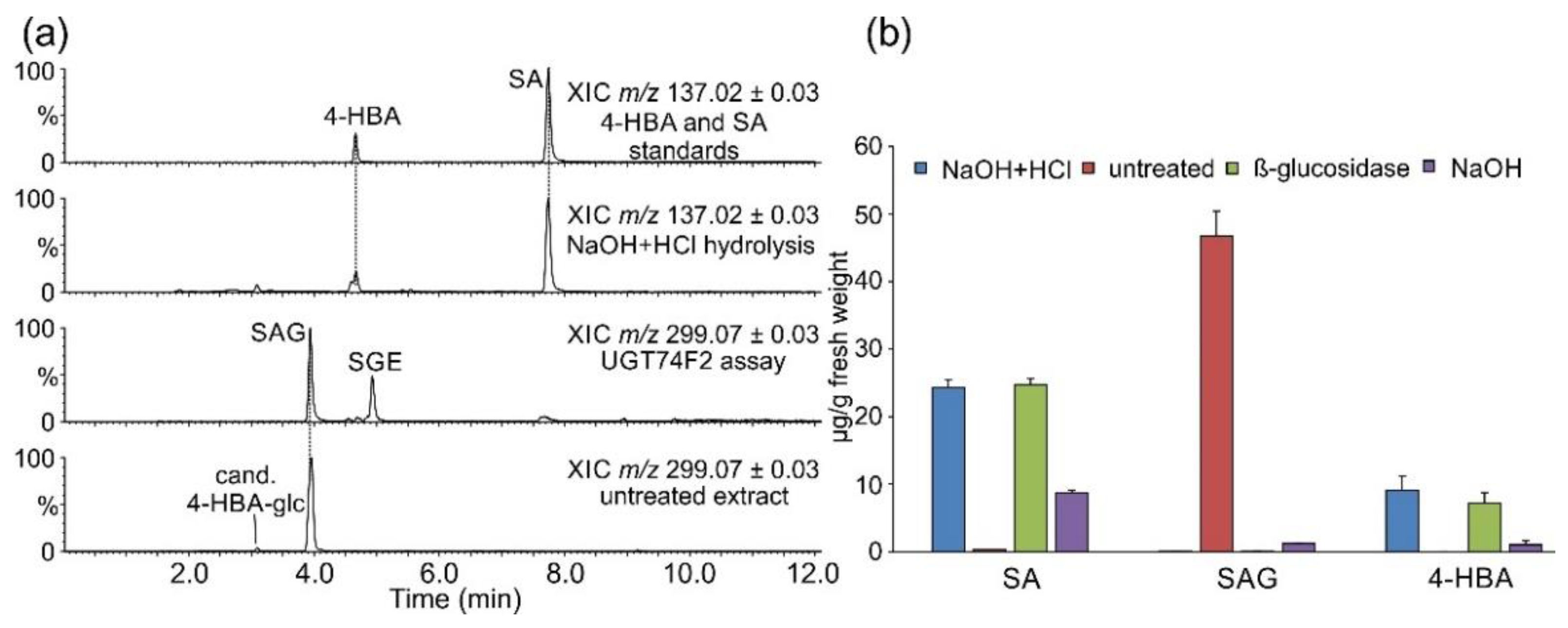
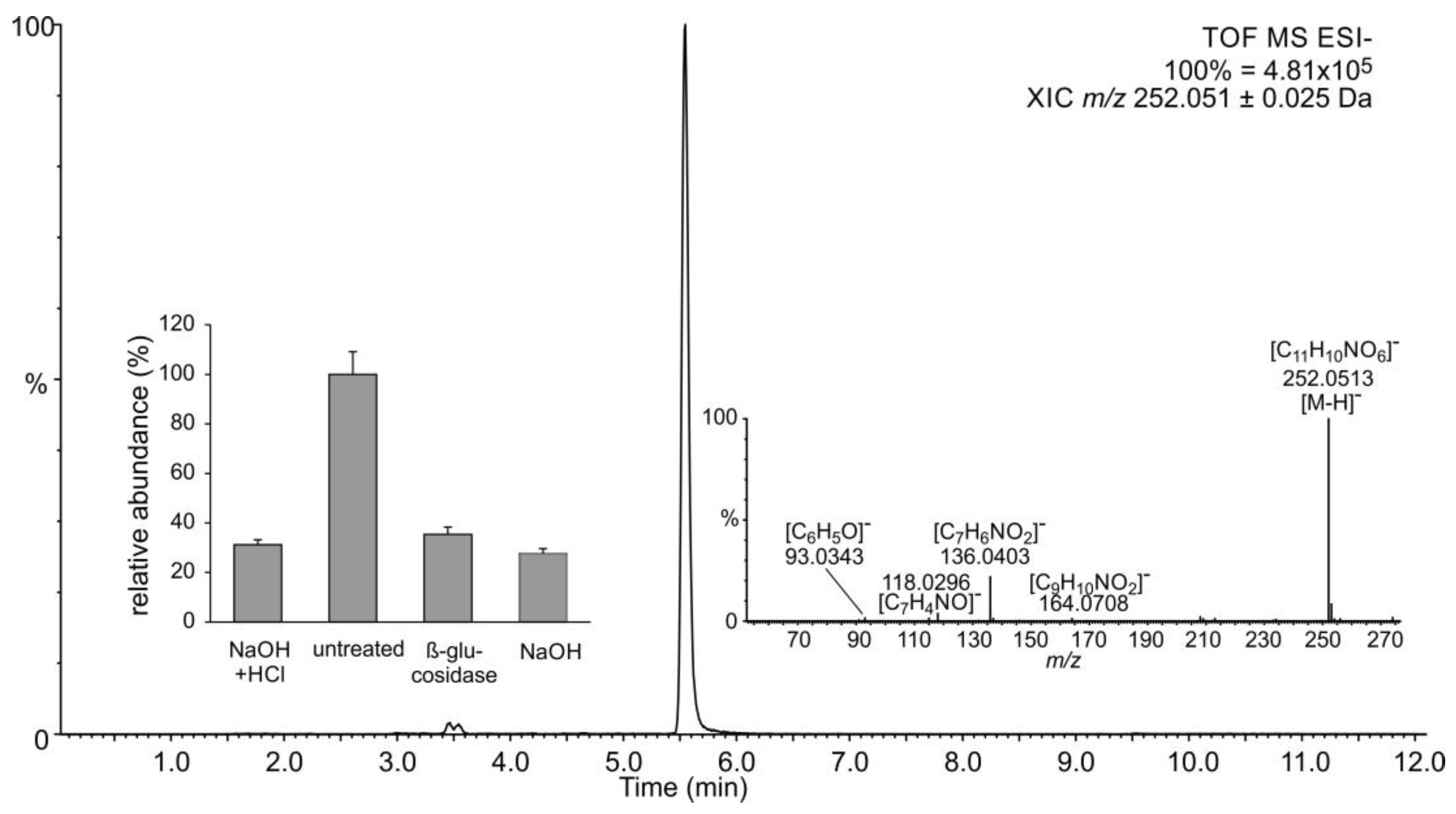
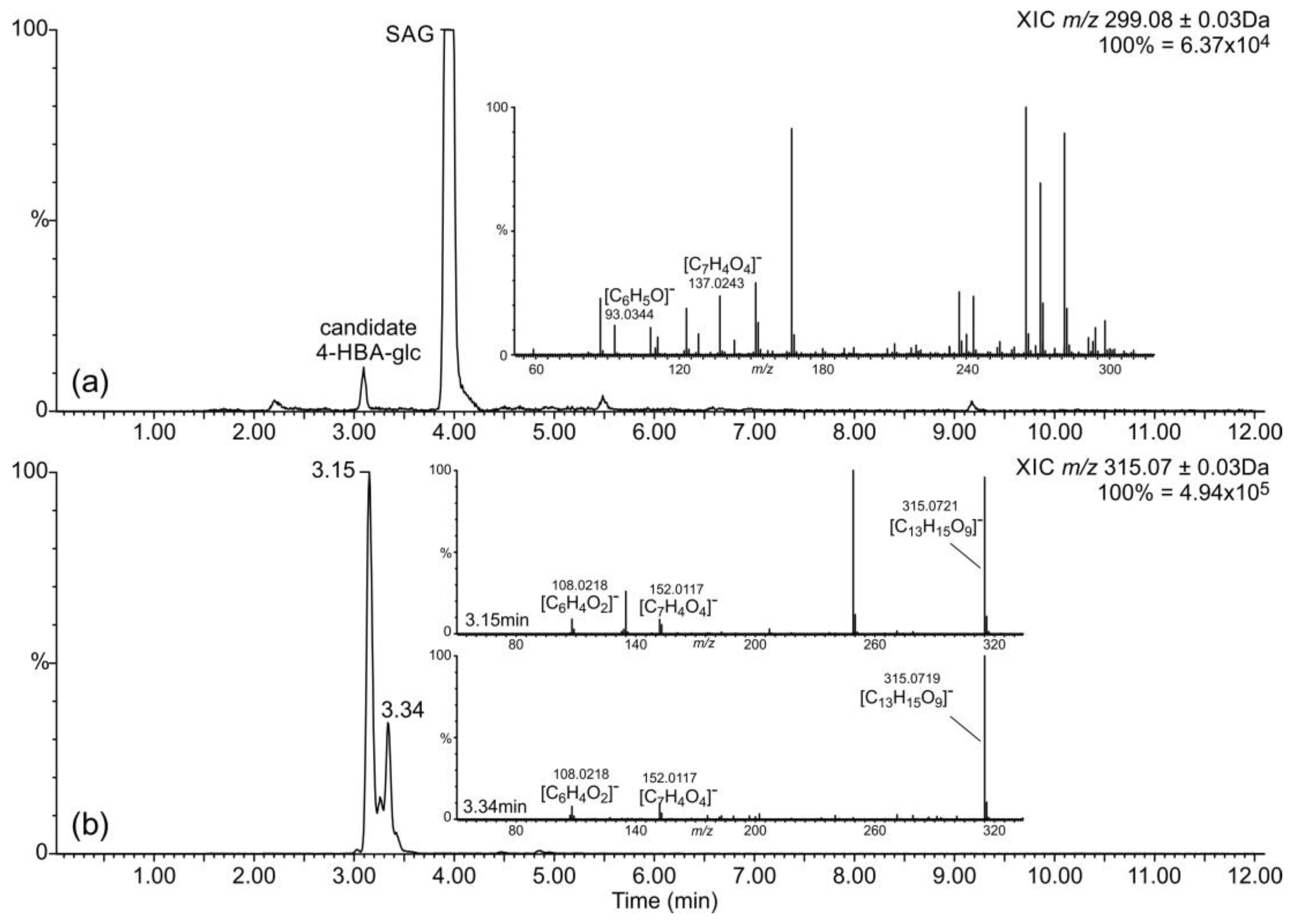
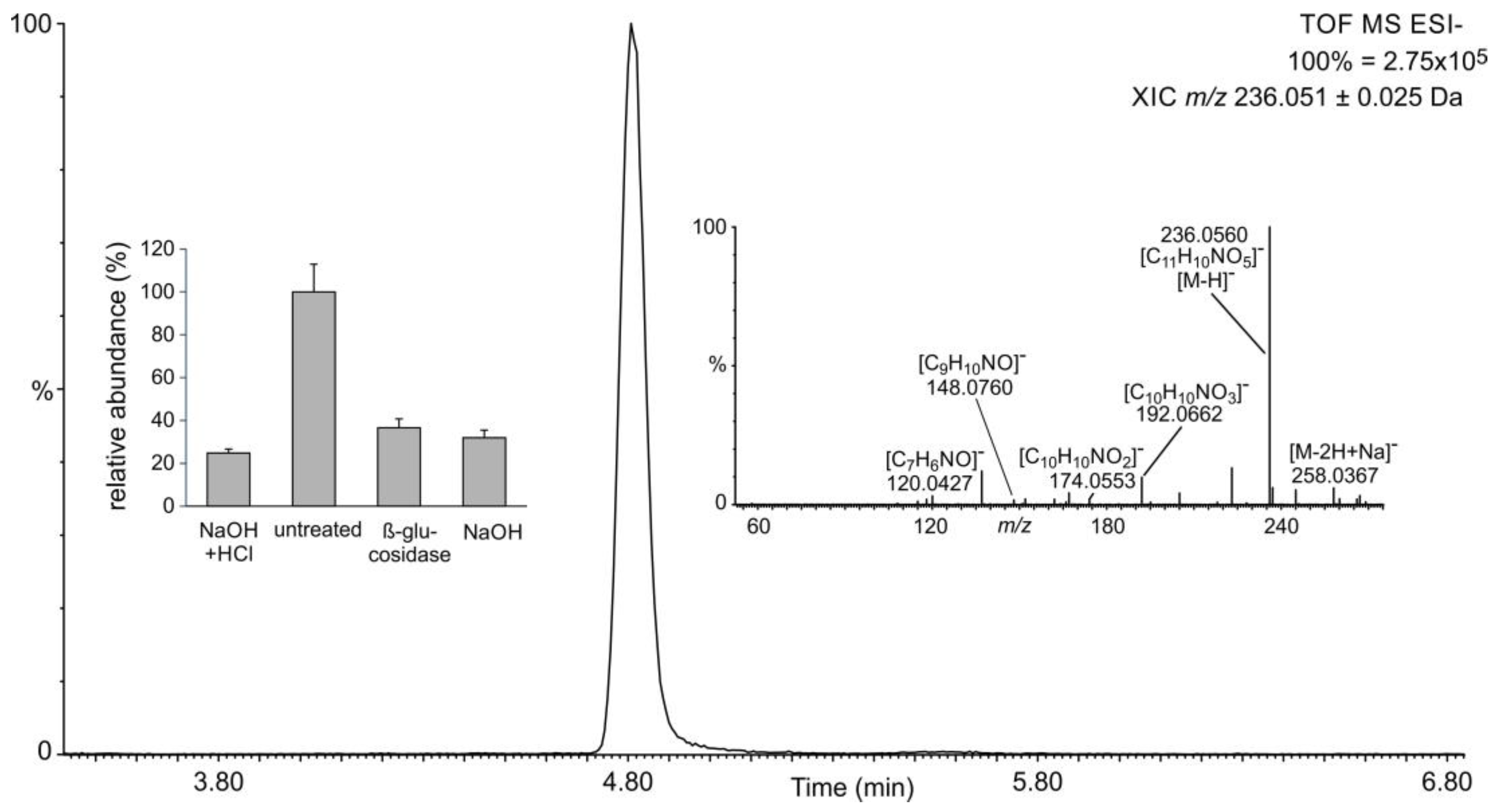
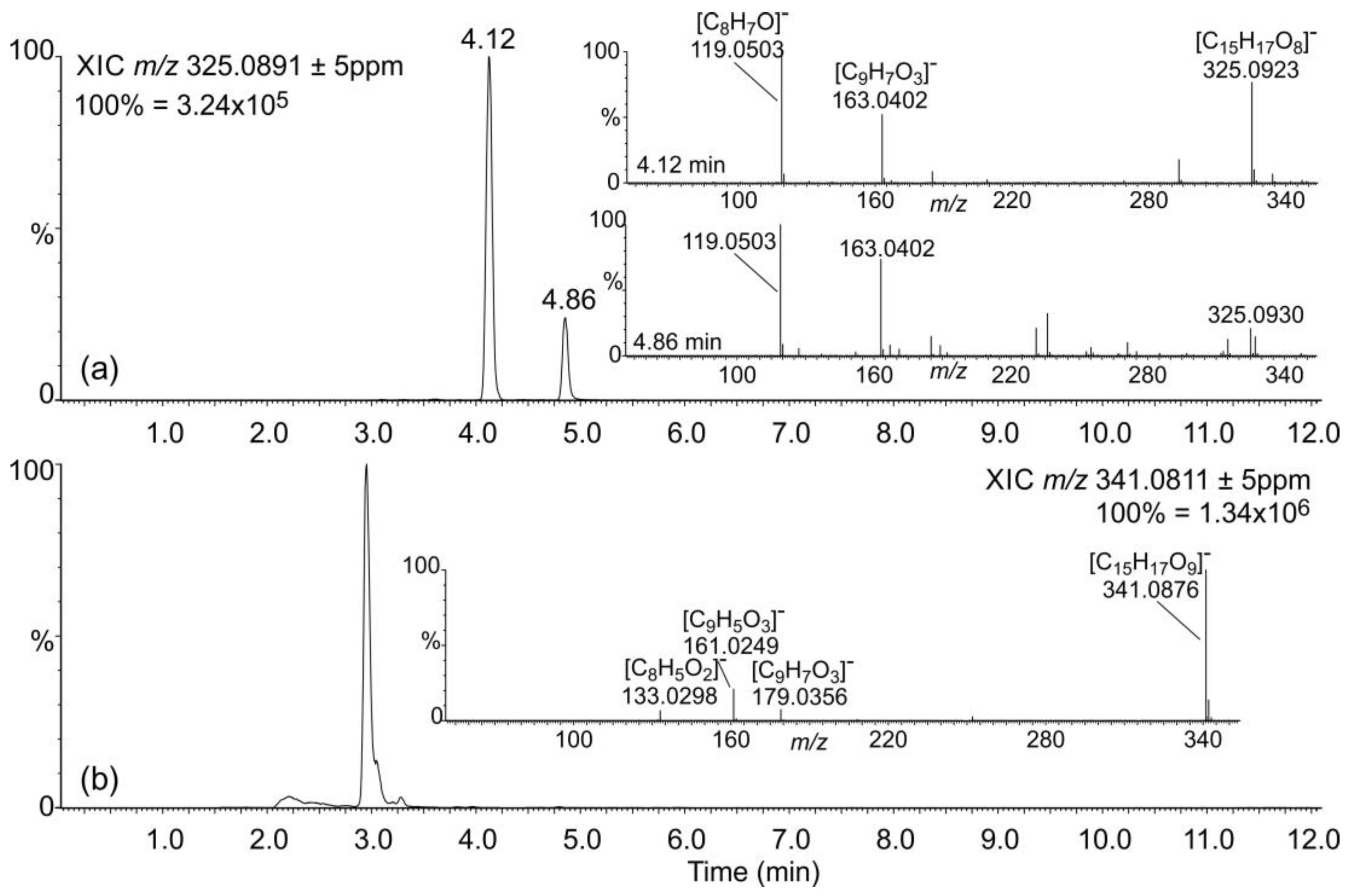
2.1. Occurrence of Salicylic Acid and Its Storage Forms
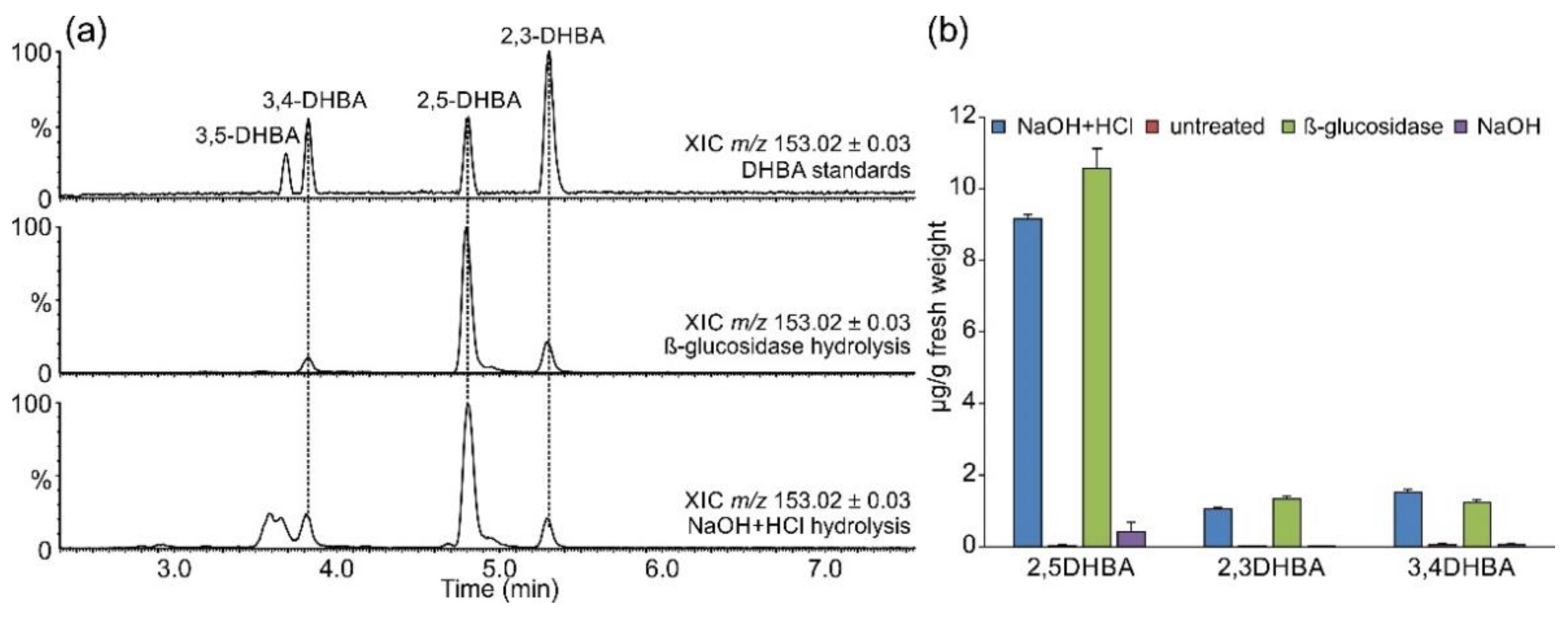
2.2. Occurrence of Related Benzoic Acids
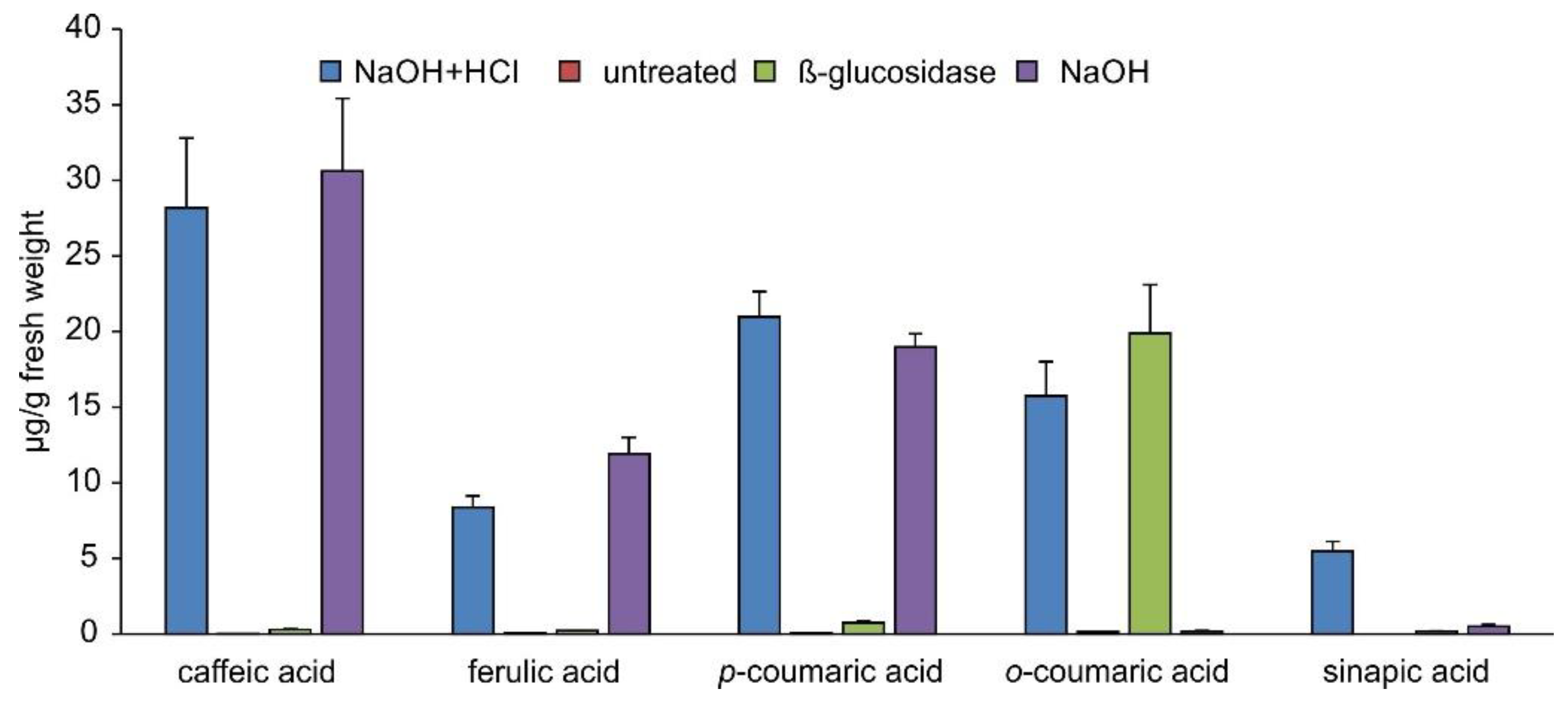
2.3. Occurrence of Phenylpropanoic Acids
2.4. Candidate SA-Modifying Proteins in S. moellendorffii Genome
3. Materials and Methods
3.1. Plant Material
3.2. General Chemicals
3.3. Extract Preparation
3.4. Analysis by UPLC-PDA-qTOF-MS
3.5. Search for Candidate SA-Modifying Proteins
3.6. Enzymatic Production of Authentic SGE
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banks, J.A. Selaginella and 400 Million Years of Separation. Annu. Rev. Plant Biol. 2009, 60, 223–238. [Google Scholar] [CrossRef]
- Shmakov, A. A community-derived classification for extant lycophytes and ferns. J. Syst. Evol. 2016, 54, 563–603. [Google Scholar] [CrossRef]
- Schmidt, A.R.; Regalado, L.; Weststrand, S.; Korall, P.; Sadowski, E.; Schneider, H.; Jansen, E.; Bechteler, J.; Krings, M.; Müller, P.; et al. Selaginella was hyperdiverse already in the Cretaceous. New Phytol. 2020, 228, 1176–1182. [Google Scholar] [CrossRef] [Green Version]
- Banks, J.A.; Nishiyama, T.; Hasebe, M.; Bowman, J.L.; Gribskov, M.; Depamphilis, C.; Albert, V.A.; Aono, N.; Aoyama, T.; Ambrose, B.A.; et al. The Selaginella Genome Identifies Genetic Changes Associated with the Evolution of Vascular Plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef] [Green Version]
- Spencer, V.; Venza, Z.N.; Harrison, C.J. What can lycophytes teach us about plant evolution and development? Modern perspectives on an ancient lineage. Evol. Dev. 2020, 23, 174–196. [Google Scholar] [CrossRef]
- Weng, J.-K.; Noel, J.P. Chemodiversity in Selaginella: A reference system for parallel and convergent metabolic evolution in terrestrial plants. Front. Plant Sci. 2013, 4, 119. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Xin, T.; Bartels, D.; Li, Y.; Gu, W.; Yao, H.; Liu, S.; Yu, H.; Pu, X.; Zhou, J.; et al. Genome Analysis of the Ancient Tracheophyte Selaginella tamariscina Reveals Evolutionary Features Relevant to the Acquisition of Desiccation Tolerance. Mol. Plant 2018, 11, 983–994. [Google Scholar] [CrossRef] [Green Version]
- VanBuren, R.; Wai, C.M.; Doug, B.; Pardo, J.; Bryant, D.; Jiang, N.; Mockler, T.C.; Edger, P.; Michael, T.P. Extreme haplotype variation in the desiccation-tolerant clubmoss Selaginella lepidophylla. Nat. Commun. 2018, 9, 13. [Google Scholar] [CrossRef] [Green Version]
- Weng, J.-K.; Li, X.; Stout, J.; Chapple, C. Independent origins of syringyl lignin in vascular plants. Proc. Natl. Acad. Sci. USA 2008, 105, 7887–7892. [Google Scholar] [CrossRef] [Green Version]
- Weng, J.-K.; Akiyama, T.; Bonawitz, N.D.; Li, X.; Ralph, J.; Chapple, C. Convergent Evolution of Syringyl Lignin Biosynthesis via Distinct Pathways in the Lycophyte Selaginella and Flowering Plants. Plant Cell 2010, 22, 1033–1045. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef]
- Vlot, A.C.; Sales, J.H.; Lenk, M.; Bauer, K.; Brambilla, A.; Sommer, A.; Chen, Y.; Wenig, M.; Nayem, S. Systemic propagation of immunity in plants. New Phytol. 2020, 229, 1234–1250. [Google Scholar] [CrossRef]
- Kachroo, A.; Kachroo, P. Mobile signals in systemic acquired resistance. Curr. Opin. Plant Biol. 2020, 58, 41–47. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic Acid Biosynthesis and Metabolism. Arab. Book 2011, 9, e0156. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, Y.; Li, S.-S.; Han, G.-Z. Insights into the Origin and Evolution of the Plant Hormone Signaling Machinery. Plant Physiol. 2015, 167, 872–886. [Google Scholar] [CrossRef] [Green Version]
- Han, G. Origin and evolution of the plant immune system. New Phytol. 2018, 222, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef]
- Park, S.-W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl Salicylate Is a Critical Mobile Signal for Plant Systemic Acquired Resistance. Science 2007, 318, 113–116. [Google Scholar] [CrossRef]
- Dempsey, D.A.; Klessig, D.F. SOS–too many signals for systemic acquired resistance? Trends Plant Sci. 2012, 17, 538–545. [Google Scholar] [CrossRef]
- Attaran, E.; Zeier, T.E.; Griebel, T.; Zeier, J. Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 2009, 21, 954–971. [Google Scholar] [CrossRef] [Green Version]
- Baek, D.; Pathange, P.; Chung, J.; Jiang, J.; Gao, L.; Oikawa, A.; Hirai, M.Y.; Saito, K.; Pare, P.W.; Shi, H. A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant Cell Environ. 2010, 33, 1383–1392. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, L.; Zhao, J.; Li, Y.; Wang, J.; Guo, R.; Gan, S.; Liu, C.-J.; Zhang, K. S5H/DMR6 Encodes a Salicylic Acid 5-Hydroxylase That Fine-Tunes Salicylic Acid Homeostasis. Plant Physiol. 2017, 175, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Halitschke, R.; Yin, C.; Liu, C.-J.; Gan, S.-S. Salicylic acid 3-hydroxylase regulatesArabidopsisleaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef] [Green Version]
- Bartsch, M.; Bednarek, P.; Vivancos, P.D.; Schneider, B.; von Roepenack-Lahaye, E.; Foyer, C.; Kombrink, E.; Scheel, D.; Parker, J.E. Accumulation of Isochorismate-derived 2,3-Dihydroxybenzoic 3-O-β-d-Xyloside in Arabidopsis Resistance to Pathogens and Ageing of Leaves. J. Biol. Chem. 2010, 285, 25654–25665. [Google Scholar] [CrossRef] [Green Version]
- Fayos, J.; Bellés, J.M.; López-Gresa, M.P.; Primo, J.; Conejero, V. Induction of gentisic acid 5-O-β-d-xylopyranoside in tomato and cucumber plants infected by different pathogens. Phytochemistry 2006, 67, 142–148. [Google Scholar] [CrossRef]
- Huang, X.-X.; Zhu, G.-Q.; Liu, Q.; Chen, L.; Li, Y.-J.; Hou, B.-K. Modulation of Plant Salicylic Acid-Associated Immune Responses via Glycosylation of Dihydroxybenzoic Acids. Plant Physiol. 2018, 176, 3103–3119. [Google Scholar] [CrossRef]
- Chen, Z.; Iyer, S.; Caplan, A.; Klessig, D.F.; Fan, B. Differential Accumulation of Salicylic Acid and Salicylic Acid-Sensitive Catalase in Different Rice Tissues. Plant Physiol. 1997, 114, 193–201. [Google Scholar] [CrossRef] [Green Version]
- Sawada, H.; Shim, I.-S.; Usui, K. Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis—Modulation by salt stress in rice seedlings. Plant Sci. 2006, 171, 263–270. [Google Scholar] [CrossRef]
- Yu, D.; Liu, Y.; Fan, B.; Klessig, D.F.; Chen, Z. Is the High Basal Level of Salicylic Acid Important for Disease Resistance in Potato? Plant Physiol. 1997, 115, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Westfall, C.S.; Sherp, A.M.; Zubieta, C.; Alvarez, S.; Schraft, E.; Marcellin, R.; Ramirez, L.; Jez, J.M. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 13917–13922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourne, D.J.; Barrow, K.D.; Milborrow, B. Salicyloylaspartate as an endogenous component in the leaves of Phaseolus vulgaris. Phytochemistry 1991, 30, 4041–4044. [Google Scholar] [CrossRef]
- Steffan, H.; Ziegler, A.; Rapp, A. N-Saliciloyl aspartic acid-a new phenolic compound in grapevines. Vitis 1988, 27, 79–86. [Google Scholar]
- Chen, Y.; Shen, H.; Wang, M.; Li, Q.; He, Z. Salicyloyl-aspartate synthesized by the acetyl-amido synthetase GH3.5 is a potential activator of plant immunity in Arabidopsis. Acta Biochim. Biophys. Sin. 2013, 45, 827–836. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Lu, S. Plastoquinone and Ubiquinone in Plants: Biosynthesis, Physiological Function and Metabolic Engineering. Front. Plant Sci. 2016, 7, 1898. [Google Scholar] [CrossRef] [Green Version]
- Yazaki, K.; Fukui, H.; Tabata, M. Accumulation of p-O-β-D-glucosylbenzoic acid and its relation to shikonin biosynthesis in Lithospermum cell cultures. Phytochemistry 1986, 25, 1629–1632. [Google Scholar] [CrossRef]
- Smith-Becker, J.; Marois, E.; Huguet, E.J.; Midland, S.L.; Sims, J.J.; Keen, N.T. Accumulation of Salicylic Acid and 4-Hydroxybenzoic Acid in Phloem Fluids of Cucumber during Systemic Acquired Resistance Is Preceded by a Transient Increase in Phenylalanine Ammonia-Lyase Activity in Petioles and Stems1. Plant Physiol. 1998, 116, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Gianfagna, T.J.; Davies, P.J. N-benzoylaspartate and N-phenylacetylaspartate from pea seeds. Phytochemistry 1980, 19, 959–961. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yamaguchi, I.; Murofushi, N.; Takahashi, N. Biological Conversion of Benzoic Acid in Lemna paucicostata 151 and its Relation to Flower Induction. Plant Cell Physiol. 1988, 29, 7512. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Sun, Q.-Y.; Yang, F.-M.; Long, C.-L.; Zhao, F.-W.; Tang, G.-H.; Niu, H.-M.; Wang, H.; Huang, Q.-Q.; Xu, J.-J.; et al. Neolignans and Caffeoyl Derivatives from Selaginella moellendorffii. Helvetica Chim. Acta 2010, 93, 2467–2477. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Harborne, J.B.; Corner, J.J.; Lim, E.-K.; Higgins, G.S.; Li, Y.; Bowles, D.J.; Ellis, B.E.; Towers, G.H.N.; Wang, L.; Garcia-Rivera, J.; et al. Plant polyphenols. 4. Hydroxycinnamic acid-sugar derivatives. Biochem. J. 1961, 81, 242–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosuge, T.; Conn, E.E. The Metabolism of Aromatic Compounds in Higher Plants. J. Biol. Chem. 1959, 234, 2133–2137. [Google Scholar] [CrossRef]
- Stoker, J.; Bellis, D. The Biosynthesis of Coumarin in Melilotus Alba. J. Biol. Chem. 1962, 237, 2303–2305. [Google Scholar] [CrossRef]
- Meuwly, P.; Métraux, J. Ortho-Anisic Acid as Internal Standard for the Simultaneous Quantitation of Salicylic Acid and Its Putative Biosynthetic Precursors in Cucumber Leaves. Anal. Biochem. 1993, 214, 500–505. [Google Scholar] [CrossRef]
- Alber, A.V.; Renault, H.; Basilio-Lopes, A.; Bassard, J.-E.; Liu, Z.; Ullmann, P.; Lesot, A.; Bihel, F.; Schmitt, M.; Werck-Reichhart, D.; et al. Evolution of coumaroyl conjugate 3-hydroxylases in land plants: Lignin biosynthesis and defense. Plant J. 2019, 99, 924–936. [Google Scholar] [CrossRef]
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Ackerveken, G.V.D. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2014, 81, 210–222. [Google Scholar] [CrossRef]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutarate-dependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- de Toledo Thomazella, D.P.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, 2479. [Google Scholar] [CrossRef]
- Hasley, J.A.R.; Navet, N.; Tian, M. CRISPR/Cas9-mediated mutagenesis of sweet basil candidate susceptibility gene ObDMR6 enhances downy mildew resistance. PLoS ONE 2021, 16, e0253245. [Google Scholar] [CrossRef]
- Ferreyra, M.L.F.; Emiliani, J.; Rodriguez, E.J.; Campos-Bermudez, V.A.; Grotewold, E.; Casati, P. The Identification of Maize and Arabidopsis Type I FLAVONE SYNTHASEs Links Flavones with Hormones and Biotic Interactions. Plant Physiol. 2015, 169, 1090–1107. [Google Scholar] [CrossRef] [Green Version]
- Righini, S.; Rodriguez, E.J.; Berosich, C.; Grotewold, E.; Casati, P.; Falcone Ferreyra, M.L. Apigenin produced by maize flavone synthase I and II protects plants against UV-B-induced damage. Plant, Cell Environ. 2019, 42, 495–508. [Google Scholar] [CrossRef]
- Mackelprang, R.; Okrent, R.A.; Wildermuth, M.C. Preference of Arabidopsis thaliana GH3.5 acyl amido synthetase for growth versus defense hormone acyl substrates is dictated by concentration of amino acid substrate aspartate. Phytochemistry 2017, 143, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate Response Locus JAR1 and Several Related Arabidopsis Genes Encode Enzymes of the Firefly Luciferase Superfamily That Show Activity on Jasmonic, Salicylic, and Indole-3-Acetic Acids in an Assay for Adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual Regulation Role of GH3.5 in Salicylic Acid and Auxin Signaling during Arabidopsis-Pseudomonas syringae Interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.-K. PBS3 and EPS1 Complete Salicylic Acid Biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okrent, R.A.; Brooks, M.D.; Wildermuth, M.C. Arabidopsis GH3.12 (PBS3) Conjugates Amino Acids to 4-Substituted Benzoates and Is Inhibited by Salicylate. J. Biol. Chem. 2009, 284, 9742–9754. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.D. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.-K.; Doucet, C.J.; Li, Y.; Elias, L.; Worrall, D.; Spencer, S.P.; Ross, J.; Bowles, D.J. The Activity of ArabidopsisGlycosyltransferases toward Salicylic Acid, 4-Hydroxybenzoic Acid, and Other Benzoates. J. Biol. Chem. 2002, 277, 586–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, S.; Mekonnen, D.W.; Hartmann, M.; Yildiz, I.; Janowski, R.; Lange, B.; Geist, B.; Zeier, J.; Schäffner, A.R. UGT76B1, a promiscuous hub of small molecule-based immune signaling, glucosylates N-hydroxypipecolic acid, and balances plant immunity. Plant Cell 2021, 33, 714–734. [Google Scholar] [CrossRef] [PubMed]
- Mohnike, L.; Rekhter, D.; Huang, W.; Feussner, K.; Tian, H.; Herrfurth, C.; Zhang, Y.; Feussner, I. The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 2021, 33, 735–749. [Google Scholar] [CrossRef]
- Holmes, E.C.; Chen, Y.-C.; Mudgett, M.B.; Sattely, E.S. Arabidopsis UGT76B1 glycosylates N-hydroxy-pipecolic acid and inactivates systemic acquired resistance in tomato. Plant Cell 2021, 33, 750–765. [Google Scholar] [CrossRef]
- Cai, J.; Jozwiak, A.; Holoidovsky, L.; Meijler, M.M.; Meir, S.; Rogachev, I.; Aharoni, A. Glycosylation of N-hydroxy-pipecolic acid equilibrates between systemic acquired resistance response and plant growth. Mol. Plant 2020, 14, 440–455. [Google Scholar] [CrossRef]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef]
- Harholt, J.; Sørensen, I.; Fangel, J.; Roberts, A.; Willats, W.G.T.; Scheller, H.V.; Petersen, B.L.; Banks, J.A.; Ulvskov, P. The Glycosyltransferase Repertoire of the Spikemoss Selaginella moellendorffii and a Comparative Study of Its Cell Wall. PLoS ONE 2012, 7, e35846. [Google Scholar] [CrossRef]
- Umemura, K.; Satou, J.; Iwata, M.; Uozumi, N.; Koga, J.; Kawano, T.; Koshiba, T.; Anzai, H.; Mitomi, M. Contribution of salicylic acid glucosyltransferase, OsSGT1, to chemically induced disease resistance in rice plants. Plant J. 2009, 57, 463–472. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, M.; Lu, M.; Wu, Y.; Jing, T.; Zhao, M.; Zhao, Y.; Feng, Y.; Wang, J.; Gao, T.; et al. Salicylic acid carboxyl glucosyltransferase UGT87E7 regulates disease resistance in Camellia sinensis. Plant Physiol. 2021, 6, 71. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.M.G.; Iancu, C.V.; Neet, K.E.; Dean, J.V.; Choe, J.-Y. Differences in salicylic acid glucose conjugations by UGT74F1 and UGT74F2 from Arabidopsis thaliana. Sci. Rep. 2017, 7, 46629. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Top BLAST NR Proteins | Top BLAST AraTh RefSeq | Top BLAST Biochemically Characterized AraTh RefSeq Protein | |||||
|---|---|---|---|---|---|---|---|
| Protein Type | SmRefSeq | Species and Accession Number | %ID/Score | Accession Number (Gene Name if Any) | %ID/Score | Accession Number (Gene Name if Any) | % ID/Score |
| ODD | XP_024534841.1 | Picea sitchensis ABK26685.1 | 47.46/338 | NP_192787.1 (DLO2) | 43.88/295 | NP_192788.1 (DLO1) | 43.60/294 |
| ODD | XP_002968368.2 | Pinus tabuliformis GA2ox11 AHW42461.1 | 51.61/363 | n/a 1 | NP_192788.1 (DLO1) | 47.14/285 | |
| ODD | XP_024525160.1 | Picea sitchensis ABK22784.1 | 48.14/312 | NP_192787.1 (DLO2) | 41.71/278 | NP_192788.1 (DLO1) | 42.42/272 |
| ODD | XP_002965938.2 | Pinus tabuliformis GA2ox11 AHW42461.1 | 53.22/373 | n/a | NP_192788.1 (DLO1) | 43.54/281 | |
| ODD | XP_002968370.1 | Pinus tabuliformis GA2ox11 AHW42461.1 | 51.17/359 | n/a | NP_192788.1 (DLO1) | 47.60/284 | |
| ODD | XP_002967783.1 | Aquilegia coerulea PIA25996.1 | 32.25/204 | n/a | NP_192788.1 (DLO1) | 33.43/182 | |
| ODD | XP_002988299.1 | Musa acuminata XP_009389864.1 | 39.45/233 | n/a | NP_197841.1 (DMR6) | 37.61/211 | |
| ODD | XP_002983067.1 | Marchantia polymorpha PTQ39739.1 | 39.63/251 | NP_192787.1 (DLO2) | 38.87/210 | NP_197841.1 (DMR6) | 39.26/207 |
| ODD | XP_002975060.1 | Tetracentron sinense KAF8411241.1 | 41.77/263 | NP_192787.1 (DLO2) | 37.61/234 | NP_197841.1 (DMR6) | 37.73/227 |
| GH3 | XP_024536354.1 | Ricinus communis XP_002533739.1 | 68.07/879 | n/a | NP_194456.1 = GH3.5/WES1 | 66.4/862 | |
| GH3 | XP_002960824.1 | Brassica carinata KAG2310584.1 | 48.99/592 | n/a | NP_200262.1 = GH3.6/DFL1 | 48.55/586 | |
| GH3 | XP_024519979.1 | Daucus carota subsp. sativus XP_017250111.1 | 45.02/504 | n/a | NP_200262.1 = GH3.6/DFL1 | 43.39/480 | |
| GH3 | XP_024529423.1 | Vigna angularis XP_017432135.1 | 43.77/503 | n/a | NP_200262.1 = GH3.6/DFL1 | 43.01/488 | |
| GH3 | XP_024516808.1 | Punica granatum XP_031407799.1 | 39.58/415 | NP_001319858.1 GH3.10/DFL2 | 36.80/393 | NP_566071.1 GH3.11/JAR1/FIN219 | 36.54/380 |
| GH3 | XP_002976207.1 | Ceratopteris richardii KAH7428824.1 | 39.89/412 | n/a | NP_566071.1 GH3.11/JAR1/FIN219 | 36.36/378 | |
| GH3 | XP_024540069.1 | Tanacetum cinerarifolium GEY79194.1 | 42.33/501 | NP_001319858.1 GH3.10/DFL2 | 40.65/465 | NP_566071.1 GH3.11/JAR1/FIN219 | 39.97/450 |
| GH3 | XP_024543479.1 (annotated as GH3.12) | Physcomytrium patens XP_024386895.1 JAR1-like | 37.69/294 | NP_001319858.1 GH3.10/DFL2 | 35.64/591 | NP_566071.1 GH3.11/JAR1/FIN219 #4: NP_001330076.1 GH3.12 | 34.89/258 35.29/249 |
| UGT | XP_024542897.1 | A. thaliana x A. arenosa KAG7593547.1 | 35.16/276 | NP_173652.1 UGT85A7 | 33.74/273 | NP_173656.1 UGT85A1 #8: NP_181910.1 UGT74F2 | 33.61/266 30.30/210 |
| UGT | XP_024532226.1 | Picea sitchensis ABR16170.1 | 35.70/299 | NP_173652.1 UGT85A7 | 33.20/275 | NP_173656.1 UGT85A1 #10: NP_187742.1 UGT76B1 | 32.12/267 32.42/221 |
| UGT | XP_002992501.2 | Ginkgo biloba ASK39407.1 | 37.86/300 | n/a | NP_173656.1 UGT85A1 #17: NP_973682.1UGT74F1 | 34.77/270 29.35/203 | |
| UGT | XP_024526026.1 | Ziziphus jujube XP_015865855.1 | 37.25/298 | n/a | NP_173656.1 UGT85A1 #15:NP_187742.1 UGT76B1 | 37.16/290 29.71/219 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berim, A.; Gang, D.R. Accumulation of Salicylic Acid and Related Metabolites in Selaginella moellendorffii. Plants 2022, 11, 461. https://doi.org/10.3390/plants11030461
Berim A, Gang DR. Accumulation of Salicylic Acid and Related Metabolites in Selaginella moellendorffii. Plants. 2022; 11(3):461. https://doi.org/10.3390/plants11030461
Chicago/Turabian StyleBerim, Anna, and David R. Gang. 2022. "Accumulation of Salicylic Acid and Related Metabolites in Selaginella moellendorffii" Plants 11, no. 3: 461. https://doi.org/10.3390/plants11030461
APA StyleBerim, A., & Gang, D. R. (2022). Accumulation of Salicylic Acid and Related Metabolites in Selaginella moellendorffii. Plants, 11(3), 461. https://doi.org/10.3390/plants11030461






