Transcriptome and Physiological Analysis of Rootstock Types and Silicon Affecting Cold Tolerance of Cucumber Seedlings
Abstract
:1. Introduction
2. Results
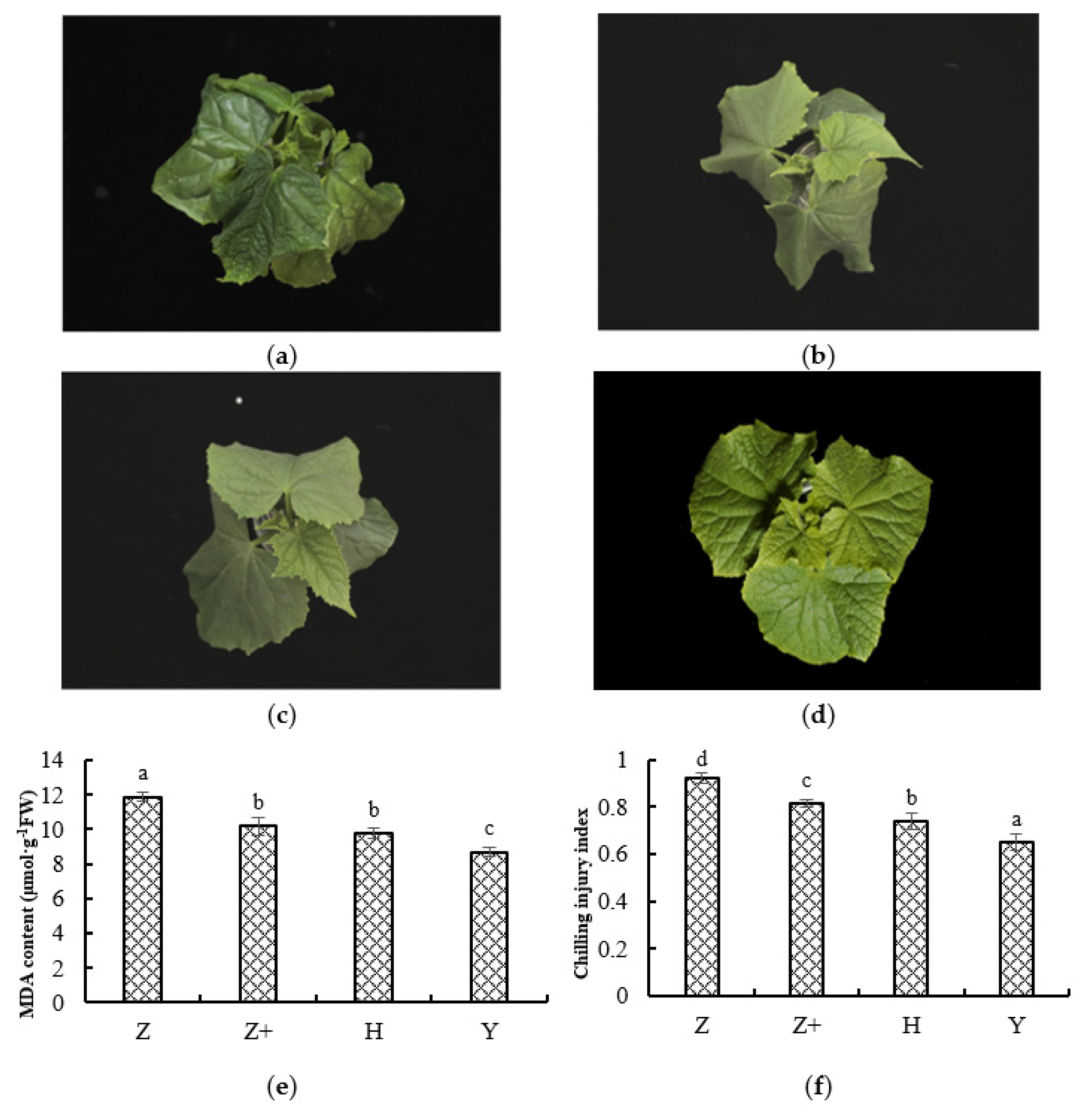
2.1. Physiological Response of Cucumber Seedlings under Cold Stress
2.2. Transcriptome Sequencing Data and Quality
2.3. Statistics of DEGs
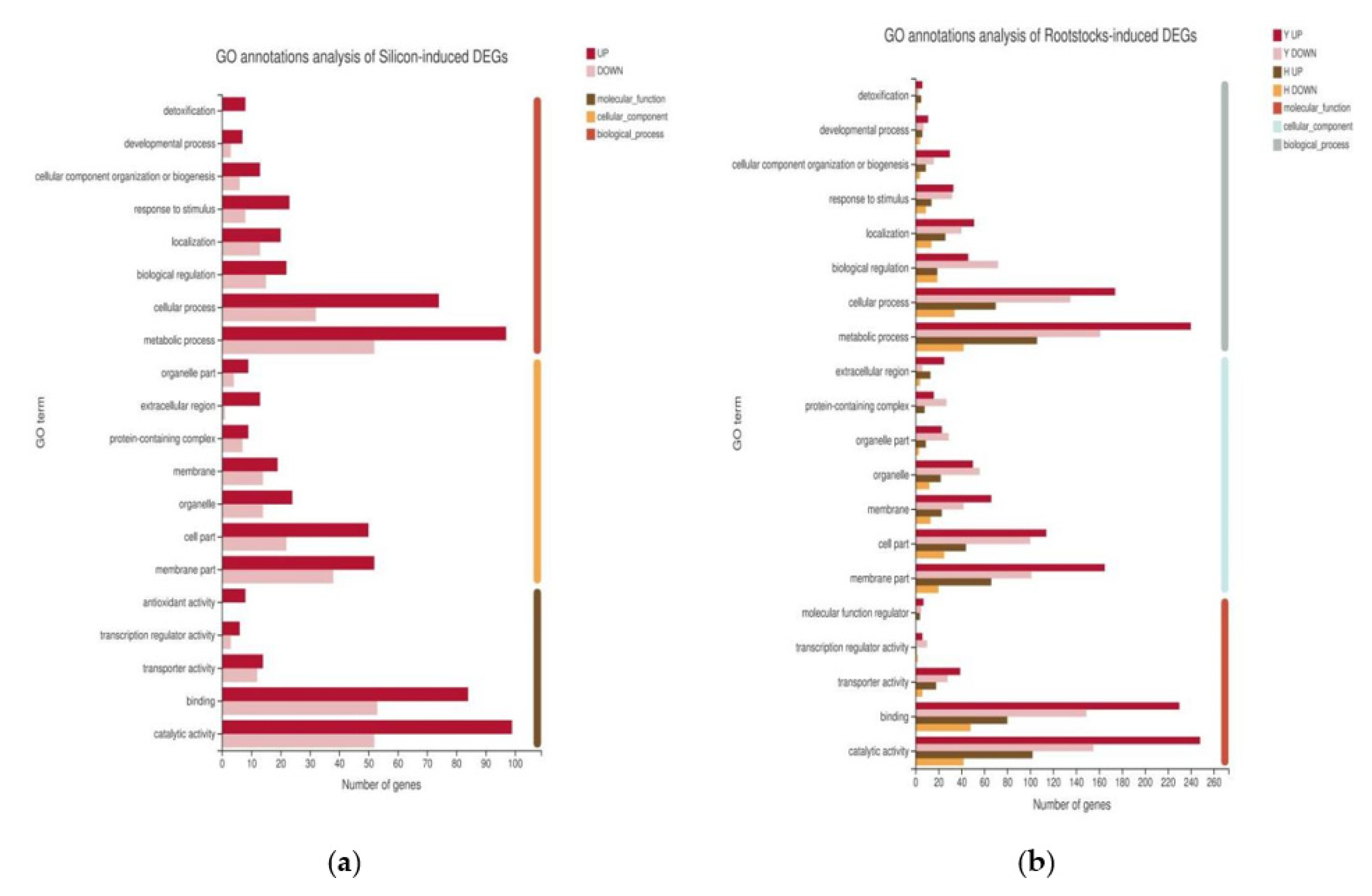
2.4. Functional Annotation of DEGs
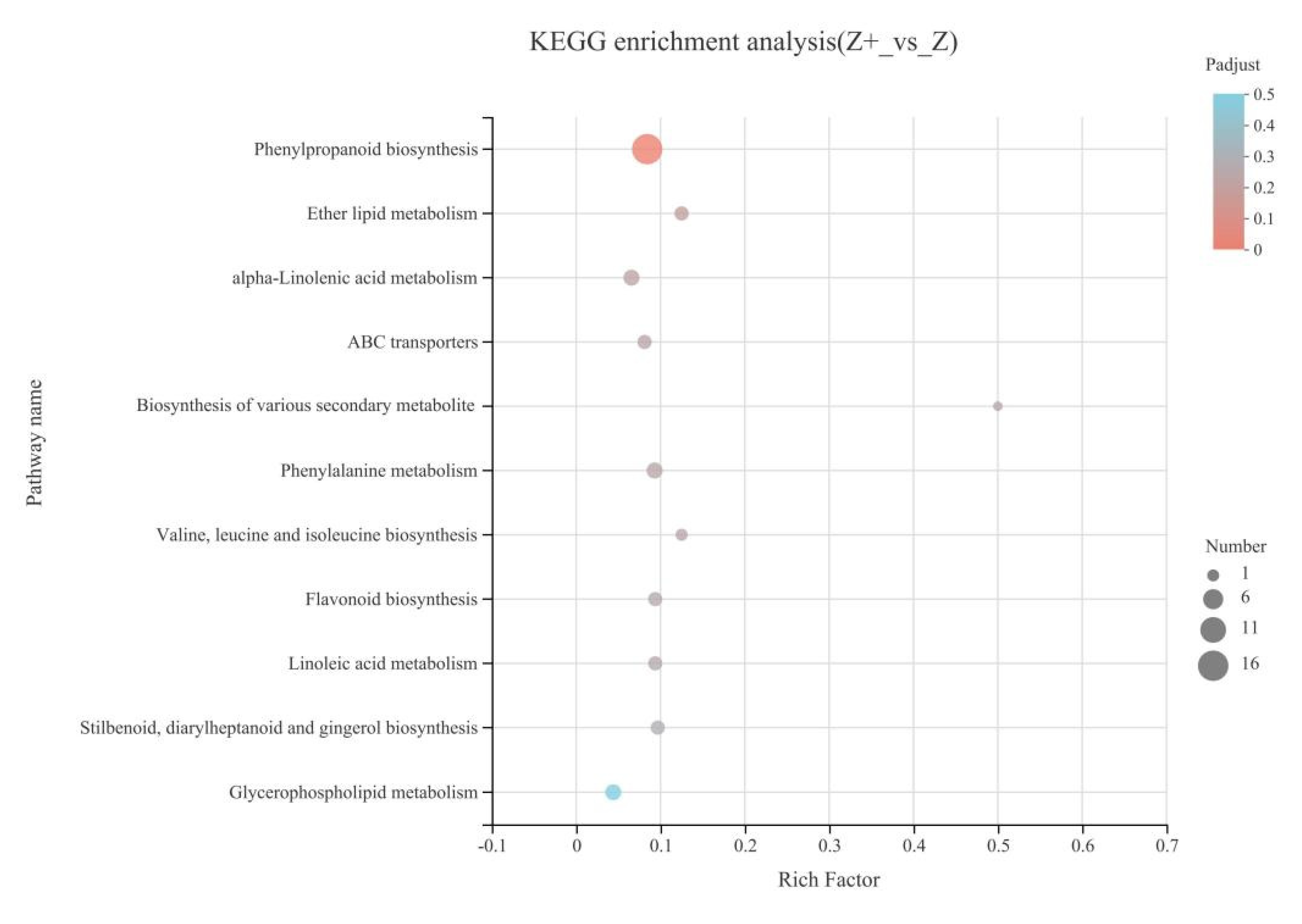
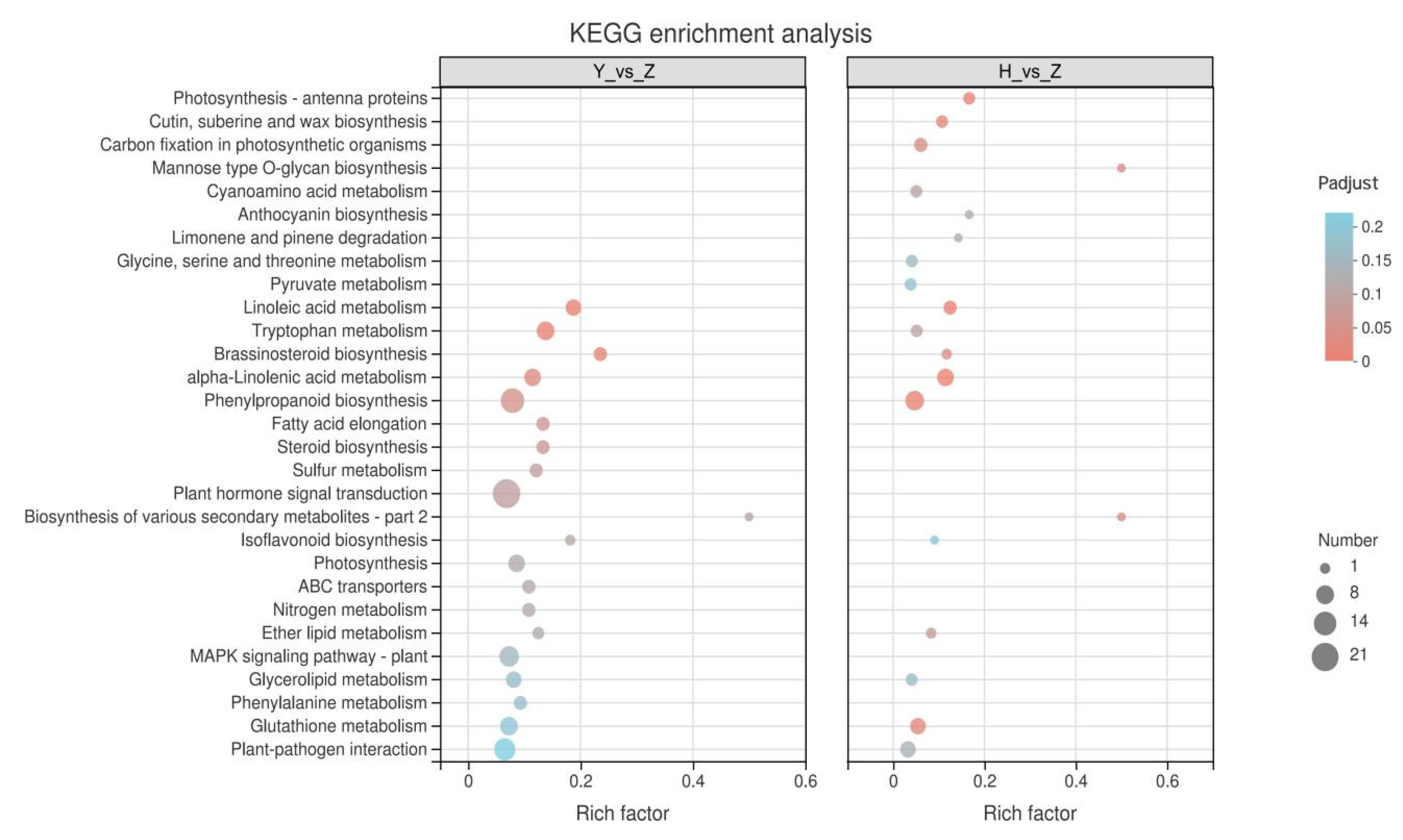
2.5. KEGG Enrichment Analysis of Silicon-Induced DEGs
2.6. KEGG Enrichment Analysis of Rootstock-Grafted-Induced DEGs
2.7. Screening for Transcription Factors
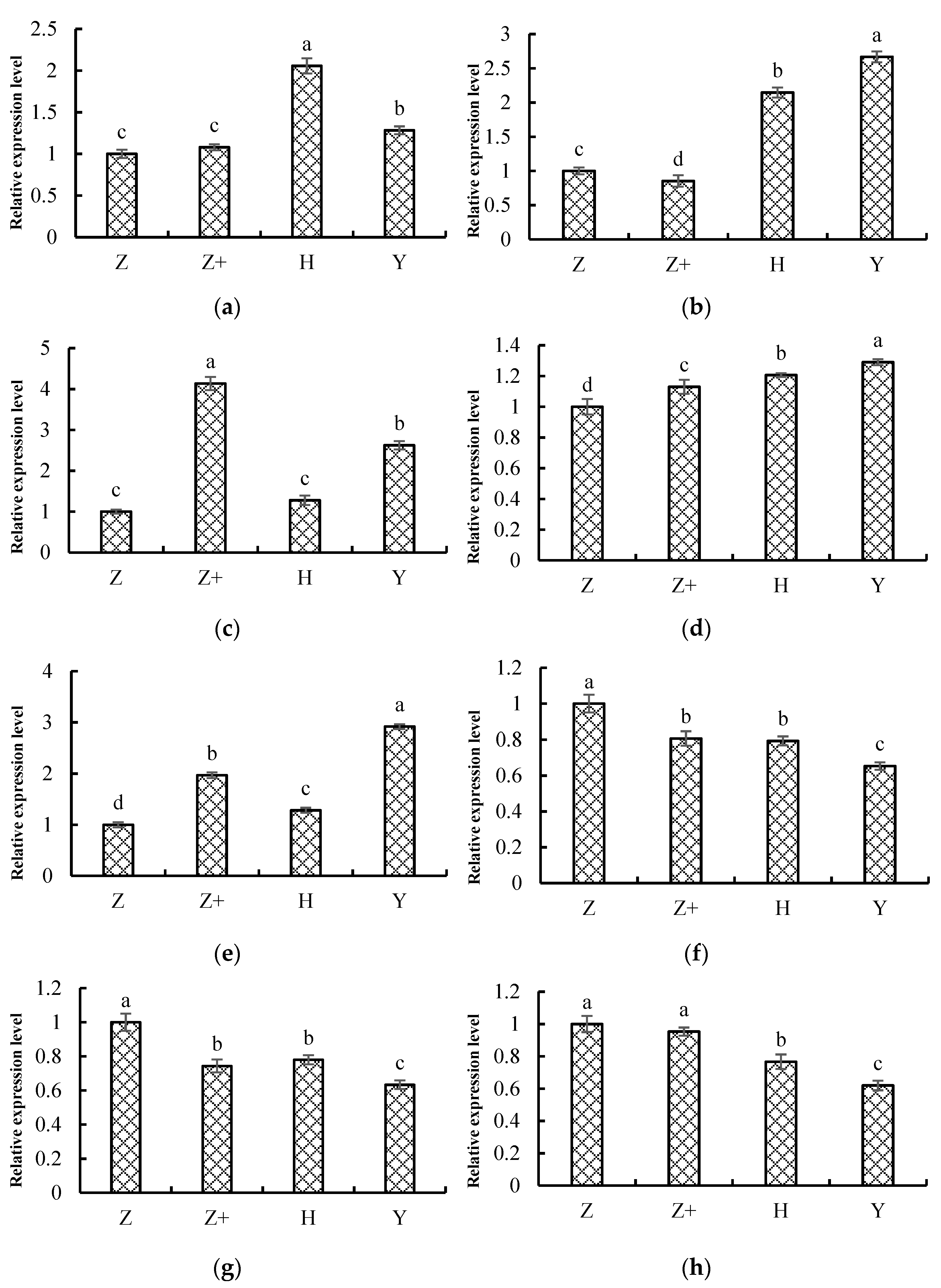
2.8. Validation of the Transcriptome Data
3. Discussion
3.1. Silicon Could Enhance the Cold-Tolerance of Cucumber Seedlings by Enhancing the Metabolism of Phenylpropanoid
3.2. Rootstock Grafting Could Boost the Scavenging of ROS and the Synthesis of Phytohormones and Maintain the Stability of Membrane Structure to Enhance the Cold-Tolerance of Cucumber Seedlings
3.3. MYB, ERF and WRKY Play Transcription Factor Family Plays a Role in Cucumber Seedlings Response to Cold Stresses
4. Materials and Methods
4.1. Plant Materials and Cold Treatment
4.2. Determination of Physiological Characteristics under Cold Stress
4.3. Preparation and Sequencing of RNA Sequence Libraries
4.4. Data Filtering and Mapping of Reads
4.5. Screening of Differentially Expressed Genes (DEGs)
4.6. Functional Annotation and Enrichment Analysis of DEGs
4.7. Transcription Factor Analysis and Screening
4.8. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabrera, R.M.; Saltveit, M.E.; Owens, K. Cucumber cultivars differ in their response to chilling temperatures. J. Am. Soc. Hortic. Sci. 1992, 117, 802–807. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Guo, S.R.; Li, H.; Sun, H.Z.; Lu, N.; Shu, S.; Sun, J. Resistance of cucumber grafting rootstock pumpkin cultivars to chilling and salinity stresses. Hortic. Sci. Technol. 2017, 35, 220–231. [Google Scholar]
- Lee, J.; Bang, H.; Ham, H. Quality of cucumber fruit as affected by rootstock. Int. Symp. Veg. Qual. Fresh Fermented Veg. 1997, 483, 117–124. [Google Scholar] [CrossRef]
- Zhou, X.; Feng, G.L.; Li, Z.H.; Liu, S.X.; Zhao, S.; Li, Y.; Wei, M. Effects of environmental conditions on absorption and distribution of silicon and formation of bloom on fruit surface of cucumber. J. Appl. Ecol. 2020, 31, 501–507. [Google Scholar]
- Zhao, J.L.; Wang, Y.L.; Yao, D.Q.; Zhu, W.Y.; Chen, L.; He, H.L.; Pan, J.S.; Cai, R. Transcriptome profiling of trichome-less reveals genes associated with multicellular trichome development in Cucumis sativus. Mol. Genet. Genom. 2015, 290, 2007–2018. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Zhou, J.; Huang, L.F.; Ding, X.T.; Shi, K.; Yu, J.Q. Grafting of Cucumis sativus onto Cucurbita ficifolia leads to improved plant growth, increased light utilization and reduced accumulation of reactive oxygen species in chilled plants. J. Plant Res. 2009, 122, 529–540. [Google Scholar] [CrossRef]
- Huang, Y.; Bie, Z.L.; Liu, Z.X.; Zhen, A.; Jiao, X.R. Improving cucumber photosynthetic capacity under NaCl stress by grafting onto two salt-tolerant pumpkin rootstocks. Biol. Plant. 2011, 55, 285–290. [Google Scholar] [CrossRef]
- Liu, Z.X.; Bie, Z.L.; Huang, Y.; Zhen, A.; Niu, M.L.; Lei, B. Rootstocks improve cucumber photosynthesis through nitrogen metabolism regulation under salt stress. Acta Physiol. Plant. 2013, 35, 2259–2267. [Google Scholar] [CrossRef]
- Rouphael, Y.; Rea, E.; Cardarelli, M.; Bitterlich, M.; Schwarz, D.; Colla, G. Can adverse effects of acidity and aluminum toxicity be alleviated by appropriate rootstock selection in cucumber? Front. Plant Sci. 2016, 7, 1283. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Tian, X.M.; Wei, M.; Shi, Q.H.; Yang, F.J.; Wang, X.F. Mechanisms of tolerance differences in cucumber seedlings grafted on rootstocks with different tolerance to low temperature and weak light stresses. Turk. J. Bot. 2015, 39, 606–614. [Google Scholar] [CrossRef]
- Samuels, A.L.; Glass, A.D.M.; Ehret, D.L.; Menzies, J.G. The effects of silicon supplementation on cucumber fruit: Changes in surface characteristics. Ann. Bot. 1993, 72, 433–440. [Google Scholar] [CrossRef]
- Seki, M.; Hotta, Y. Effect of bloomless stock cultivar on the growth and mineral uptake of cucumber [Cucumis sativus] plants. Res. Bull. Aichi-Ken Agric. Res. Cent. 1997, 29, 127–133. [Google Scholar]
- Ranjan, A.; Sinha, R.; Bala, M.; Pareek, A.; Singla-Pareek, S.L.; Singh, A.K. Silicon-mediated abiotic and biotic stress mitigation in plants: Underlying mechanisms and potential for stress resilient agriculture. Plant Physiol. Biochem. 2021, 163, 15–25. [Google Scholar] [CrossRef]
- Kaniuga, Z. Chilling response of plants: Importance of galactolipase, free fatty acids and free radicals. Plant Biol. 2008, 10, 171–184. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, C.J.; Ren, J.Y.; Dong, J.L.; Shi, X.L.; Zhao, X.H.; Wang, X.G.; Wang, J.; Zhong, C.; Zhao, S.L.; et al. An advanced lipid metabolism system revealed by transcriptomic and lipidomic analyses plays a central role in peanut cold tolerance. Front. Plant Sci. 2020, 11, 1110. [Google Scholar] [CrossRef]
- Li, F.M.; Lu, X.P.; Duan, P.F.; Liang, Y.J.; Cui, J. Integrating transcriptome and metabolome analyses of the response to cold stress in pumpkin (Cucurbita maxima). PLoS ONE 2021, 16, e0249108. [Google Scholar] [CrossRef]
- Wang, B.; Wu, C.S.; Wang, G.; He, J.M.; Zhu, S.J. Transcriptomic analysis reveals a role of phenylpropanoid pathway in the enhancement of chilling tolerance by pre-storage cold acclimation in cucumber fruit. Sci. Hortic. 2021, 288, 110282. [Google Scholar] [CrossRef]
- Jiang, W.; Li, Z.; Wan, M.; Wu, T.; Bian, M.; Huang, K.; Zhang, X. Overexpression of OsETR4, aputative ethylene receptor increases cold tolerance in rice. Int. J. Agric. Biol. 2020, 24, 969–978. [Google Scholar]
- Agarwal, P.K.; Jha, B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 2010, 54, 201–212. [Google Scholar] [CrossRef]
- Ntatsi, G.; Savvas, D.; Papasotiropoulos, V.; Katsileros, A.; Zrenner, R.M.; Hincha, D.K.; Zuther, E.; Schwarz, D. Rootstock sub-optimal temperature tolerance determines transcriptomic responses after long-term root cooling in rootstocks and scions of grafted tomato plants. Front. Plant Sci. 2017, 8, 911. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.H.; Zhang, M.; Liu, G.; Yang, X.P.; Hou, X.L. Comparative transcriptome profiling of chilling stress responsiveness in grafted watermelon seedlings. Plant Physiol. Biochem. 2016, 109, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, J.; Zhou, X.H.; Liu, S.Y.; Zhuang, Y. Transcriptomics analysis unravels the response to low temperature in sensitive and tolerant eggplants. Sci. Hortic. 2020, 271, 109468. [Google Scholar] [CrossRef]
- Kleiber, T. The role of silicon in plant tolerance to abiotic stress. In Plant Nutrients and Abiotic Stress Tolerance; Springer: Singapore, 2018; pp. 253–267. [Google Scholar]
- Zhang, J.; Yang, J.J.; Yang, Y.; Luo, J.; Zheng, X.Y.; Wen, C.L.; Xu, Y. Transcription factor CsWIN1 regulates pericarp bloom biosynthesis in cucumber grafted on pumpkin. Front. Plant Sci. 2019, 10, 1564. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Yin, J.L.; Liang, Y.F.; Liu, J.Q.; Jia, J.H.; Huo, H.Q.; Wu, Z.F.; Yang, R.L.; Gong, H.J. Transcriptomic dynamics provide an insight into the mechanism for silicon-mediated alleviation of salt stress in cucumber plants. Ecotoxicol. Environ. Saf. 2019, 174, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kumar, S.; Rani, A.; Gulati, A.; Gulati, A.; Ahuja, P.S. Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct. Integr. Genom. 2009, 9, 125–134. [Google Scholar] [CrossRef]
- Logan, G. Photoprotection of the Photosynthetic Apparatus || Energy Dissipation and Radical Scavenging by the Plant Phenylpropanoid Pathway. Philos. Trans. Biol. Sci. 2000, 355, 1499–1510. [Google Scholar]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broun, P.; Poindexter, P.; Osborne, E.; Jiang, C.Z.; Riechmann, J.L. WIN1, a transcriptional activator of epidermal bloom accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 4706–4711. [Google Scholar] [CrossRef] [Green Version]
- Park, C.S.; Go, Y.S.; Suh, M.C. Cuticular bloom biosynthesis is positively regulated by WRINKLED 4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 2016, 88, 257–270. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Song, C.B.; Yang, Y.Y.; Yang, T.W.; Ba, L.J.; Zhang, H.; Han, Y.C.; Xiao, Y.Y.; Shan, W.; Kuang, J.F.; Chen, J.Y.; et al. MaMYB4 recruits histone deacetylase MaHDA2 and modulates the expression of ω-3 fatty acid desaturase genes during cold stress response in banana fruit. Plant Cell Physiol. 2019, 60, 2410–2422. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA 2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.M.; Zhou, M.L.; Xiao, X.G.; Tang, Y.X.; Wu, Y.M. Genome-wide analysis of AP2/ERF family genes from Lotus corniculatus shows LcERF054 enhances salt tolerance. Funct. Integr. Genom. 2014, 14, 453–466. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef] [Green Version]
- Hodges, D.M.; Delong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Zhang, X.W.; Zhang, Y.Y.; Xu, C.X.; Liu, K.; Bi, H.G.; Ai, X.Z. H2O2 Functions as a Downstream Signal of IAA to Mediate H2S-Induced Chilling Tolerance in Cucumber. Int. J. Mol. Sci. 2021, 22, 12910. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
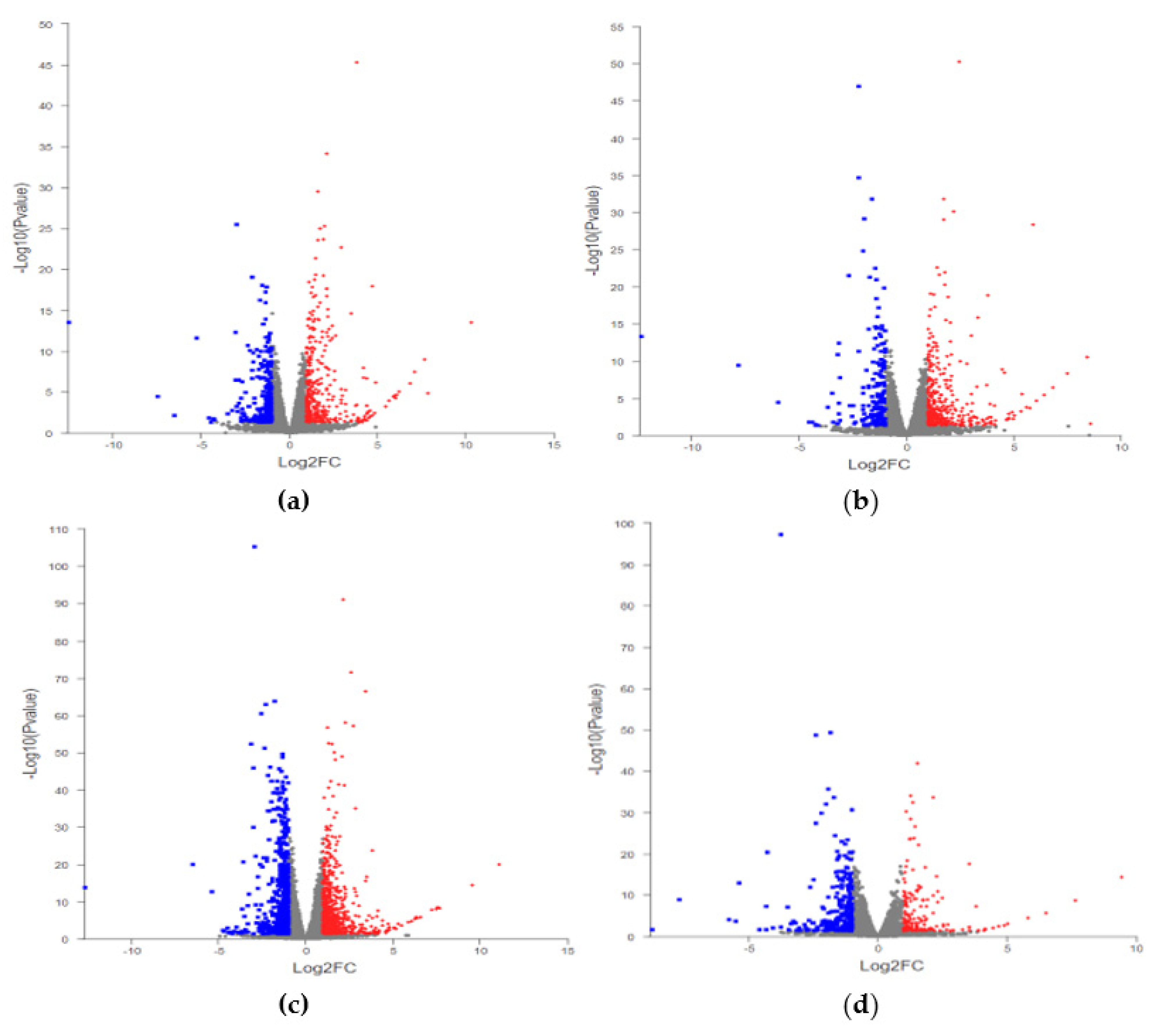
| Compare | Total DEGs | Up | Down |
|---|---|---|---|
| Z+_vs_Z | 433 | 274 | 159 |
| H_vs_Z | 412 | 264 | 148 |
| Y_vs_Z | 1217 | 679 | 538 |
| Y_vs_H | 305 | 107 | 198 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luan, H.; Niu, C.; Nie, X.; Li, Y.; Wei, M. Transcriptome and Physiological Analysis of Rootstock Types and Silicon Affecting Cold Tolerance of Cucumber Seedlings. Plants 2022, 11, 445. https://doi.org/10.3390/plants11030445
Luan H, Niu C, Nie X, Li Y, Wei M. Transcriptome and Physiological Analysis of Rootstock Types and Silicon Affecting Cold Tolerance of Cucumber Seedlings. Plants. 2022; 11(3):445. https://doi.org/10.3390/plants11030445
Chicago/Turabian StyleLuan, Heng, Chenxu Niu, Xinmiao Nie, Yan Li, and Min Wei. 2022. "Transcriptome and Physiological Analysis of Rootstock Types and Silicon Affecting Cold Tolerance of Cucumber Seedlings" Plants 11, no. 3: 445. https://doi.org/10.3390/plants11030445
APA StyleLuan, H., Niu, C., Nie, X., Li, Y., & Wei, M. (2022). Transcriptome and Physiological Analysis of Rootstock Types and Silicon Affecting Cold Tolerance of Cucumber Seedlings. Plants, 11(3), 445. https://doi.org/10.3390/plants11030445





