The Novel Cucurbitaceae miRNA ClmiR86 Is Involved in Grafting-Enhanced Phosphate Utilization and Phosphate Starvation Tolerance in Watermelon
Abstract
:1. Introduction
2. Results
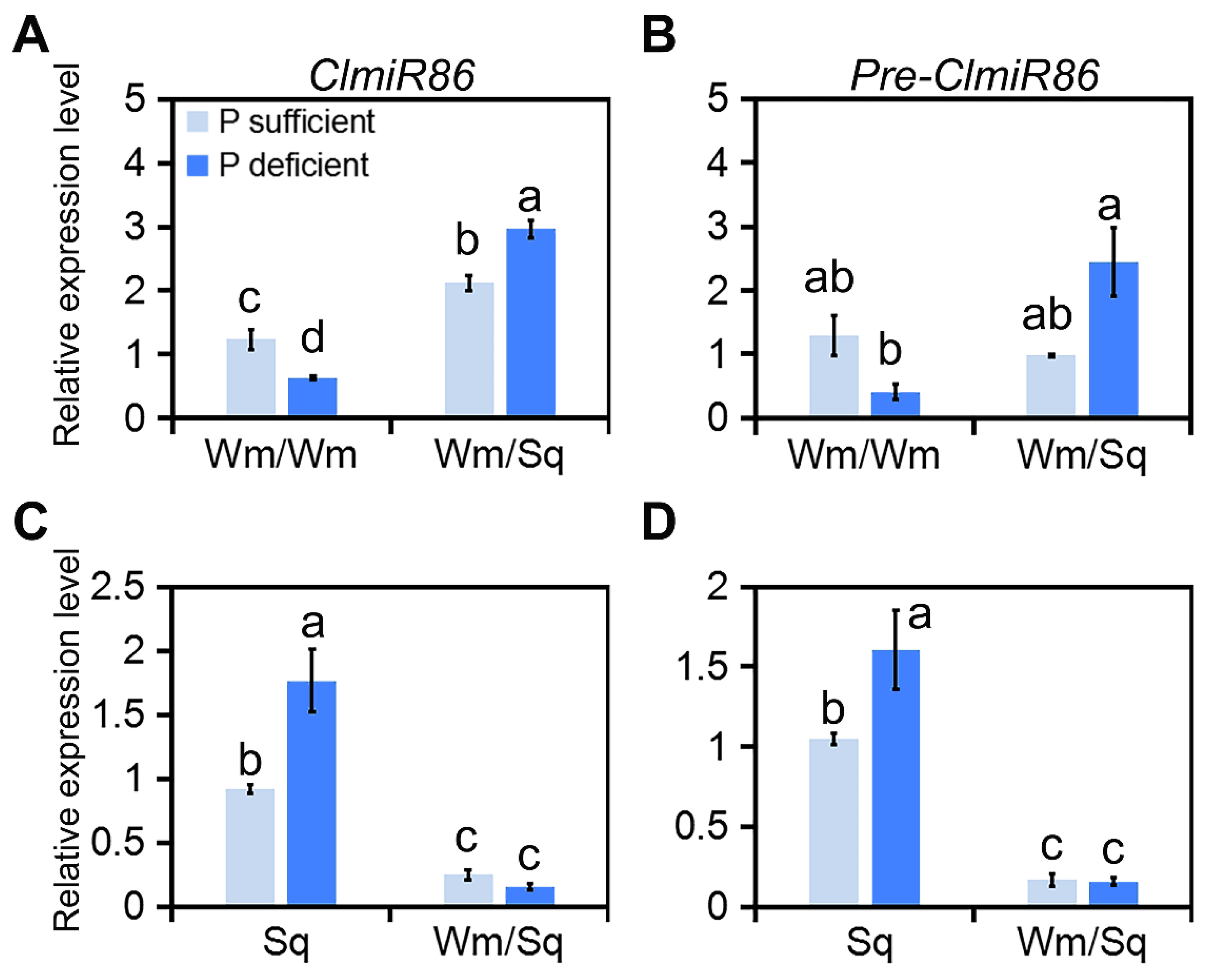
2.1. ClmiR86 May Enhance PUE of Watermelon
2.2. ClmiR86 Suppresses ClCIPK5, AtCIPK5, and AtCIPK25 at the Post-Transcription Level
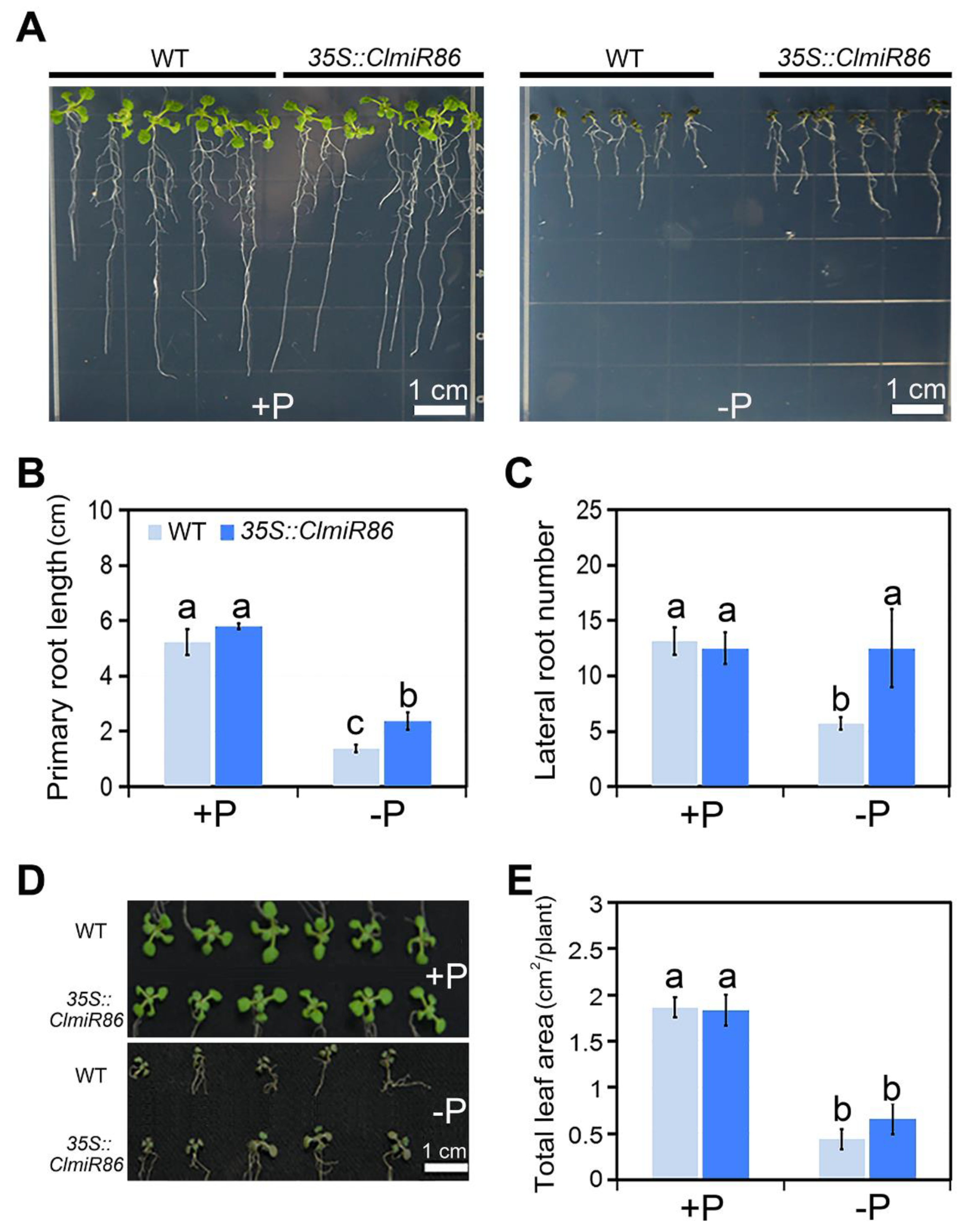
2.3. Ectopic ClmiR86 Expression Enhanced Phosphate Starvation Tolerance
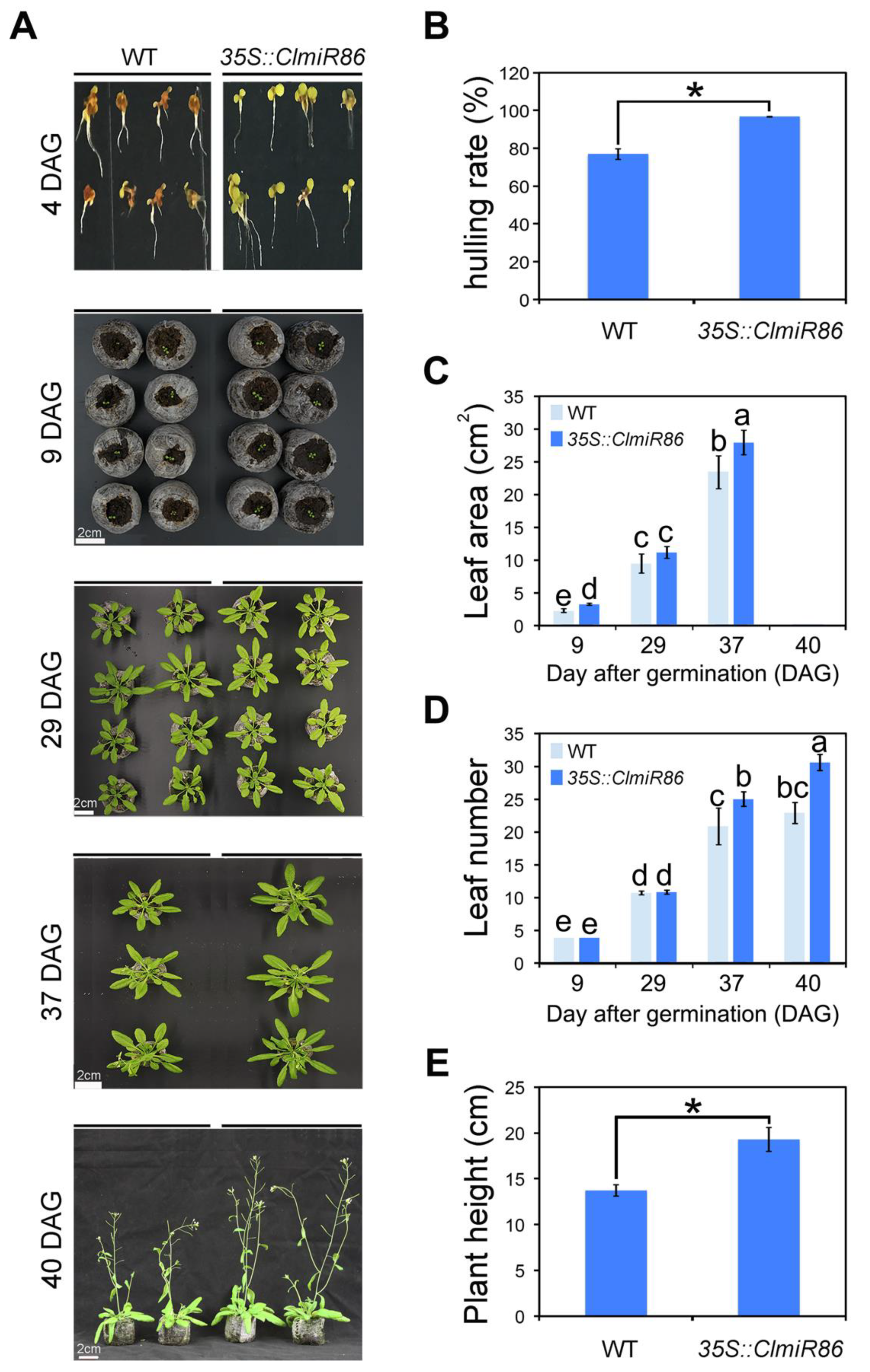
2.4. Ectopic ClmiR86 Expression Enhanced Plant Growth under Normal Conditions
3. Discussion
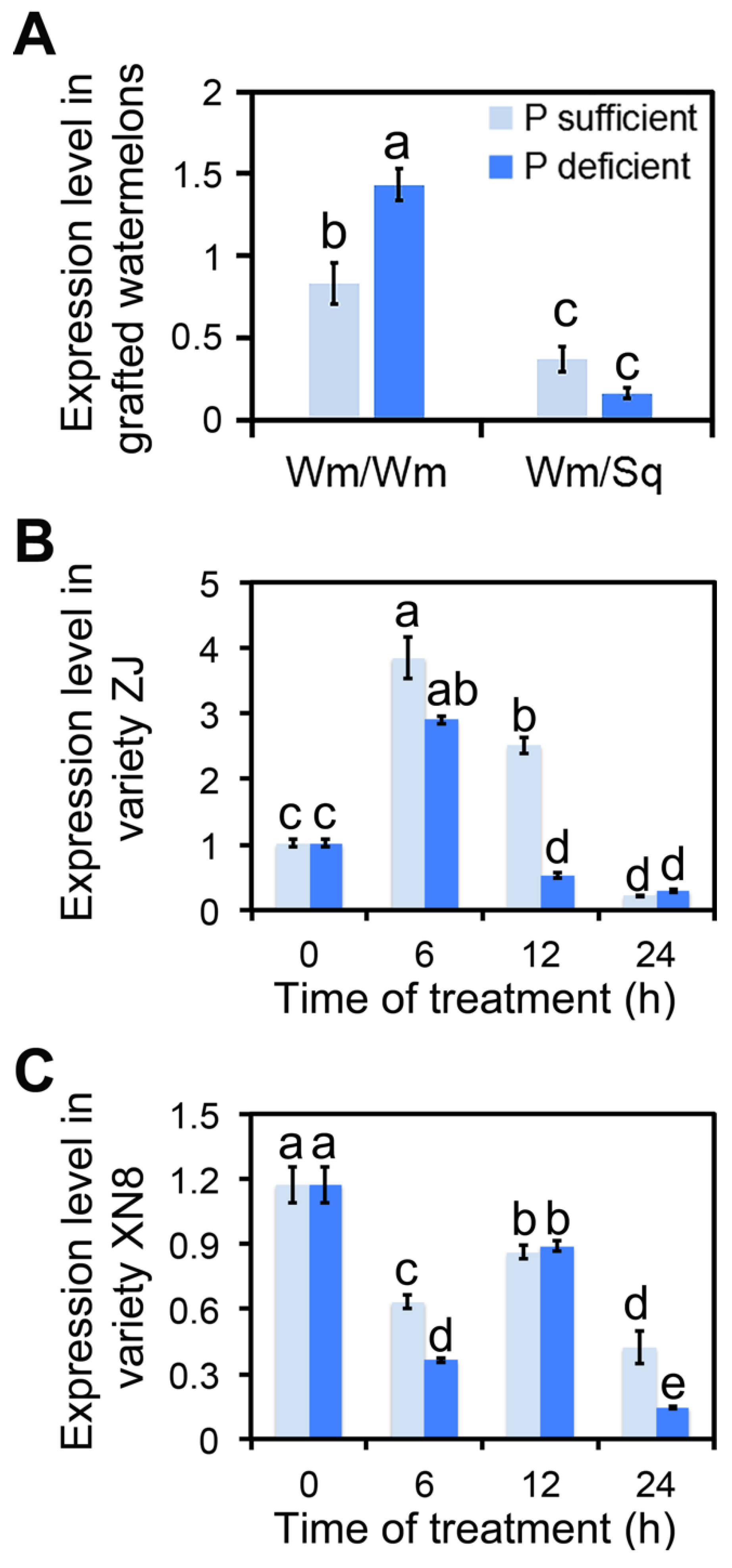
3.1. Squash Grafting Enhanced Watermelon Phosphate Utilization
3.2. ClmiR86 Is Involved in Phosphate Starvation Response and Phosphate Utilization in Watermelon
3.3. CIPK5 Works Downstream of ClmiR86 under Phosphate Starvation and Normal Conditions
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatments
4.2. Vector Construction
4.3. Arabidopsis Transformation
4.4. Target Gene Prediction
4.5. Phylogenetic Tree Construction
4.6. PUE Measurement
4.7. RNA Isolation, cDNA Synthesis, and qRT-PCR
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Liu, N.; Yang, J.H.; Guo, S.G.; Xu, Y.; Zhang, M. Genome-wide identification and comparative analysis of conserved and novel microRNAs in grafted watermelon by high-throughput sequencing. PLoS ONE 2013, 8, e57359. [Google Scholar] [CrossRef]
- Schwarz, D.; Öztekin, G.B.; Tüzel, Y.; Brückner, B.; Krumbein, A. Rootstocks can enhance tomato growth and quality characteristics at low potassium supply. Sci. Hortic. 2013, 149, 70–79. [Google Scholar] [CrossRef]
- Schwarz, D.; Rouphael, Y.; Colla, G.; Venema, J.H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010, 127, 162–171. [Google Scholar] [CrossRef]
- Bhatt, R.M.; Upreti, K.K.; Divya, M.; Bhat, S.; Pavithra, C.; Sadashiva, A. Interspecific grafting to enhance physiological resilience to flooding stress in tomato (Solanum lycopersicum L.). Sci. Hortic. 2015, 182, 8–17. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wang, Z.; Guo, X.; Wang, F.; Xia, X.; Zhou, J.; Shi, K.; Yu, J.; Zhou, Y. Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 2016, 39, 1790–1804. [Google Scholar] [CrossRef] [Green Version]
- Tateishi, K. Grafting watermelon onto pumpkin. J. Jpn. Hortic. 1927, 39, 5–8. (In Japanese) [Google Scholar]
- Sato, N.; Takamatsu, T. Grafting culture of watermelon. Nogyo Sekai. 1930, 25, 24–28. (In Japanese) [Google Scholar]
- Rivero, R.M.; Ruíz, J.M.; Romero, L. Iron Metabolism in Tomato and Watermelon Plants: Influence of Grafting. J. Plant Nutr. 2005, 27, 2221–2234. [Google Scholar] [CrossRef]
- Zhang, L.; Meng, X.; Liu, N.; Yang, J.; Zhang, M. Effects of grafting on phosphorus uptake and utilization of watermelon at early stage under low phosphorus stress. J. Fruit Sci. 2012, 29, 120–124. [Google Scholar]
- Yetisir, H.; Özdemir, A.E.; Aras, V.; Candır, E.; Aslan, Ö. Rootstocks effect on plant nutrition concentration in different organ of grafted watermelon. Agric. Sci. 2013, 04, 230–237. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Jiao, Y.; Nawaz, M.A.; Chen, C.; Liu, L.; Lu, Z.; Kong, Q.; Cheng, F.; Bie, Z. Improving magnesium uptake, photosynthesis and antioxidant enzyme activities of watermelon by grafting onto pumpkin rootstock under low magnesium. Plant Soil 2016, 409, 229–246. [Google Scholar] [CrossRef]
- Nawaz, M.A.; Imtiaz, M.; Kong, Q.; Cheng, F.; Ahmed, W.; Huang, Y.; Bie, Z. Grafting: A Technique to Modify Ion Accumulation in Horticultural Crops. Front. Plant Sci. 2016, 7, 1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marschner, H. Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995; p. 889. ISBN 0-12-473543-6. [Google Scholar]
- Gilbert, N. Environment: The disappearing nutrient. Nature 2009, 461, 716–718. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate Nutrition: Improving Low-Phosphate Tolerance in Crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Williamson, L.C.; Ribrioux, S.P.; Fitter, A.H.; Leyser, H.O. Phosphate Availability Regulates Root System Architecture in Arabidopsis. Plant Physiol. 2001, 126, 875–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Bucio, J.; Hernández-Abreu, E.; Sánchez-Calderón, L.; Nieto-Jacobo, M.F.; Simpson, J.; Herrera-Estrella, L.R. Phosphate Availability Alters Architecture and Causes Changes in Hormone Sensitivity in the Arabidopsis Root System. Plant Physiol. 2002, 129, 244–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Rouached, H.; Arpat, A.B.; Poirier, Y. Regulation of Phosphate Starvation Responses in Plants: Signaling Players and Cross-Talks. Mol. Plant 2010, 3, 288–299. [Google Scholar] [CrossRef]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.-K. Endogenous siRNAs Derived from a Pair of Natural cis-Antisense Transcripts Regulate Salt Tolerance in Arabidopsis. Cell 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309. [Google Scholar] [CrossRef]
- Kruszka, K.; Pieczynski, M.; Windels, D.; Bielewicz, D.; Jarmolowski, A.; Szweykowska-Kulinska, Z.; Vazquez, F. Role of microRNAs and other sRNAs of plants in their changing environments. J. Plant Physiol. 2012, 169, 1664–1672. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.-R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, L.-C.; Lin, S.-I.; Shih, A.C.-C.; Chen, J.-W.; Lin, W.-Y.; Tseng, C.-Y.; Li, W.-H.; Chiou, T.-J. Uncovering Small RNA-Mediated Responses to Phosphate Deficiency in Arabidopsis by Deep Sequencing. Plant Physiol. 2009, 151, 2120–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulcheski, F.R.; Correa, R.; Gomes, I.A.; De Lima, J.C.; Margis, R. NPK macronutrients and microRNA homeostasis. Front. Plant Sci. 2015, 6, 451. [Google Scholar] [CrossRef] [Green Version]
- Fujii, H.; Chiou, T.-J.; Lin, S.-I.; Aung, K.; Zhu, J.-K. A miRNA Involved in Phosphate-Starvation Response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043. [Google Scholar] [CrossRef] [Green Version]
- Aung, K.; Lin, S.-I.; Wu, C.-C.; Huang, Y.-T.; Su, C.-L.; Chiou, T.-J. pho2, a Phosphate Overaccumulator, Is Caused by a Nonsense Mutation in a MicroRNA399 Target Gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef] [Green Version]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W.-R. PHO2, MicroRNA399, and PHR1 Define a Phosphate-Signaling Pathway in Plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiou, T.-J.; Aung, K.; Lin, S.-I.; Wu, C.-C.; Chiang, S.-F.; Su, C.-L. Regulation of Phosphate Homeostasis by MicroRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batistic, O. Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 2004, 219, 915–924. [Google Scholar] [CrossRef]
- Cheong, Y.H.; Pandey, G.K.; Grant, J.J.; Batistic, O.; Li, L.; Kim, B.-G.; Lee, S.-C.; Kudla, J.; Luan, S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007, 52, 223–239. [Google Scholar] [CrossRef]
- Hu, H.-C.; Wang, Y.-Y.; Tsay, Y.-F. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef]
- Vert, G.; Chory, J. A Toggle Switch in Plant Nitrate Uptake. Cell 2009, 138, 1064–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Ren, F.; Zhou, L.; Wang, Q.-Q.; Zhong, H.; Li, X.-B. The Brassica napus Calcineurin B-Like 1/CBL-interacting protein kinase 6 (CBL1/CIPK6) component is involved in the plant response to abiotic stress and ABA signalling. J. Exp. Bot. 2012, 63, 6211–6222. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-N.; Cheong, Y.H.; Grant, J.J.; Pandey, G.K.; Luan, S. CIPK3, a Calcium Sensor–Associated Protein Kinase That Regulates Abscisic Acid and Cold Signal Transduction in Arabidopsis. Plant Cell 2003, 15, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Pandey, G.K.; Cheong, Y.H.; Kim, K.-N.; Grant, J.J.; Li, L.; Hung, W.; D’Angelo, C.; Weinl, S.; Kudla, J.; Luan, S. The Calcium Sensor Calcineurin B-Like 9 Modulates Abscisic Acid Sensitivity and Biosynthesis in Arabidopsis. Plant Cell 2004, 16, 1912–1924. [Google Scholar] [CrossRef] [Green Version]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.-G.; Li, L.G.; Luan, S. Calcineurin-B-Like Protein CBL9 Interacts with Target Kinase CIPK3 in the Regulation of ABA Response in Seed Germination. Mol. Plant 2008, 1, 238–248. [Google Scholar] [CrossRef]
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a Putative Calcium Sensor, Interacts with the Protein Kinase SOS2 to Protect Arabidopsis Shoots from Salt Stress. Plant Cell 2007, 19, 1415–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Kim, B.-G.; Cheong, Y.H.; Pandey, G.K.; Luan, S. A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 12625–12630. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, H.-D.; Chen, L.-Q.; Wang, Y.; Liu, L.-L.; He, L.; Wu, W.-H. A Protein Kinase, Interacting with Two Calcineurin B-like Proteins, Regulates K+ Transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef] [Green Version]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Liu, N.; Zhang, L.; Yang, J.; Zhang, M. Genotypic differences in phosphorus uptake and utilization of watermelon under low phosphorus stress. J. Plant Nutr. 2014, 37, 312–326. [Google Scholar] [CrossRef]
- Kim, K.-N.; Cheong, Y.H.; Gupta, R.; Luan, S. Interaction Specificity of Arabidopsis Calcineurin B-Like Calcium Sensors and Their Target Kinases. Plant Physiol. 2000, 124, 1844–1853. [Google Scholar] [CrossRef] [Green Version]
- Albacete, A.; Martínez-Andújar, C.; Ghanem, M.E.; Acosta, M.; Sánchez-Bravo, J.; Asins, M.J.; Cuartero, J.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Rootstock-mediated changes in xylem ionic and hormonal status are correlated with delayed leaf senescence, and increased leaf area and crop productivity in salinized tomato. Plant, Cell Environ. 2009, 32, 928–938. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Hua, B.; Liu, Z.; Fan, M.; Bie, Z. Grafting onto different rootstocks as a means to improve watermelon tolerance to low potassium stress. Sci. Hortic. 2013, 149, 80–85. [Google Scholar] [CrossRef]
- Zeng, H.; Wang, G.; Hu, X.; Wang, H.; Du, L.; Zhu, Y. Role of microRNAs in plant responses to nutrient stress. Plant Soil 2014, 374, 1005–1021. [Google Scholar] [CrossRef]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of Root Cap Formation by MicroRNA-Targeted Auxin Response Factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [Green Version]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef] [Green Version]
- Vidal, E.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutierrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Mai, Y.-X.; Zhang, Y.-C.; Luo, Q.; Yang, H.-Q. MicroRNA171c-Targeted SCL6-II, SCL6-III, and SCL6-IV Genes Regulate Shoot Branching in Arabidopsis. Mol. Plant 2010, 3, 794–806. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Ding, H.; Zhu, J.-K.; Zhang, F.; Li, W. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; He, H.; Yu, D. Identification of Nitrogen Starvation-Responsive MicroRNAs in Arabidopsis thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Liang, G.; Li, Y.; Wang, F.; Yu, D. Two Young MicroRNAs Originating from Target Duplication Mediate Nitrogen Starvation Adaptation via Regulation of Glucosinolate Synthesis in Arabidopsis thaliana. Plant Physiol. 2014, 164, 853–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Okumura, S.; Mitsukawa, N.; Shirano, Y.; Shibata, D. Phosphate Transporter Gene Family of Arabidopsis thaliana. DNA Res. 1998, 5, 261–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poirier, Y.; Thoma, S.; Somerville, C.; Schiefelbein, J. Mutant of Arabidopsis Deficient in Xylem Loading of Phosphate. Plant Physiol. 1991, 97, 1087–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamburger, D.; Rezzonico, E.; Macdonald-Comber Petetot, J.; Somerville, C.; Poirier, Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant. Cell 2002, 14, 889–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kant, S.; Peng, M.; Rothstein, S.J. Genetic Regulation by NLA and MicroRNA827 for Maintaining Nitrate-Dependent Phosphate Homeostasis in Arabidopsis. PLoS Genet. 2011, 7, e1002021. [Google Scholar] [CrossRef] [Green Version]
- Huang, T.-K.; Han, C.-L.; Lin, S.-I.; Chen, Y.-J.; Tsai, Y.-C.; Chen, Y.-R.; Chen, J.-W.; Lin, W.-Y.; Chen, P.-M.; Liu, T.-Y.; et al. Identification of Downstream Components of Ubiquitin-Conjugating Enzyme PHOSPHATE2 by Quantitative Membrane Proteomics in Arabidopsis Roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef] [Green Version]
- Park, B.S.; Seo, J.S.; Chua, N.-H. Nitrogen limitation adaptation recruits phosphate2 to Target the Phosphate Transporter PT2 for Degradation during the Regulation of Arabidopsis Phosphate Homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Yu, X.; Bai, L.; He, C.; Li, Y. Responses of miRNAs and their target genes to nitrogen- or phosphorus-deficiency in grafted cucumber seedlings. Hortic. Environ. Biotechnol. 2016, 57, 97–112. [Google Scholar] [CrossRef]
- Weinl, S.; Kudla, J. The CBL–CIPK Ca 2+ -decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.-S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The Putative Plasma Membrane Na+/H+ Antiporter SOS1 Controls Long-Distance Na+ Transport in Plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, R.-J.; Zhao, F.-G.; Garcia, V.; Kleist, T.J.; Yang, L.; Zhang, H.-X.; Luan, S. Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3134–3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Léran, S.; Edel, K.H.; Pervent, M.; Hashimoto, K.; Corratgé-Faillie, C.; Offenborn, J.N.; Tillard, P.; Gojon, A.; Kudla, J.; Lacombe, B. Nitrate sensing and uptake in Arabidopsisare enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Sci. Signal. 2015, 8, ra43. [Google Scholar] [CrossRef]
- Tripathi, V.; Parasuraman, B.; Laxmi, A.; Chattopadhyay, D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009, 58, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Kolukisaoglu, U.; Weinl, S.; Blazevic, D.; Batistic, O.; Kudla, J. Calcium Sensors and Their Interacting Protein Kinases: Genomics of the Arabidopsis and Rice CBL-CIPK Signaling Networks. Plant Physiol. 2004, 134, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Ying, Y.; Yue, W.; Wang, S.; Li, S.; Wang, M.; Zhao, Y.; Wang, C.; Mao, C.; Whelan, J.; Shou, H. Two h-Type Thioredoxins Interact with the E2 Ubiquitin Conjugase PHO2 to Fine-Tune Phosphate Homeostasis in Rice. Plant Physiol. 2017, 173, 812–824. [Google Scholar] [CrossRef] [Green Version]
- Curtis, M.D.; Grossniklaus, U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Chapman, H.D.; Pratt, P.F. Phosphorus. Methods of Analysis for Soils, Plants and Waters; University of California: Berkeley, CA, USA, 1961; pp. 160–170. [Google Scholar]
- Siddiqi, M.Y.; Glass, A.D. Utilization index: A modified approach to the estimation and comparison of nutrient utilization efficiency in plants. J. Plant Nutr. 1981, 4, 289–302. [Google Scholar] [CrossRef]
- Wu, W.; Deng, Q.; Shi, P.; Yang, J.; Hu, Z.; Zhang, M. Identification of Appropriate Reference Genes for Normalization of miRNA Expression in Grafted Watermelon Plants under Different Nutrient Stresses. PLoS ONE 2016, 11, e0164725. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, W.; Zhao, H.; Deng, Q.; Yang, H.; Guan, X.; Qi, R.; Shi, P.; Yang, J.; Zhang, M.; Hu, Z. The Novel Cucurbitaceae miRNA ClmiR86 Is Involved in Grafting-Enhanced Phosphate Utilization and Phosphate Starvation Tolerance in Watermelon. Plants 2021, 10, 2133. https://doi.org/10.3390/plants10102133
Wu W, Zhao H, Deng Q, Yang H, Guan X, Qi R, Shi P, Yang J, Zhang M, Hu Z. The Novel Cucurbitaceae miRNA ClmiR86 Is Involved in Grafting-Enhanced Phosphate Utilization and Phosphate Starvation Tolerance in Watermelon. Plants. 2021; 10(10):2133. https://doi.org/10.3390/plants10102133
Chicago/Turabian StyleWu, Weifang, Haoshun Zhao, Qin Deng, Haiyang Yang, Xiaoxiao Guan, Rui Qi, Pibiao Shi, Jinghua Yang, Mingfang Zhang, and Zhongyuan Hu. 2021. "The Novel Cucurbitaceae miRNA ClmiR86 Is Involved in Grafting-Enhanced Phosphate Utilization and Phosphate Starvation Tolerance in Watermelon" Plants 10, no. 10: 2133. https://doi.org/10.3390/plants10102133





